The Winding Road of Cardiac Regeneration—Stem Cell Omics in the Spotlight
Abstract
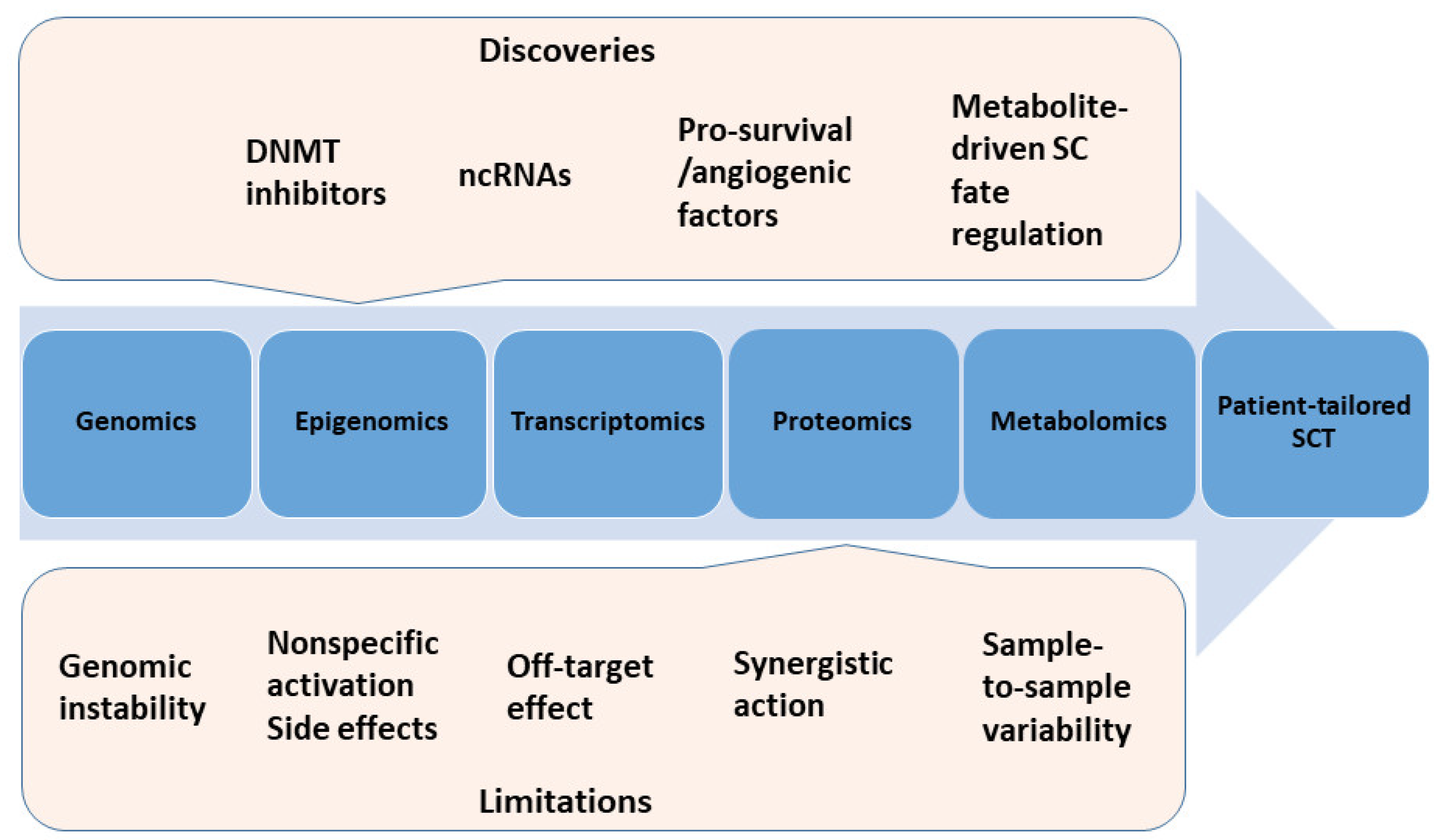
1. Introduction
2. Genomics
3. Epigenomics
3.1. DNA Methylation
3.2. Histones Modification
4. Transcriptomics
4.1. MicroRNAs
4.2. Long Non-Coding RNAs
5. Proteomics
5.1. Secreted Factors
5.2. Genetic Modification of SCs
6. Metabolomics
7. Exosomics
8. Challenges in Omics Data Management
9. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Nunez, G.A.; Sanz-Ruiz, R.; Fernandez, S.M.E.; Fernandez-Aviles, F. “Second-generation” stem cells for cardiac repair. World J. Stem Cell 2015, 7, 352–367. [Google Scholar]
- Fernandez-Aviles, F.; Sanz-Ruiz, R.; Climent, A.M.; Badimon, L.; Bolli, R.; Charron, D.; Fuster, V.; Janssens, S.; Kastrup, J.; Kim, H.S.; et al. Global position paper on cardiovascular regenerative medicine. Eur. Heart J. 2017, 38, 2532–2546. [Google Scholar] [CrossRef] [PubMed]
- Micheu, M.M.; Dorobantu, M. Fifteen years of bone marrow mononuclear cell therapy in acute myocardial infarction. World J. Stem Cell 2017, 9, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Dorobantu, M.; Popa-Fotea, N.M.; Popa, M.; Rusu, I.; Micheu, M.M. Pursuing meaningful end-points for stem cell therapy assessment in ischemic cardiac disease. World J. Stem Cell 2017, 9, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Strauer, B.E.; Brehm, M.; Zeus, T.; Gattermann, N.; Hernandez, A.; Sorg, R.V.; Kogler, G.; Wernet, P. Intracoronary, human autologous stem cell transplantation for myocardial regeneration following myocardial infarction. Deut. Med. Wochenschr. 2001, 126, 932–938. [Google Scholar] [CrossRef]
- Assou, S.; Bouckenheimer, J.; De Vos, J. Concise Review: Assessing the Genome Integrity of Human Induced Pluripotent Stem Cells: What Quality Control Metrics? Stem Cells 2018, 36, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, M.; Hayashizaki, Y.; Murakawa, Y. Genomic Instability of iPSCs: Challenges Towards Their Clinical Applications. Stem Cell Rev. 2017, 13, 7–16. [Google Scholar] [CrossRef]
- Taapken, S.M.; Nisler, B.S.; Newton, M.A.; Sampsell-Barron, T.L.; Leonhard, K.A.; McIntire, E.M.; Montgomery, K.D. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat. Biotechnol. 2011, 29, 313–314. [Google Scholar] [CrossRef]
- Mayshar, Y.; Ben-David, U.; Lavon, N.; Biancotti, J.C.; Yakir, B.; Clark, A.T.; Plath, K.; Lowry, W.E.; Benvenisty, N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell 2010, 7, 521–531. [Google Scholar] [CrossRef]
- Rebuzzini, P.; Zuccotti, M.; Redi, C.A.; Garagna, S. Chromosomal Abnormalities in Embryonic and Somatic Stem Cells. Cytogenet. Genome Res. 2015, 147, 1–9. [Google Scholar] [CrossRef]
- Tosca, L.; Feraud, O.; Magniez, A.; Bas, C.; Griscelli, F.; Bennaceur-Griscelli, A.; Tachdjian, G. Genomic instability of human embryonic stem cell lines using different passaging culture methods. Mol. Cytogenet. 2015, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Geens, M.; Mertzanidou, A.; Jacobs, K.; Heirman, C.; Breckpot, K.; Spits, C. Gain of 20q11.21 in human embryonic stem cells improves cell survival by increased expression of Bcl-xL. Mol. Hum. Reprod. 2014, 20, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Lefort, N.; Feyeux, M.; Bas, C.; Feraud, O.; Bennaceur-Griscelli, A.; Tachdjian, G.; Peschanski, M.; Perrier, A.L. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat. Biotechnol. 2008, 26, 1364–1366. [Google Scholar] [CrossRef] [PubMed]
