Comparative and Expression Analysis of Ubiquitin Conjugating Domain-Containing Genes in Two Pyrus Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval and Identification of UBC Genes
2.2. Phylogenetic Analysis and Gene Duplication
2.3. Gene Structure and Motif Analysis
2.4. Expression Analysis
2.5. Expression Correlation of Orthologous UBC Genes in Two Pyrus Species
2.6. Availability of Data and Materials
3. Results
3.1. Identification of UBC Genes in Two Pyrus Species
3.2. Phylogenetic Analysis of UBC Genes in Two Pyrus Species
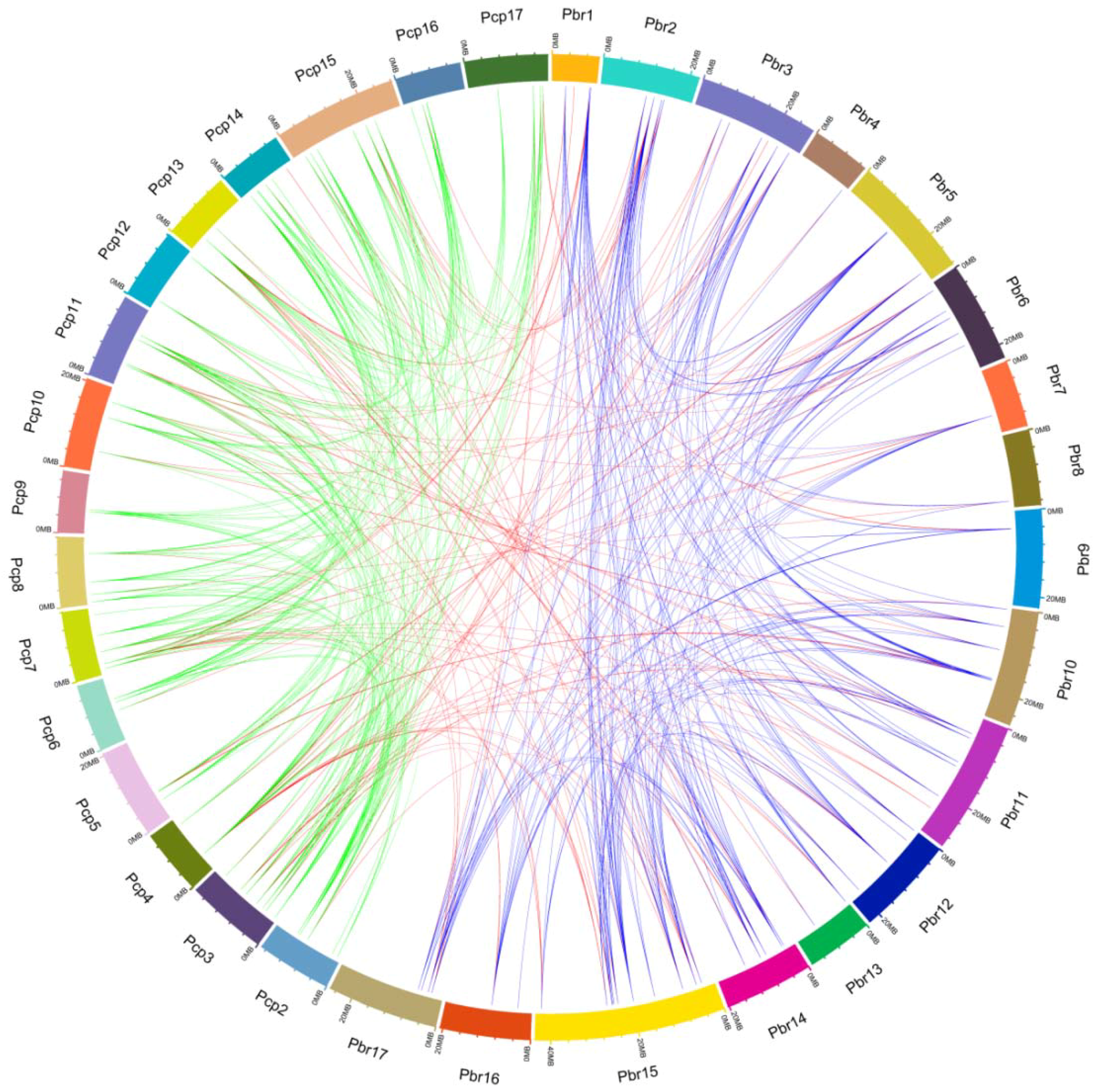
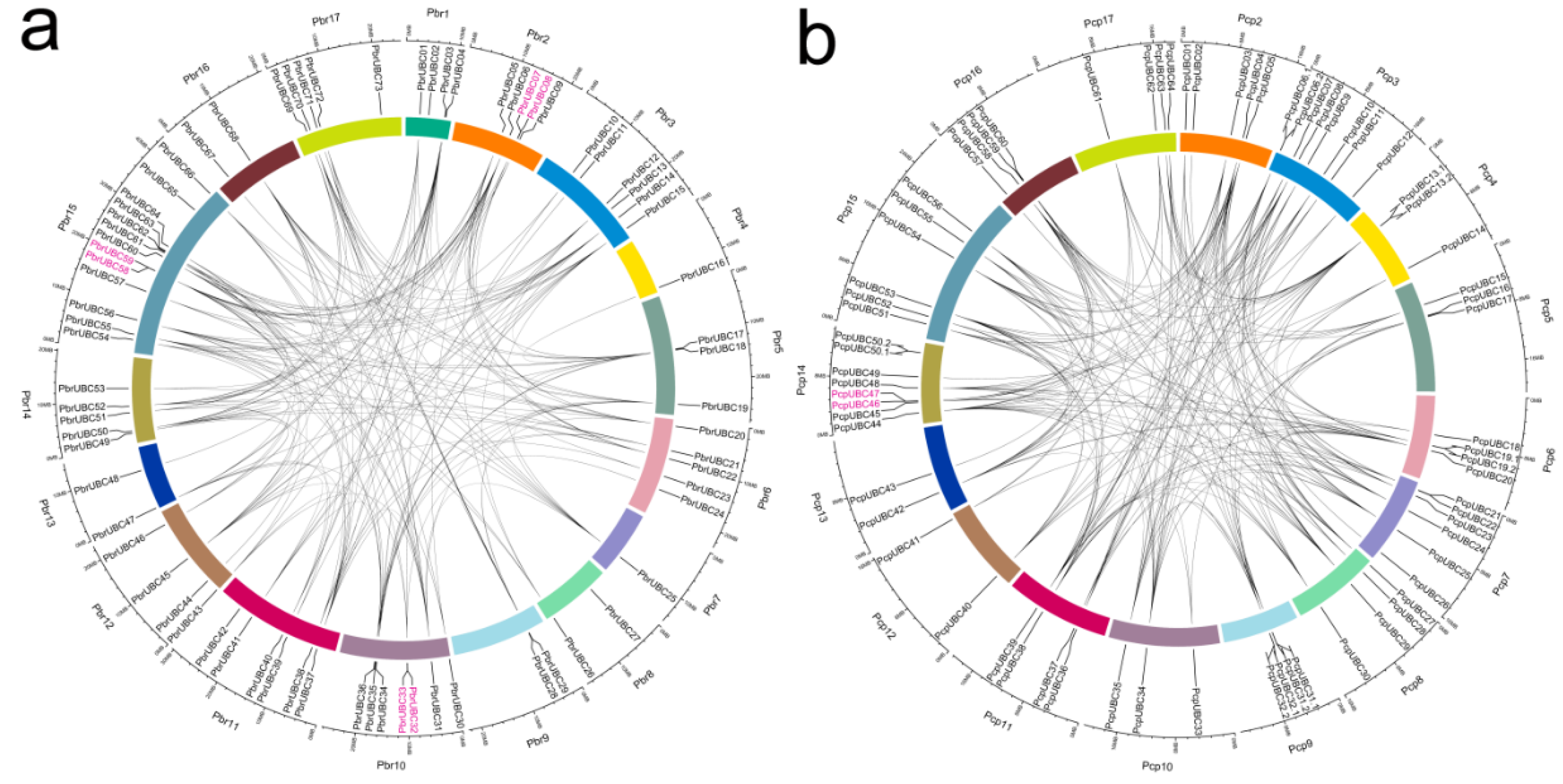
3.3. Chromosomal Distribution and Gene Duplication of UBC Genes in Two Pyrus Species
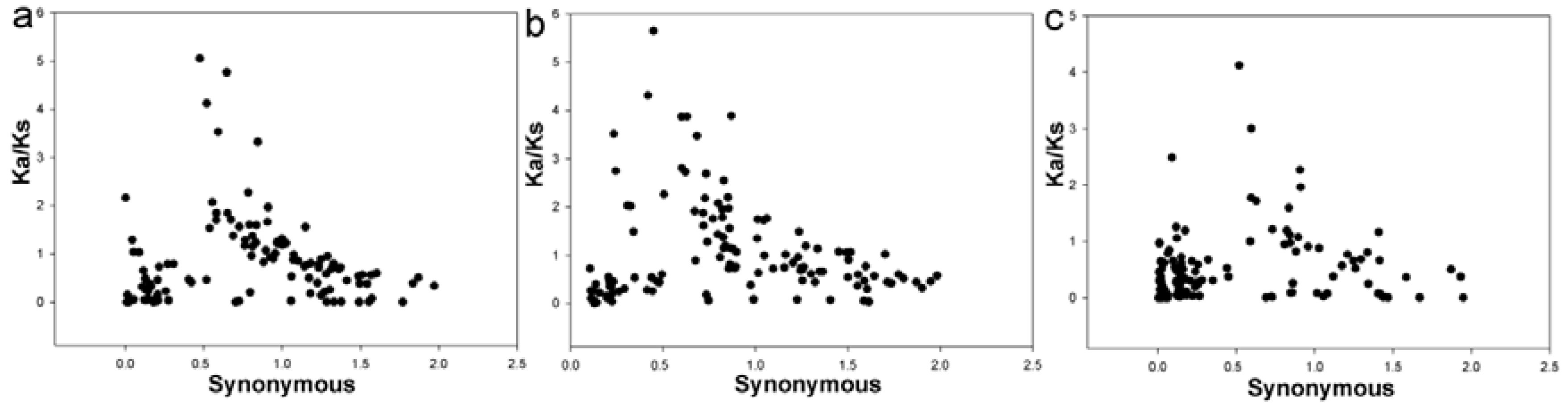
3.4. Evolutionary Patterns in Two Pyrus Species
3.5. Expression Profiles of UBC Genes in Pyrus Fruit Development
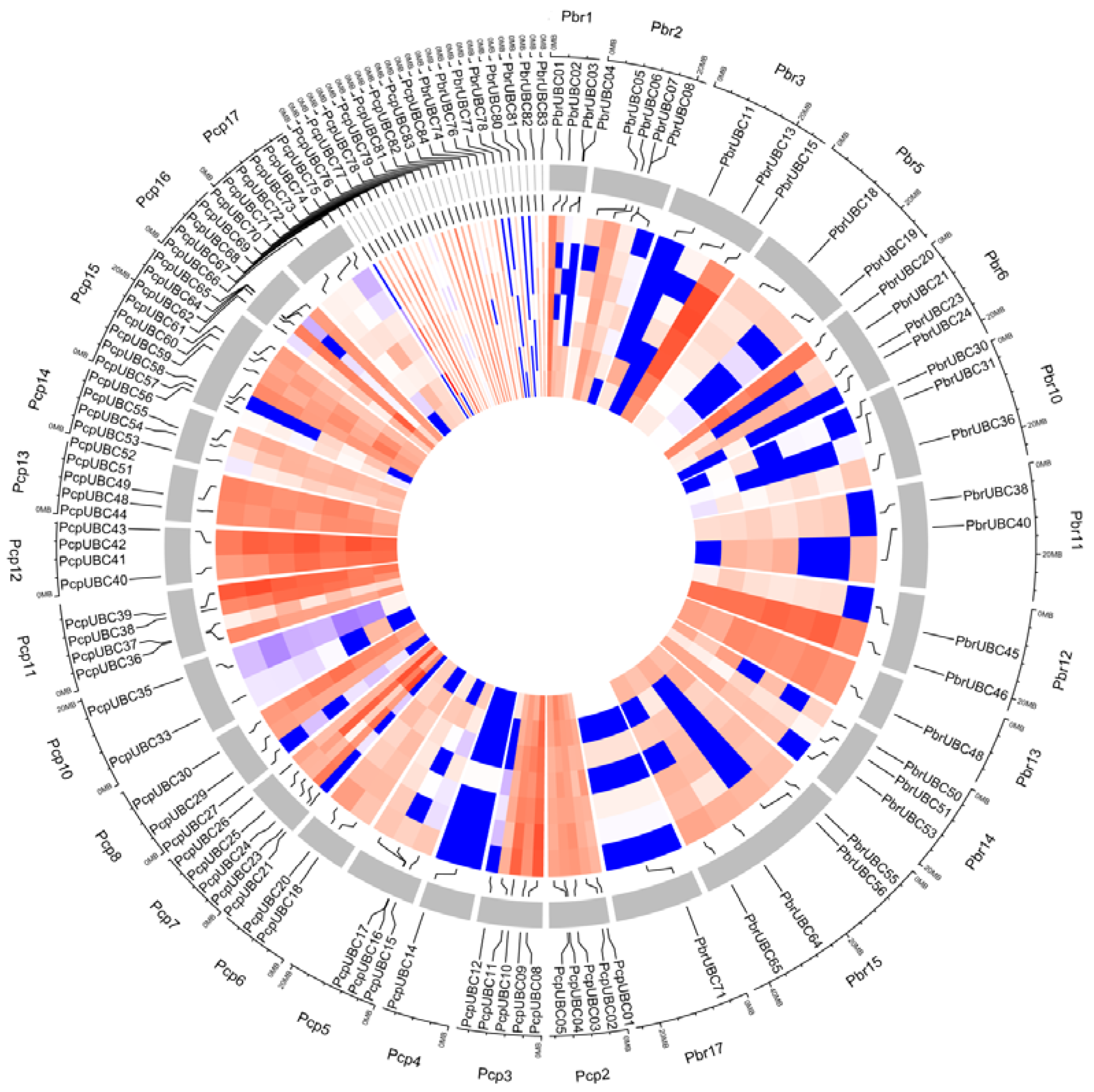
3.6. Comparison of the Expression Patterns of UBC Genes in Two Pyrus Species
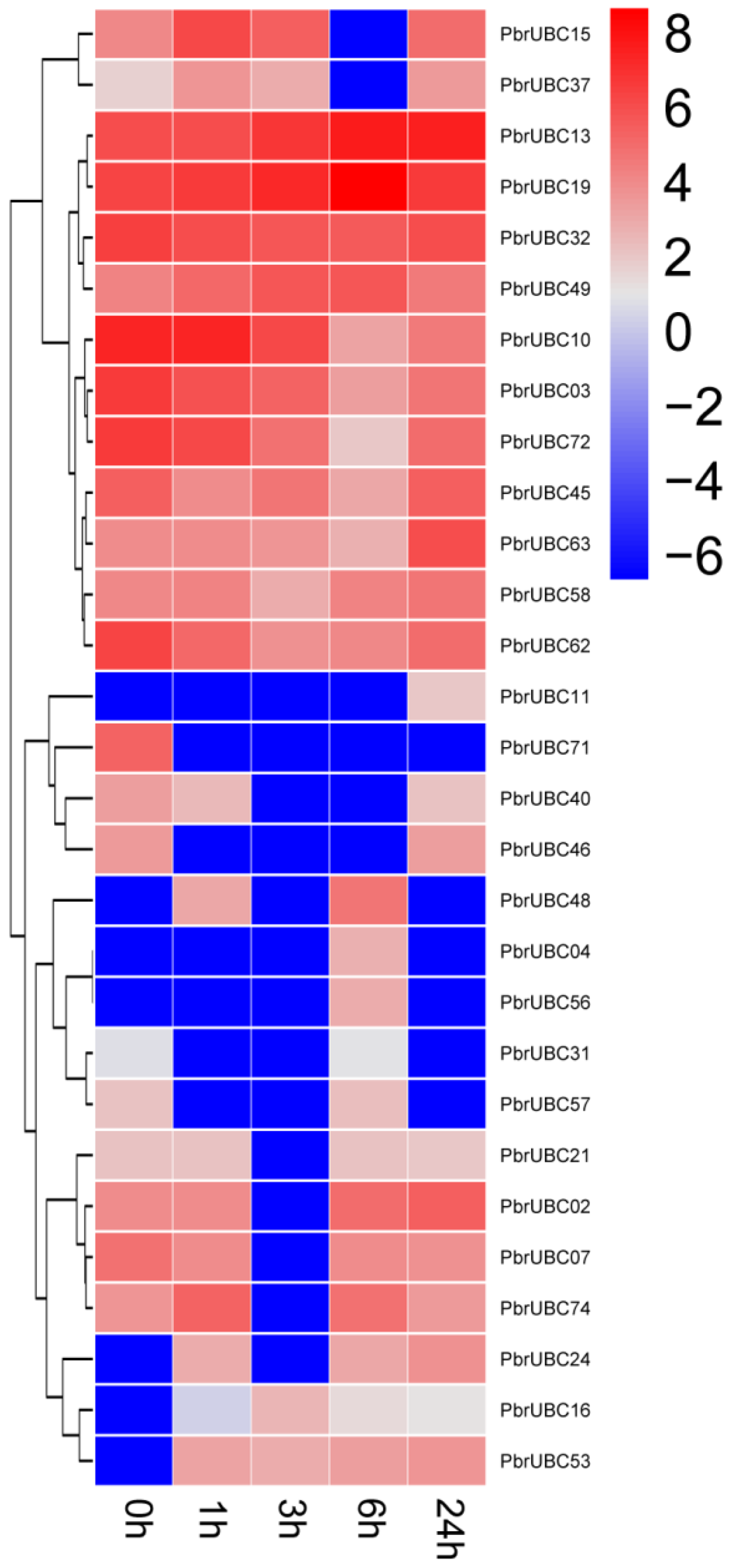
3.7. Expression Profiles of PbrUBC Genes Respond to Drought Stress
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethics Approval and Consent to Participate
References
- Kraft, E.; Stone, S.L.; Ma, L.; Su, N.; Gao, Y.; Lau, O.-S.; Deng, X.-W.; Callis, J. Genome analysis and functional characterization of the e2 and ring-type e3 ligase ubiquitination enzymes of arabidopsis. Plant Physiol. 2005, 139, 1597–1611. [Google Scholar] [CrossRef] [PubMed]
- Callis, J.; Vierstra, R.D. Protein degradation in signaling. Curr. Opin. Plant Boil. 2000, 3, 381–386. [Google Scholar] [CrossRef]
- Peters, J.-M.; Harris, J.R.; Finley, D. Ubiquitin and the Biology of the Cell; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Glickman, M.H.; Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002, 82, 373–428. [Google Scholar] [CrossRef] [PubMed]
- Van Wijk, S.J.; Timmers, H.M. The family of ubiquitin-conjugating enzymes (E2s): Deciding between life and death of proteins. FASEB J. 2010, 24, 981–993. [Google Scholar] [CrossRef] [PubMed]
- Michelle, C.; Vourc’h, P.; Mignon, L.; Andres, C.R. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? J. Mol. Evol. 2009, 68, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.; Crowe, E.; Stevens, T.A.; Candido, E.P.M. Functional and phylogenetic analysis of the ubiquitylation system in Caenorhabditis elegans: Ubiquitin-conjugating enzymes, ubiquitin-activating enzymes, and ubiquitin-like proteins. Genome Boil. 2001, 3, research0002.1–0002.15. [Google Scholar]