- Spits, C.; Mateizel, I.; Geens, M.; Mertzanidou, A.; Staessen, C.; Vandeskelde, Y.; Van der Elst, J.; Liebaers, I.; Sermon, K. Recurrent chromosomal abnormalities in human embryonic stem cells. Nat. Biotechnol. 2008, 26, 1361–1363. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Mayshar, Y.; Benvenisty, N. Large-scale analysis reveals acquisition of lineage-specific chromosomal aberrations in human adult stem cells. Cell Stem Cell 2011, 9, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Li, T.S.; Marban, E. Physiological levels of reactive oxygen species are required to maintain genomic stability in stem cells. Stem Cell. 2010, 28, 1178–1185. [Google Scholar] [CrossRef] [PubMed]
- Li, T.S.; Cheng, K.; Malliaras, K.; Matsushita, N.; Sun, B.; Marban, L.; Zhang, Y.; Marban, E. Expansion of human cardiac stem cells in physiological oxygen improves cell production efficiency and potency for myocardial repair. Cardiovasc. Res. 2011, 89, 157–165. [Google Scholar] [CrossRef]
- Oliveira, P.H.; da Silva, C.L.; Cabral, J.M. Concise review: Genomic instability in human stem cells: Current status and future challenges. Stem Cell. 2014, 32, 2824–2832. [Google Scholar] [CrossRef]
- Laurent, L.C.; Ulitsky, I.; Slavin, I.; Tran, H.; Schork, A.; Morey, R.; Lynch, C.; Harness, J.V.; Lee, S.; Barrero, M.J.; et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell 2011, 8, 106–118. [Google Scholar] [CrossRef]
- Cai, J.; Miao, X.; Li, Y.; Smith, C.; Tsang, K.; Cheng, L.; Wang, Q.F. Whole-genome sequencing identifies genetic variances in culture-expanded human mesenchymal stem cells. Stem Cell Rep. 2014, 3, 227–233. [Google Scholar] [CrossRef]
- Kim, M.; Rhee, J.K.; Choi, H.; Kwon, A.; Kim, J.; Lee, G.D.; Jekarl, D.W.; Lee, S.; Kim, Y.; Kim, T.M. Passage-dependent accumulation of somatic mutations in mesenchymal stromal cells during in vitro culture revealed by whole genome sequencing. Sci. Rep. 2017, 7, 14508. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Z.; Chi, Y.; Zhang, Q.; Xu, F.; Yang, Z.; Meng, L.; Yang, S.; Yan, S.; Mao, A.; et al. Long-term cultured mesenchymal stem cells frequently develop genomic mutations but do not undergo malignant transformation. Cell Death Dis. 2013, 4, e950. [Google Scholar] [CrossRef] [PubMed]
- Menasche, P.; Vanneaux, V.; Hagege, A.; Bel, A.; Cholley, B.; Cacciapuoti, I.; Parouchev, A.; Benhamouda, N.; Tachdjian, G.; Tosca, L.; et al. Human embryonic stem cell-derived cardiac progenitors for severe heart failure treatment: First clinical case report. Eur. Heart J. 2015, 36, 2011–2017. [Google Scholar] [CrossRef] [PubMed]
- Godini, R.; Lafta, H.Y.; Fallahi, H. Epigenetic modifications in the embryonic and induced pluripotent stem cells. Gene Expr. Patterns. 2018, 29, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dupont, C.; Armant, D.R.; Brenner, C.A. Epigenetics: Definition, mechanisms and clinical perspective. Semin. Reprod. Med. 2009, 27, 351–357. [Google Scholar] [CrossRef]
- Klungland, A.; Robertson, A.B. Oxidized C5-methyl cytosine bases in DNA: 5-Hydroxymethylcytosine; 5-formylcytosine; and 5-carboxycytosine. Free Radic. Biol. Med. 2017, 107, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.; Kiskinis, E.; Verstappen, G.; Gu, H.; Boulting, G.; Smith, Z.D.; Ziller, M.; Croft, G.F.; Amoroso, M.W.; Oakley, D.H.; et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 2011, 144, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Muller, F.; Liao, J.; Zhang, Y.; Gu, H.; Bock, C.; Boyle, P.; Epstein, C.B.; Bernstein, B.E.; Lengauer, T.; et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011, 7, e1002389. [Google Scholar] [CrossRef] [PubMed]
- Antonitsis, P.; Ioannidou-Papagiannaki, E.; Kaidoglou, A.; Charokopos, N.; Kalogeridis, A.; Kouzi-Koliakou, K.; Kyriakopoulou, I.; Klonizakis, I.; Papakonstantinou, C. Cardiomyogenic potential of human adult bone marrow mesenchymal stem cells in vitro. Thorac. Cardiovasc. Surg. 2008, 56, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.S.; Chen, J.; Luo, G.A.; Zhang, R.L.; Zhao, Y.F.; Wang, Y.M. Proteomic profiling of rat bone marrow mesenchymal stem cells induced by 5-azacytidine. Stem Cells Dev. 2006, 15, 665–676. [Google Scholar] [CrossRef]
- Antonitsis, P.; Ioannidou-Papagiannaki, E.; Kaidoglou, A.; Papakonstantinou, C. In vitro cardiomyogenic differentiation of adult human bone marrow mesenchymal stem cells. The role of 5-azacytidine. Int. Cardiovasc. Thorac. Surg. 2007, 6, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Ye, N.S.; Zhang, R.L.; Zhao, Y.F.; Feng, X.; Wang, Y.M.; Luo, G.A. Effect of 5-azacytidine on the protein expression of porcine bone marrow mesenchymal stem cells in vitro. Genom. Proteom. Bioinform. 2006, 4, 18–25. [Google Scholar] [CrossRef]
- Martin-Rendon, E.; Sweeney, D.; Lu, F.; Girdlestone, J.; Navarrete, C.; Watt, S.M. 5-Azacytidine-treated human mesenchymal stem/progenitor cells derived from umbilical cord, cord blood and bone marrow do not generate cardiomyocytes in vitro at high frequencies. Vox Sang. 2008, 95, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Song, J.; Liu, W.; Wan, Y.; Chen, X.; Hu, C. Growth and differentiation of rat bone marrow stromal cells: Does 5-azacytidine trigger their cardiomyogenic differentiation? Cardiovasc. Res. 2003, 58, 460–468. [Google Scholar] [CrossRef]
- Rosca, A.M.; Burlacu, A. Effect of 5-azacytidine: Evidence for alteration of the multipotent ability of mesenchymal stem cells. Stem Cells Dev. 2011, 20, 1213–1221. [Google Scholar] [CrossRef] [PubMed]
- Naeem, N.; Haneef, K.; Kabir, N.; Iqbal, H.; Jamall, S.; Salim, A. DNA methylation inhibitors, 5-azacytidine and zebularine potentiate the transdifferentiation of rat bone marrow mesenchymal stem cells into cardiomyocytes. Cardiovasc. Ther. 2013, 31, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Horrillo, A.; Pezzolla, D.; Fraga, M.F.; Aguilera, Y.; Salguero-Aranda, C.; Tejedo, J.R.; Martin, F.; Bedoya, F.J.; Soria, B.; Hmadcha, A. Zebularine regulates early stages of mESC differentiation: Effect on cardiac commitment. Cell Death Dis. 2013, 4, e570. [Google Scholar] [CrossRef] [PubMed]
- Berdasco, M.; Esteller, M. DNA methylation in stem cell renewal and multipotency. Stem Cell Res. Ther. 2011, 2, 42. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Jin, Y. Role of Oct4 in maintaining and regaining stem cell pluripotency. Stem Cell Res. Ther. 2010, 1, 39. [Google Scholar] [CrossRef] [PubMed]
- Lagarkova, M.A.; Volchkov, P.Y.; Lyakisheva, A.V.; Philonenko, E.S.; Kiselev, S.L. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle 2006, 5, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Fouse, S.D.; Shen, Y.; Pellegrini, M.; Cole, S.; Meissner, A.; Van Neste, L.; Jaenisch, R.; Fan, G. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell 2008, 2, 160–169. [Google Scholar] [CrossRef]
- Barrand, S.; Collas, P. Chromatin states of core pluripotency-associated genes in pluripotent, multipotent and differentiated cells. Biochem. Biophys. Res. Commun. 2010, 391, 762–767. [Google Scholar] [CrossRef]
- Yang, J.Y.; Wang, Q.; Wang, W.; Zeng, L.F. Histone deacetylases and cardiovascular cell lineage commitment. World J. Stem Cells 2015, 7, 852–858. [Google Scholar] [CrossRef]