- Bae, H.; Kim, W.T. Classification and interaction modes of 40 rice E2 ubiquitin-conjugating enzymes with 17 rice ARM-U-box E3 ubiquitin ligases. Biochem. Biophys. Res. Commun. 2014, 444, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.; Hu, H.; Jue, D.; Zhao, Q.; Chen, H.; Xie, J.; Jia, L. The banana e2 gene family: Genomic identification, characterization, expression profiling analysis. Plant Sci. 2016, 245, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Jue, D.; Sang, X.; Lu, S.; Dong, C.; Zhao, Q.; Chen, H.; Jia, L. Genome-wide identification, phylogenetic and expression analyses of the ubiquitin-conjugating enzyme gene family in maize. PLoS ONE 2015, 10, e0143488. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, W.; Cai, J.; Zhang, Y.; Qin, G.; Tian, S. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Boil. 2014, 15, 548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jue, D.; Sang, X.; Shu, B.; Liu, L.; Wang, Y.; Jia, Z.; Zou, Y.; Shi, S. Characterization and expression analysis of genes encoding ubiquitin conjugating domain-containing enzymes in Carica papaya. PLoS ONE 2017, 12, e0171357. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ménard, R.; Berr, A.; Fuchs, J.; Cognat, V.; Meyer, D.; Shen, W.H. The E2 ubiquitin-conjugating enzymes, Atubc1 and Atubc2, play redundant roles and are involved in activation of Flc expression and repression of flowering in Arabidopsis thaliana. Plant J. 2009, 57, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Liu, L.; Zhao, Q.; Zhang, Z.; Li, Q.; Lin, B.; Wu, Y.; Tang, S.; Xie, Q. Arabidopsis ubiquitin conjugase UBC32 is an ERAD component that functions in brassinosteroid-mediated salt stress tolerance. Plant Cell 2012, 24, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.-A.; Chang, R.-Z.; Qiu, L.-J. Overexpression of soybean ubiquitin-conjugating enzyme gene GmUBC2 confers enhanced drought and salt tolerance through modulating abiotic stress-responsive gene expression in Arabidopsis. Plant Mol. Boil. 2010, 72, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.; Mo, A.; Liu, S.; Yang, L.; Li, L. Constitutive expression of a peanut ubiquitin-conjugating enzyme gene in Arabidopsis confers improved water-stress tolerance through regulation of stress-responsive gene expression. J. Biosci. Bioeng. 2011, 111, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Chung, E.; Cho, C.-W.; So, H.-A.; Kang, J.-S.; Chung, Y.S.; Lee, J.-H. Overexpression of VrUBC1, a mung bean e2 ubiquitin-conjugating enzyme, enhances osmotic stress tolerance in Arabidopsis. PLoS ONE 2013, 8, e66056. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Schmidt, W. A lysine-63-linked ubiquitin chain-forming conjugase, UBC13, promotes the developmental responses to iron deficiency in Arabidopsis roots. Plant J. 2010, 62, 330–343. [Google Scholar] [CrossRef] [PubMed]
- Wen, R.; Newton, L.; Li, G.; Wang, H.; Xiao, W. Arabidopsis thaliana UBC13: Implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Mol. Boil. 2006, 61, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Zolman, B.K.; Monroe-Augustus, M.; Silva, I.D.; Bartel, B. Identification and functional characterization of arabidopsis peroxin4 and the interacting protein peroxin22. Plant Cell 2005, 17, 3422–3435. [Google Scholar] [CrossRef] [PubMed]
- E, Z.; Zhang, Y.; Li, T.; Wang, L.; Zhao, H. Characterization of the ubiquitin-conjugating enzyme gene family in rice and evaluation of expression profiles under abiotic stresses and hormone treatments. PLoS ONE 2015, 10, e0122621. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. Myb transcription factors in Chinese pear (Pyrus bretschneideri rehd.): Genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 2016, 7, 577. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H. The genome of the pear (Pyrus bretschneideri rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Finn, R.D.; Eddy, S.R.; Bateman, A.; Punta, M. Challenges in homology search: Hmmer3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 2013, 41, e121. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2011, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chagné, D.; Crowhurst, R.N.; Pindo, M.; Thrimawithana, A.; Deng, C.; Ireland, H.; Fiers, M.; Dzierzon, H.; Cestaro, A.; Fontana, P.; et al. The draft genome sequence of European pear (Pyrus communis L. ‘Bartlett’). PLoS ONE 2014, 9, e92644. [Google Scholar]
- Letunic, I.; Doerks, T.; Bork, P. Smart 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012, 40, D302–D305. [Google Scholar] [CrossRef] [PubMed]
- Zdobnov, E.M.; Apweiler, R. Interproscan—An integration platform for the signature-recognition methods in interpro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Ficklin, S.P.; Lee, T.; Cheng, C.H.; Blenda, A.; Zheng, P.; Yu, J.; Bombarely, A.; Cho, I.; Ru, S. The genome database for rosaceae (GDR): Year 10 update. Nucleic Acids Res. 2014, 42, 1237–1244. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Boil. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H. Mcscanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Xia, R.; Chen, H.; He, Y. Tbtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv 2018. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The meme suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed]