- Zeng, L.; Xiao, Q.; Margariti, A.; Zhang, Z.; Zampetaki, A.; Patel, S.; Capogrossi, M.C.; Hu, Y.; Xu, Q. HDAC3 is crucial in shear- and VEGF-induced stem cell differentiation toward endothelial cells. J. Cell Biol. 2006, 174, 1059–1069. [Google Scholar] [CrossRef]
- Dovey, O.M.; Foster, C.T.; Cowley, S.M. Histone deacetylase 1 (HDAC1), but not HDAC2, controls embryonic stem cell differentiation. Proc. Natl. Acad. Sci. USA 2010, 107, 8242–8247. [Google Scholar] [CrossRef]
- Lu, D.F.; Yao, Y.; Su, Z.Z.; Zeng, Z.H.; Xing, X.W.; He, Z.Y.; Zhang, C. Downregulation of HDAC1 is involved in the cardiomyocyte differentiation from mesenchymal stem cells in a myocardial microenvironment. PLoS ONE 2014, 9, e93222. [Google Scholar] [CrossRef]
- Lu, D.F.; Wang, Y.; Su, Z.Z.; Zeng, Z.H.; Xing, X.W.; He, Z.Y.; Zhang, C. Knockdown of the HDAC1 promotes the directed differentiation of bone mesenchymal stem cells into cardiomyocytes. PLoS ONE 2014, 9, e92179. [Google Scholar] [CrossRef]
- Liu, Z.; Li, T.; Liu, Y.; Jia, Z.; Li, Y.; Zhang, C.; Chen, P.; Ma, K.; Affara, N.; Zhou, C. WNT signaling promotes Nkx2.5 expression and early cardiomyogenesis via downregulation of Hdac1. Biochim. Biophys. Acta 2009, 1793, 300–311. [Google Scholar] [CrossRef]
- Hoxha, E.; Lambers, E.; Wasserstrom, J.A.; Mackie, A.; Ramirez, V.; Abramova, T.; Verma, S.K.; Krishnamurthy, P.; Kishore, R. Elucidation of a novel pathway through which HDAC1 controls cardiomyocyte differentiation through expression of SOX-17 and BMP2. PLoS ONE 2012, 7, e45046. [Google Scholar] [CrossRef]
- Hoxha, E.; Lambers, E.; Xie, H.; De Andrade, A.; Krishnamurthy, P.; Wasserstrom, J.A.; Ramirez, V.; Thal, M.; Verma, S.K.; Soares, M.B.; et al. Histone deacetylase 1 deficiency impairs differentiation and electrophysiological properties of cardiomyocytes derived from induced pluripotent cells. Stem Cell. 2012, 30, 2412–2422. [Google Scholar] [CrossRef]
- Hasin, Y.; Seldin, M.; Lusis, A. Multi-omics approaches to disease. Genome Biol. 2017, 18, 83. [Google Scholar] [CrossRef]
- Billing, A.M.; Ben Hamidane, H.; Dib, S.S.; Cotton, R.J.; Bhagwat, A.M.; Kumar, P.; Hayat, S.; Yousri, N.A.; Goswami, N.; Suhre, K.; et al. Comprehensive transcriptomic and proteomic characterization of human mesenchymal stem cells reveals source specific cellular markers. Sci. Rep. 2016, 6, 21507. [Google Scholar] [CrossRef]
- Medina, R.J.; Barber, C.L.; Sabatier, F.; Dignat-George, F.; Melero-Martin, J.M.; Khosrotehrani, K.; Ohneda, O.; Randi, A.M.; Chan, J.K.Y.; Yamaguchi, T.; et al. Endothelial Progenitors: A Consensus Statement on Nomenclature. Stem Cells Transl. Med. 2017, 6, 1316–1320. [Google Scholar] [CrossRef]
- Medina, R.J.; O’Neill, C.L.; Sweeney, M.; Guduric-Fuchs, J.; Gardiner, T.A.; Simpson, D.A.; Stitt, A.W. Molecular analysis of endothelial progenitor cell (EPC) subtypes reveals two distinct cell populations with different identities. BMC Med. Genom. 2010, 3, 18. [Google Scholar] [CrossRef]
- Steinle, H.; Behring, A.; Schlensak, C.; Wendel, H.P.; Avci-Adali, M. Concise Review: Application of In Vitro Transcribed Messenger RNA for Cellular Engineering and Reprogramming: Progress and Challenges. Stem Cell. 2017, 35, 68–79. [Google Scholar] [CrossRef]
- Zangi, L.; Lui, K.O.; von Gise, A.; Ma, Q.; Ebina, W.; Ptaszek, L.M.; Spater, D.; Xu, H.; Tabebordbar, M.; Gorbatov, R.; et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013, 31, 898–907. [Google Scholar] [CrossRef]
- Seeger, F.H.; Zeiher, A.M.; Dimmeler, S. MicroRNAs in stem cell function and regenerative therapy of the heart. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1739–1746. [Google Scholar] [CrossRef]