- Gordon, A.; Hannon, G. Fastx-Toolkit. Fastq/a Short-Reads Pre-Processing Tools. Unpublished work. 2003. [Google Scholar]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Boil. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with Tophat and Cufflinks. Nat. Protoc. 2012, 7, 562. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 2004, 16, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Yim, W.C.; Lee, B.-M.; Jang, C.S. Expression diversity and evolutionary dynamics of rice duplicate genes. Mol. Genet. Genom. 2009, 281, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maher, C.; Stein, L.; Ware, D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006, 16, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Meng, D.; Abdullah, M.; Jin, Q.; Lin, Y.; Cai, Y. Genome wide identification, evolutionary, and expression analysis of VQ genes from two Pyrus species. Genes 2018, 9, 224. [Google Scholar] [CrossRef] [PubMed]
- Fawcett, J.A.; Maere, S.; Van de Peer, Y. Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. USA 2009, 106, 5737–5742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jin, Q.; Lin, Y.; Cai, Y. Structural, evolutionary, and functional analysis of the class iii peroxidase gene family in Chinese Pear (Pyrus bretschneideri). Front. Plant Sci. 2016, 7, 1874. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Meng, D.; Li, G.; Li, D.; Abdullah, M.; Jin, Q.; Lin, Y.; Cai, Y. Genome-wide analysis suggests the relaxed purifying selection affect the evolution of WOX genes in Pyrus bretschneideri, Prunus persica, Prunus mume, and Fragaria vesca. Front. Genet. 2017, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Meng, D.; Abdullah, M.; Li, D.; Jin, Q.; Lin, Y.; Cai, Y. Systematic analysis and comparison of the PHD-Finger gene family in Chinese pear (Pyrus bretschneideri) and its role in fruit development. Funct. Integr. Genom. 2018, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jeon, E.H.; Pak, J.H.; Kim, M.J.; Kim, H.J.; Shin, S.H.; Lee, J.H.; Kim, D.H.; Oh, J.S.; Oh, B.-J.; Jung, H.W. Ectopic expression of ubiquitin-conjugating enzyme gene from wild rice, OgUBC1, confers resistance against UV-B radiation and Botrytis infection in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2012, 427, 309–314. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Gene Identifier | Chromosme | 5′ End | 3′ End | Protein Size (aa) |
|---|---|---|---|---|---|
| PcpUBC01 | PCP011547.1 | chr2 | 908379 | 915288 | 378 |
| PcpUBC02 | PCP008190.1 | chr2 | 2303235 | 2304455 | 406 |
| PcpUBC03 | PCP026226.1 | chr2 | 9163625 | 9165622 | 161 |
| PcpUBC04 | PCP025062.1 | chr2 | 11398030 | 11400462 | 160 |
| PcpUBC05 | PCP029820.1 | chr2 | 11983749 | 11985588 | 154 |
| PcpUBC06.1 | PCP020092.1 | chr3 | 547762 | 556678 | 986 |
| PcpUBC06.2 | PCP041697.1 | chr3 | 551276 | 556678 | 387 |
| PcpUBC07 | PCP000885.1 | chr3 | 3236140 | 3247151 | 841 |
| PcpUBC08 | PCP013747.1 | chr3 | 4266095 | 4267481 | 148 |
| PcpUBC09 | PCP031109.1 | chr3 | 6092388 | 6096394 | 192 |