- Kuppusamy, K.T.; Sperber, H.; Ruohola-Baker, H. MicroRNA regulation and role in stem cell maintenance, cardiac differentiation and hypertrophy. Curr. Mol. Med. 2013, 13, 757–764. [Google Scholar] [CrossRef]
- Kane, N.M.; Thrasher, A.J.; Angelini, G.D.; Emanueli, C. Concise review: MicroRNAs as modulators of stem cells and angiogenesis. Stem Cell. 2014, 32, 1059–1066. [Google Scholar] [CrossRef]
- Li, B.; Meng, X.; Zhang, L. microRNAs and cardiac stem cells in heart development and disease. Drug Discov. Today. 2018, in press. [Google Scholar] [CrossRef]
- Prathipati, P.; Nandi, S.S.; Mishra, P.K. Stem Cell-Derived Exosomes, Autophagy, Extracellular Matrix Turnover, and miRNAs in Cardiac Regeneration during Stem Cell Therapy. Stem Cell Rev. 2017, 13, 79–91. [Google Scholar] [CrossRef]
- Wojciechowska, A.; Braniewska, A.; Kozar-Kaminska, K. MicroRNA in cardiovascular biology and disease. Adv. Clin. Exp. Med. 2017, 26, 865–874. [Google Scholar] [CrossRef]
- Jakob, P.; Doerries, C.; Briand, S.; Mocharla, P.; Krankel, N.; Besler, C.; Mueller, M.; Manes, C.; Templin, C.; Baltes, C.; et al. Loss of angiomiR-126 and 130a in angiogenic early outgrowth cells from patients with chronic heart failure: Role for impaired in vivo neovascularization and cardiac repair capacity. Circulation 2012, 126, 2962–2975. [Google Scholar] [CrossRef]
- Chen, J.J.; Zhou, S.H. Mesenchymal stem cells overexpressing MiR-126 enhance ischemic angiogenesis via the AKT/ERK-related pathway. Cardiol. J. 2011, 18, 675–681. [Google Scholar] [CrossRef]
- Huang, F.; Zhu, X.; Hu, X.Q.; Fang, Z.F.; Tang, L.; Lu, X.L.; Zhou, S.H. Mesenchymal stem cells modified with miR-126 release angiogenic factors and activate Notch ligand Delta-like-4, enhancing ischemic angiogenesis and cell survival. Int. J. Mol. Med. 2013, 31, 484–492. [Google Scholar] [CrossRef]
- Wang, S.; Aurora, A.B.; Johnson, B.A.; Qi, X.; McAnally, J.; Hill, J.A.; Richardson, J.A.; Bassel-Duby, R.; Olson, E.N. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 2008, 15, 261–271. [Google Scholar] [CrossRef]
- Hu, S.; Huang, M.; Nguyen, P.K.; Gong, Y.; Li, Z.; Jia, F.; Lan, F.; Liu, J.; Nag, D.; Robbins, R.C.; et al. Novel microRNA prosurvival cocktail for improving engraftment and function of cardiac progenitor cell transplantation. Circulation 2011, 124 (Suppl. 11), S27–S34. [Google Scholar] [CrossRef]
- Choong, O.K.; Lee, D.S.; Chen, C.Y.; Hsieh, P.C. The roles of non-coding RNAs in cardiac regenerative medicine. Non-Coding RNA Res. 2017, 2, 100–110. [Google Scholar] [CrossRef]
- Gu, G.L.; Xu, X.L.; Sun, X.T.; Zhang, J.; Guo, C.F.; Wang, C.S.; Sun, B.; Guo, G.L.; Ma, K.; Huang, Y.Y.; et al. Cardioprotective Effect of MicroRNA-21 in Murine Myocardial Infarction. Cardiovasc. Ther. 2015, 33, 109–117. [Google Scholar] [CrossRef]
- Gupta, S.K.; Itagaki, R.; Zheng, X.; Batkai, S.; Thum, S.; Ahmad, F.; Van Aelst, L.N.; Sharma, A.; Piccoli, M.T.; Weinberger, F.; et al. miR-21 promotes fibrosis in an acute cardiac allograft transplantation model. Cardiovasc. Res. 2016, 110, 215–226. [Google Scholar] [CrossRef]
- Glass, C.; Singla, D.K. MicroRNA-1 transfected embryonic stem cells enhance cardiac myocyte differentiation and inhibit apoptosis by modulating the PTEN/Akt pathway in the infarcted heart. Am. J. Physiol. Heart Circ. Physiol. 2011, 301, H2038–H2049. [Google Scholar] [CrossRef]
- Ivey, K.N.; Muth, A.; Arnold, J.; King, F.W.; Yeh, R.F.; Fish, J.E.; Hsiao, E.C.; Schwartz, R.J.; Conklin, B.R.; Bernstein, H.S.; et al. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell 2008, 2, 219–229. [Google Scholar] [CrossRef]
- Wilson, K.D.; Hu, S.; Venkatasubrahmanyam, S.; Fu, J.D.; Sun, N.; Abilez, O.J.; Baugh, J.J.; Jia, F.; Ghosh, Z.; Li, R.A.; et al. Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: Role for miR-499. Circ. Cardiovasc. Genet. 2010, 3, 426–435. [Google Scholar] [CrossRef]
- Sluijter, J.P.; van Mil, A.; van Vliet, P.; Metz, C.H.; Liu, J.; Doevendans, P.A.; Goumans, M.J. MicroRNA-1 and -499 Regulate Differentiation and Proliferation in Human-Derived Cardiomyocyte Progenitor Cells. Arteriosc. Throm. Vasc. Biol. 2010, 30, 859–868. [Google Scholar] [CrossRef]
- Eulalio, A.; Mano, M.; Dal Ferro, M.; Zentilin, L.; Sinagra, G.; Zacchigna, S.; Giacca, M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature 2012, 492, 376–381. [Google Scholar] [CrossRef]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef]
- Flynn, R.A.; Chang, H.Y. Long noncoding RNAs in cell-fate programming and reprogramming. Cell Stem Cell 2014, 14, 752–761. [Google Scholar] [CrossRef]
- Di Mauro, V.; Barandalla-Sobrados, M.; Catalucci, D. The noncoding-RNA landscape in cardiovascular health and disease. Non. Coding RNA Res. 2018, 3, 12–19. [Google Scholar] [CrossRef]
- Klattenhoff, C.A.; Scheuermann, J.C.; Surface, L.E.; Bradley, R.K.; Fields, P.A.; Steinhauser, M.L.; Ding, H.; Butty, V.L.; Torrey, L.; Haas, S.; et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 2013, 152, 570–583. [Google Scholar] [CrossRef]
- Ounzain, S.; Micheletti, R.; Arnan, C.; Plaisance, I.; Cecchi, D.; Schroen, B.; Reverter, F.; Alexanian, M.; Gonzales, C.; Ng, S.Y.; et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J. Mol. Cell. Cardiol. 2015, 89, 98–112. [Google Scholar] [CrossRef]
- Loewer, S.; Cabili, M.N.; Guttman, M.; Loh, Y.H.; Thomas, K.; Park, I.H.; Garber, M.; Curran, M.; Onder, T.; Agarwal, S.; et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat. Genet. 2010, 42, 1113–1117. [Google Scholar] [CrossRef]
- Shi, B.; Wang, Y.; Zhao, R.; Long, X.; Deng, W.; Wang, Z. Bone marrow mesenchymal stem cell-derived exosomal miR-21 protects C-kit+ cardiac stem cells from oxidative injury through the PTEN/PI3K/Akt axis. PLoS ONE 2018, 13, e0191616. [Google Scholar] [CrossRef]
- Zamani, P.; Fereydouni, N.; Butler, A.E.; Navashenaq, J.G.; Sahebkar, A. The therapeutic and diagnostic role of exosomes in cardiovascular diseases. Trends Cardiovasc. Med. 2018, in press. [Google Scholar] [CrossRef]
- Feng, Y.; Huang, W.; Wani, M.; Yu, X.; Ashraf, M. Ischemic preconditioning potentiates the protective effect of stem cells through secretion of exosomes by targeting Mecp2 via miR-22. PLoS ONE 2014, 9, e88685. [Google Scholar] [CrossRef]
- Gong, M.; Yu, B.; Wang, J.; Wang, Y.; Liu, M.; Paul, C.; Millard, R.W.; Xiao, D.S.; Ashraf, M.; Xu, M. Mesenchymal stem cells release exosomes that transfer miRNAs to endothelial cells and promote angiogenesis. Oncotarget 2017, 8, 45200–45212. [Google Scholar] [CrossRef]
- De Couto, G.; Gallet, R.; Cambier, L.; Jaghatspanyan, E.; Makkar, N.; Dawkins, J.F.; Berman, B.P.; Marban, E. Exosomal MicroRNA Transfer into Macrophages Mediates Cellular Postconditioning. Circulation 2017, 136, 200–214. [Google Scholar] [CrossRef]
- Ibrahim, A.G.; Cheng, K.; Marban, E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Rep. 2014, 2, 606–619. [Google Scholar] [CrossRef]
- Wang, X.; Ha, T.; Liu, L.; Zou, J.; Zhang, X.; Kalbfleisch, J.; Gao, X.; Williams, D.; Li, C. Increased expression of microRNA-146a decreases myocardial ischaemia/reperfusion injury. Cardiovasc. Res. 2013, 97, 432–442. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; Cui, J.; Sun, M.; Pu, Z.; Wang, C.; Du, W.; Liu, X.; Wu, J.; Hou, J.; et al. miR199a-3p regulates P53 by targeting CABLES1 in mouse cardiac c-kit(+) cells to promote proliferation and inhibit apoptosis through a negative feedback loop. Stem Cell Res. Ther. 2017, 8, 127. [Google Scholar] [CrossRef]