| PcpUBC10 | PCP029016.1 | chr3 | 9480497 | 9483572 | 146 |
| PcpUBC11 | PCP029037.1 | chr3 | 11156660 | 11161769 | 769 |
| PcpUBC12 | PCP008664.1 | chr3 | 16102272 | 16105426 | 277 |
| PcpUBC13.1 | PCP006596.1 | chr4 | 3944944 | 3952328 | 304 |
| PcpUBC13.2 | PCP033076.1 | chr4 | 3944944 | 3946278 | 148 |
| PcpUBC14 | PCP032557.1 | chr4 | 13065481 | 13065759 | 92 |
| PcpUBC15 | PCP004584.1 | chr5 | 4531710 | 4535205 | 920 |
| PcpUBC16 | PCP000343.1 | chr5 | 6602829 | 6603942 | 120 |
| PcpUBC17 | PCP000325.1 | chr5 | 6785831 | 6791593 | 1148 |
| PcpUBC18 | PCP013561.1 | chr6 | 7061585 | 7063890 | 183 |
| PcpUBC19.1 | PCP003994.1 | chr6 | 9051182 | 9054610 | 490 |
| PcpUBC19.2 | PCP038680.1 | chr6 | 9053326 | 9054610 | 152 |
| PcpUBC20 | PCP006677.1 | chr6 | 10001882 | 10012807 | 890 |
| PcpUBC21 | PCP025848.1 | chr7 | 1250213 | 1253437 | 172 |
| PcpUBC22 | PCP025852.1 | chr7 | 1277835 | 1279055 | 189 |
| PcpUBC23 | PCP027463.1 | chr7 | 2394123 | 2396438 | 148 |
| PcpUBC24 | PCP039252.1 | chr7 | 5395929 | 5399115 | 181 |
| PcpUBC25 | PCP041398.1 | chr7 | 9425098 | 9426267 | 78 |
| PcpUBC26 | PCP023269.1 | chr7 | 14434633 | 14435892 | 419 |
| PcpUBC27 | PCP018282.1 | chr8 | 526845 | 534958 | 439 |
| PcpUBC28 | PCP024977.1 | chr8 | 2320747 | 2321922 | 189 |
| PcpUBC29 | PCP014622.1 | chr8 | 5359408 | 5371794 | 989 |
| PcpUBC30 | PCP013024.1 | chr8 | 12182295 | 12185615 | 188 |
| PcpUBC31.1 | PCP025013.1 | chr9 | 5181126 | 5187890 | 423 |
| PcpUBC31.2 | PCP042586.1 | chr9 | 5181126 | 5183531 | 195 |
| PcpUBC32.1 | PCP029211.1 | chr9 | 5697502 | 5704326 | 356 |
| PcpUBC32.2 | PCP037499.1 | chr9 | 5697502 | 5700526 | 237 |
| PcpUBC33 | PCP022975.1 | chr10 | 4876099 | 4877045 | 137 |
| PcpUBC34 | PCP006043.1 | chr10 | 13347113 | 13347565 | 150 |
| PcpUBC35 | PCP019803.1 | chr10 | 17163601 | 17167148 | 921 |
| PcpUBC36 | PCP037747.1 | chr11 | 5179080 | 5182742 | 153 |
| PcpUBC37 | PCP003934.1 | chr11 | 5615653 | 5620782 | 686 |
| PcpUBC38 | PCP014945.1 | chr11 | 12048835 | 12052442 | 227 |
| PcpUBC39 | PCP007359.1 | chr11 | 13280818 | 13282231 | 148 |
| PcpUBC40 | PCP041407.1 | chr12 | 2420398 | 2421551 | 148 |
| PcpUBC41 | PCP043134.1 | chr12 | 15435377 | 15437609 | 146 |
| PcpUBC42 | PCP003304.1 | chr13 | 3769096 | 3771160 | 152 |
| PcpUBC43 | PCP028885.1 | chr13 | 7446155 | 7448170 | 373 |
| PcpUBC44 | PCP010847.1 | chr14 | 1973506 | 1975549 | 195 |
| PcpUBC45 | PCP000493.1 | chr14 | 3899314 | 3905194 | 754 |
| PcpUBC46 | PCP006717.1 | chr14 | 4144989 | 4159895 | 1373 |
| PcpUBC47 | PCP033096.1 | chr14 | 4155864 | 4157211 | 152 |
| PcpUBC48 | PCP017973.1 | chr14 | 6488805 | 6490257 | 144 |
| PcpUBC49 | PCP031605.1 | chr14 | 8511513 | 8512945 | 265 |
| PcpUBC50.1 | PCP001680.1 | chr14 | 12856983 | 12862739 | 444 |