- Lee, S.; Yu, K.R.; Ryu, Y.S.; Oh, Y.S.; Hong, I.S.; Kim, H.S.; Lee, J.Y.; Kim, S.; Seo, K.W.; Kang, K.S. miR-543 and miR-590-3p regulate human mesenchymal stem cell aging via direct targeting of AIMP3/p18. Age 2014, 36, 9724. [Google Scholar] [CrossRef]
- Kim, H.W.; Haider, H.K.; Jiang, S.; Ashraf, M. Ischemic preconditioning augments survival of stem cells via miR-210 expression by targeting caspase-8-associated protein 2. J. Biol. Chem. 2009, 284, 33161–33168. [Google Scholar] [CrossRef]
- Khan, M.; Nickoloff, E.; Abramova, T.; Johnson, J.; Verma, S.K.; Krishnamurthy, P.; Mackie, A.R.; Vaughan, E.; Garikipati, V.N.; Benedict, C.; et al. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ. Res. 2015, 117, 52–64. [Google Scholar] [CrossRef]
- Wu, F.; Yang, Z.; Li, G. Role of specific microRNAs for endothelial function and angiogenesis. Biochem Biophys. Res. Commun. 2009, 386, 549–553. [Google Scholar] [CrossRef]
- Hou, J.; Long, H.; Zhou, C.; Zheng, S.; Wu, H.; Guo, T.; Wu, Q.; Zhong, T.; Wang, T. Long noncoding RNA Braveheart promotes cardiogenic differentiation of mesenchymal stem cells in vitro. Stem Cell Res. Ther. 2017, 8, 4. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Z.; Jiang, J.; Xu, C.; Kang, J.; Xiao, L.; Wu, M.; Xiong, J.; Guo, X.; Liu, H. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev. Cell. 2013, 25, 69–80. [Google Scholar] [CrossRef]
- Wyles, S.P.; Faustino, R.S.; Li, X.; Terzic, A.; Nelson, T.J. Systems-based technologies in profiling the stem cell molecular framework for cardioregenerative medicine. Stem Cell Rev. 2015, 11, 501–510. [Google Scholar] [CrossRef]
- Prudhomme, W.; Daley, G.Q.; Zandstra, P.; Lauffenburger, D.A. Multivariate proteomic analysis of murine embryonic stem cell self-renewal versus differentiation signaling. Proc. Natl. Acad. Sci. USA 2004, 101, 2900–2905. [Google Scholar] [CrossRef]
- Abazova, N.; Krijgsveld, J. Advances in stem cell proteomics. Curr. Opin. Genet. Dev. 2017, 46, 149–155. [Google Scholar] [CrossRef]
- Kim, M.S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575–581. [Google Scholar] [CrossRef]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-spectrometry-based draft of the human proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef]
- Tao, Z.H.; Han, Z.C.; Li, Z. Proangiogenic Features of Mesenchymal Stem Cells and Their Therapeutic Applications. Stem Cell. Int. 2016. [Google Scholar] [CrossRef]
- Pankajakshan, D.; Agrawal, D.K. Mesenchymal Stem Cell Paracrine Factors in Vascular Repair and Regeneration. J. Biomed. Technol. Res. 2014, 1. [Google Scholar] [CrossRef]
- Hastings, C.L.; Roche, E.T.; Ruiz-Hernandez, E.; Schenke-Layland, K.; Walsh, C.J.; Duffy, G.P. Drug and cell delivery for cardiac regeneration. Adv. Drug Deliv. Rev. 2015, 84, 85–106. [Google Scholar] [CrossRef]
- Ge, Q.; Zhang, H.; Hou, J.; Wan, L.; Cheng, W.; Wang, X.; Dong, D.; Chen, C.; Xia, J.; Guo, J.; et al. VEGF secreted by mesenchymal stem cells mediates the differentiation of endothelial progenitor cells into endothelial cells via paracrine mechanisms. Mol. Med. Rep. 2018, 17, 1667–1675. [Google Scholar] [CrossRef]
- Stastna, M.; Abraham, M.R.; Van Eyk, J.E. Cardiac stem/progenitor cells, secreted proteins, and proteomics. FEBS Lett. 2009, 583, 1800–1807. [Google Scholar] [CrossRef]
- Razban, V.; Lotfi, A.S.; Soleimani, M.; Ahmadi, H.; Massumi, M.; Khajeh, S.; Ghaedi, M.; Arjmand, S.; Najavand, S.; Khoshdel, A. HIF-1alpha Overexpression Induces Angiogenesis in Mesenchymal Stem Cells. Bio. Res. Open Access 2012, 1, 174–183. [Google Scholar] [CrossRef]