| PcpUBC50.2 | PCP032125.1 | chr14 | 12860593 | 12862739 | 146 |
| PcpUBC51 | PCP022610.1 | chr15 | 2571489 | 2577399 | 467 |
| PcpUBC52 | PCP032902.1 | chr15 | 3995850 | 3996286 | 78 |
| PcpUBC53 | PCP013613.1 | chr15 | 5801442 | 5811234 | 828 |
| PcpUBC54 | PCP005846.1 | chr15 | 15546784 | 15549513 | 160 |
| PcpUBC55 | PCP020991.1 | chr15 | 18866853 | 18872463 | 319 |
| PcpUBC56 | PCP014540.1 | chr15 | 21362811 | 21364686 | 161 |
| PcpUBC57 | PCP006806.1 | chr16 | 1486195 | 1494553 | 341 |
| PcpUBC58 | PCP026158.1 | chr16 | 4290275 | 4292359 | 152 |
| PcpUBC59 | PCP021281.1 | chr16 | 5293770 | 5294891 | 373 |
| PcpUBC60 | PCP021299.1 | chr16 | 5419315 | 5421158 | 307 |
| PcpUBC61 | PCP010786.1 | chr17 | 6829215 | 6832764 | 267 |
| PcpUBC62 | PCP042470.1 | chr17 | 15393404 | 15402997 | 596 |
| PcpUBC63 | PCP012277.1 | chr17 | 17062013 | 17063062 | 349 |
| PcpUBC64 | PCP007143.1 | chr17 | 17728444 | 17733945 | 621 |
| PcpUBC65 | PCP018399.1 | scaffold00612 | 73468 | 83219 | 651 |
| PcpUBC66 | PCP021641.1 | scaffold00634 | 76924 | 79227 | 148 |
| PcpUBC67 | PCP007398.1 | scaffold00805 | 133398 | 134716 | 174 |
| PcpUBC68 | PCP018521.1 | scaffold00852 | 13601 | 20410 | 278 |
| PcpUBC69 | PCP004264.1 | scaffold00983 | 16646 | 19316 | 148 |
| PcpUBC70 | PCP021954.1 | scaffold01394 | 34467 | 36817 | 191 |
| PcpUBC71 | PCP007629.1 | scaffold01465 | 20581 | 22904 | 291 |
| PcpUBC72 | PCP002761.1 | scaffold01522 | 35381 | 36867 | 160 |
| PcpUBC73 | PCP020361.1 | scaffold01593 | 68663 | 70923 | 191 |
| PcpUBC74 | PCP045045.1 | scaffold01774 | 29298 | 33191 | 754 |
| PcpUBC75 | PCP001255.1 | scaffold01881 | 39525 | 44855 | 1147 |
| PcpUBC76 | PCP004531.1 | scaffold01923 | 15838 | 21766 | 337 |
| PcpUBC77 | PCP028469.1 | scaffold02358 | 32352 | 36077 | 463 |
| PcpUBC78 | PCP022164.1 | scaffold02454 | 7209 | 11865 | 389 |
| PcpUBC79 | PCP012568.1 | scaffold02548 | 27443 | 29342 | 178 |
| PcpUBC80 | PCP043259.1 | scaffold04878 | 3046 | 6910 | 134 |
| PcpUBC81 | PCP001444.1 | scaffold05041 | 9798 | 11719 | 183 |
| PcpUBC82 | PCP022346.1 | scaffold17014 | 829 | 2135 | 180 |
| PcpUBC83 | PCP040042.1 | scaffold23907 | 470 | 1254 | 125 |
| PcpUBC84 | PCP011188.1 | scaffold27287 | 92 | 1717 | 232 |
| PbrUBC01 | Pbr021045.1 | Chr1 | 3279060 | 3282148 | 149 |
| PbrUBC02 | Pbr022046.1 | Chr1 | 5355491 | 5357926 | 192 |
| PbrUBC03 | Pbr018716.1 | Chr1 | 9129424 | 9132884 | 149 |
| PbrUBC04 | Pbr013632.1 | Chr1 | 9364933 | 9367901 | 149 |
| PbrUBC05 | Pbr029889.2 | Chr2 | 11205065 | 11207039 | 195 |
| PbrUBC06 | Pbr025178.1 | Chr2 | 13123810 | 13126801 | 161 |
| PbrUBC07 | Pbr022865.1 | Chr2 | 15059120 | 15062337 | 184 |
| PbrUBC08 | Pbr022866.1 | Chr2 | 15070696 | 15073441 | 182 |
| PbrUBC09 | Pbr040498.1 | Chr2 | 15603251 | 15605586 | 162 |
| PbrUBC10 | Pbr024232.2 | Chr3 | 7098302 | 7101956 | 160 |
| PbrUBC11 | Pbr027637.1 | Chr3 | 9231735 | 9233562 | 181 |
| PbrUBC12 | Pbr023139.1 | Chr3 | 17780615 | 17782004 | 149 |
| PbrUBC13 | Pbr000740.1 | Chr3 | 19467052 | 19469759 | 170 |
| PbrUBC14 | Pbr013150.2 | Chr3 | 22108425 | 22111475 | 149 |
| PbrUBC15 | Pbr034016.1 | Chr3 | 25287937 | 25291916 | 168 |
| PbrUBC16 | Pbr030934.3 | Chr4 | 12531057 | 12533684 | 161 |
| PbrUBC17 | Pbr027417.1 | Chr5 | 12936909 | 12946872 | 201 |
| PbrUBC18 | Pbr027395.1 | Chr5 | 13135775 | 13142057 | 1149 |
| PbrUBC19 | Pbr000361.1 | Chr5 | 25903450 | 25906904 | 853 |
| PbrUBC20 | Pbr011471.1 | Chr6 | 1539205 | 1541448 | 153 |
| PbrUBC21 | Pbr009129.1 | Chr6 | 7334609 | 7336195 | 77 |
| PbrUBC22 | Pbr014124.1 | Chr6 | 9284322 | 9290606 | 296 |
| PbrUBC23 | Pbr018194.1 | Chr6 | 13523851 | 13526524 | 184 |
| PbrUBC24 | Pbr040529.1 | Chr6 | 16795923 | 16797553 | 181 |
| PbrUBC25 | Pbr032353.1 | Chr7 | 10867139 | 10869902 | 224 |
| PbrUBC26 | Pbr006183.1 | Chr8 | 15645575 | 15647047 | 190 |
| PbrUBC27 | Pbr004154.1 | Chr8 | 4690292 | 4691740 | 129 |
| PbrUBC28 | Pbr032653.1 | Chr9 | 4250106 | 4253822 | 461 |
| PbrUBC29 | Pbr032645.1 | Chr9 | 4153561 | 4154900 | 279 |
| PbrUBC30 | Pbr031810.1 | Chr10 | 268749 | 273571 | 458 |
| PbrUBC31 | Pbr016259.1 | Chr10 | 4489977 | 4494846 | 922 |
| PbrUBC32 | Pbr009080.1 | Chr10 | 10152737 | 10153189 | 151 |
| PbrUBC33 | Pbr009081.1 | Chr10 | 10158759 | 10159211 | 151 |
| PbrUBC34 | Pbr020740.1 | Chr10 | 17324391 | 17325467 | 149 |
| PbrUBC35 | Pbr020719.1 | Chr10 | 17573570 | 17574635 | 149 |
| PbrUBC36 | Pbr020703.1 | Chr10 | 17785964 | 17790523 | 891 |
| PbrUBC37 | Pbr038220.3 | Chr11 | 4285552 | 4289283 | 190 |
| PbrUBC38 | Pbr038323.1 | Chr11 | 5463176 | 5468616 | 589 |
| PbrUBC39 | Pbr017901.1 | Chr11 | 11843306 | 11844937 | 181 |
| PbrUBC40 | Pbr031559.1 | Chr11 | 13000377 | 13002724 | 149 |
| PbrUBC41 | Pbr041320.1 | Chr11 | 21287679 | 21289091 | 152 |
| PbrUBC42 | Pbr017298.1 | Chr11 | 24728725 | 24731067 | 149 |
| PbrUBC43 | Pbr028474.1 | Chr12 | 194166 | 195789 | 149 |
| PbrUBC44 | Pbr016440.1 | Chr12 | 3192332 | 3192601 | 90 |
| PbrUBC45 | Pbr039044.1 | Chr12 | 10209302 | 10211625 | 309 |
| PbrUBC46 | Pbr015391.1 | Chr12 | 19750784 | 19753479 | 147 |
| PbrUBC47 | Pbr010810.1 | Chr13 | 289004 | 291284 | 149 |
| PbrUBC48 | Pbr011958.2 | Chr13 | 9713060 | 9715379 | 231 |
| PbrUBC49 | Pbr010372.1 | Chr14 | 2425473 | 2427613 | 147 |
| PbrUBC50 | Pbr010424.1 | Chr14 | 2933078 | 2935217 | 147 |
| PbrUBC51 | Pbr038166.1 | Chr14 | 7236972 | 7240490 | 273 |
| PbrUBC52 | Pbr026720.1 | Chr14 | 8739052 | 8742374 | 169 |
| PbrUBC53 | Pbr027115.1 | Chr14 | 12937009 | 12939516 | 196 |
| PbrUBC54 | Pbr005908.1 | Chr15 | 2962026 | 2964448 | 162 |
| PbrUBC55 | Pbr009224.1 | Chr15 | 4281248 | 4283960 | 158 |
| PbrUBC56 | Pbr019673.1 | Chr15 | 7696112 | 7697156 | 100 |
| PbrUBC57 | Pbr016945.1 | Chr15 | 14705287 | 14708021 | 524 |
| PbrUBC58 | Pbr017248.1 | Chr15 | 19933740 | 19935641 | 141 |
| PbrUBC59 | Pbr017249.1 | Chr15 | 19937558 | 19939328 | 182 |
| PbrUBC60 | Pbr015294.2 | Chr15 | 23696990 | 23705032 | 371 |
| PbrUBC61 | Pbr024308.1 | Chr15 | 24667752 | 24669854 | 153 |
| PbrUBC62 | Pbr024286.1 | Chr15 | 24961413 | 24963994 | 190 |
| PbrUBC63 | Pbr024279.1 | Chr15 | 25164896 | 25167005 | 178 |
| PbrUBC64 | Pbr017425.1 | Chr15 | 26387149 | 26392677 | 702 |
| PbrUBC65 | Pbr040652.1 | Chr15 | 36951735 | 36952300 | 112 |
| PbrUBC66 | Pbr020836.2 | Chr15 | 42195161 | 42198738 | 228 |
| PbrUBC67 | Pbr012108.1 | Chr16 | 3304638 | 3306350 | 202 |
| PbrUBC68 | Pbr013690.1 | Chr16 | 9644295 | 9646021 | 188 |
| PbrUBC69 | Pbr022472.1 | Chr17 | 2772442 | 2779627 | 468 |
| PbrUBC70 | Pbr026816.1 | Chr17 | 3678331 | 3681687 | 454 |
| PbrUBC71 | Pbr034051.1 | Chr17 | 5335721 | 5339074 | 454 |
| PbrUBC72 | Pbr008641.1 | Chr17 | 6164722 | 6165771 | 350 |
| PbrUBC73 | Pbr040232.1 | Chr17 | 20655869 | 20656618 | 106 |
| PbrUBC74 | Pbr003941.1 | scaffold1182.0 | 19987 | 21731 | 175 |
| PbrUBC75 | Pbr005003.1 | scaffold1241.0 | 889 | 1709 | 133 |
| PbrUBC76 | Pbr005004.1 | scaffold1241.0 | 11548 | 13331 | 182 |
| PbrUBC77 | Pbr006049.1 | scaffold1301.0 | 22029 | 24744 | 149 |
| PbrUBC78 | Pbr009005.1 | scaffold1564.0 | 5159 | 7986 | 149 |
| PbrUBC79 | Pbr028213.1 | scaffold467.0 | 324186 | 326069 | 190 |
| PbrUBC80 | Pbr028219.1 | scaffold467.0 | 348935 | 352574 | 172 |
| PbrUBC81 | Pbr032413.2 | scaffold581.0.1 | 30262 | 34773 | 126 |
| PbrUBC82 | Pbr034367.1 | scaffold640.0 | 26565 | 30347 | 154 |
| PbrUBC83 | Pbr042566.1 | scaffold992.0 | 77372 | 83394 | 147 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Meng, D.; Chen, Y.; Abdullah, M.; Jin, Q.; Lin, Y.; Cai, Y. Comparative and Expression Analysis of Ubiquitin Conjugating Domain-Containing Genes in Two Pyrus Species. Cells 2018, 7, 77. https://doi.org/10.3390/cells7070077
Cao Y, Meng D, Chen Y, Abdullah M, Jin Q, Lin Y, Cai Y. Comparative and Expression Analysis of Ubiquitin Conjugating Domain-Containing Genes in Two Pyrus Species. Cells. 2018; 7(7):77. https://doi.org/10.3390/cells7070077
Chicago/Turabian StyleCao, Yunpeng, Dandan Meng, Yu Chen, Muhammad Abdullah, Qing Jin, Yi Lin, and Yongping Cai. 2018. "Comparative and Expression Analysis of Ubiquitin Conjugating Domain-Containing Genes in Two Pyrus Species" Cells 7, no. 7: 77. https://doi.org/10.3390/cells7070077