- Urbich, C.; Aicher, A.; Heeschen, C.; Dernbach, E.; Hofmann, W.K.; Zeiher, A.M.; Dimmeler, S. Soluble factors released by endothelial progenitor cells promote migration of endothelial cells and cardiac resident progenitor cells. J. Mol. Cell Cardiol. 2005, 39, 733–742. [Google Scholar] [CrossRef]
- Bartunek, J.; Behfar, A.; Dolatabadi, D.; Vanderheyden, M.; Ostojic, M.; Dens, J.; El Nakadi, B.; Banovic, M.; Beleslin, B.; Vrolix, M.; et al. Cardiopoietic stem cell therapy in heart failure: The C-CURE (Cardiopoietic stem Cell therapy in heart failURE) multicenter randomized trial with lineage-specified biologics. J. Am. Coll. Cardiol. 2013, 61, 2329–2338. [Google Scholar] [CrossRef]
- Bartunek, J.; Terzic, A.; Davison, B.A.; Filippatos, G.S.; Radovanovic, S.; Beleslin, B.; Merkely, B.; Musialek, P.; Wojakowski, W.; Andreka, P.; et al. Cardiopoietic cell therapy for advanced ischaemic heart failure: Results at 39 weeks of the prospective, randomized, double blind, sham-controlled CHART-1 clinical trial. Eur. Heart J. 2017, 38, 648–660. [Google Scholar]
- Chung, E.S.; Miller, L.; Patel, A.N.; Anderson, R.D.; Mendelsohn, F.O.; Traverse, J.; Silver, K.H.; Shin, J.; Ewald, G.; Farr, M.J.; et al. Changes in ventricular remodelling and clinical status during the year following a single administration of stromal cell-derived factor-1 non-viral gene therapy in chronic ischaemic heart failure patients: The STOP-HF randomized Phase II trial. Eur. Heart J. 2015, 36, 2228–2238. [Google Scholar] [CrossRef]
- Oliveira, M.S.; Saldanha-Araujo, F.; Goes, A.M.; Costa, F.F.; de Carvalho, J.L. Stem cells in cardiovascular diseases: Turning bad days into good ones. Drug Dis. Today 2017, 22, 1730–1739. [Google Scholar] [CrossRef]
- Abdelwahid, E.; Kalvelyte, A.; Stulpinas, A.; de Carvalho, K.A.; Guarita-Souza, L.C.; Foldes, G. Stem cell death and survival in heart regeneration and repair. Apoptosis 2016, 21, 252–268. [Google Scholar] [CrossRef]
- Li, L.; Chen, X.; Wang, W.E.; Zeng, C. How to Improve the Survival of Transplanted Mesenchymal Stem Cell in Ischemic Heart? Stem Cell. Int. 2016, 2016, 9682757. [Google Scholar] [CrossRef]
- Der Sarkissian, S.; Levesque, T.; Noiseux, N. Optimizing stem cells for cardiac repair: Current status and new frontiers in regenerative cardiology. World J. Stem Cell. 2017, 9, 9–25. [Google Scholar] [CrossRef]
- Matsumoto, R.; Omura, T.; Yoshiyama, M.; Hayashi, T.; Inamoto, S.; Koh, K.R.; Ohta, K.; Izumi, Y.; Nakamura, Y.; Akioka, K.; et al. Vascular endothelial growth factor-expressing mesenchymal stem cell transplantation for the treatment of acute myocardial infarction. Arteriosc. Throm. Vascul. Biol. 2005, 25, 1168–1173. [Google Scholar] [CrossRef]
- Yi, F.; Guo, W.Y.; Lu, A.L.; Wang, H.C.; Li, H.; Li, W.J.; Liu, B.; Zhang, D.X.; Luan, R.H.; Cheng, H.X.; et al. Vascular endothelial growth factor expressing mesenchymal stem cells improves cardiac function in chronic myocardial infarction in pigs. Chin. Med. J. 2006, 119, 1664–1668. [Google Scholar]
- Mangi, A.A.; Noiseux, N.; Kong, D.; He, H.; Rezvani, M.; Ingwall, J.S.; Dzau, V.J. Mesenchymal stem cells modified with Akt prevent remodeling and restore performance of infarcted hearts. Nat. Med. 2003, 9, 1195–1201. [Google Scholar] [CrossRef]
- Noiseux, N.; Gnecchi, M.; Lopez-Ilasaca, M.; Zhang, L.; Solomon, S.D.; Deb, A.; Dzau, V.J.; Pratt, R.E. Mesenchymal stem cells overexpressing Akt dramatically repair infarcted myocardium and improve cardiac function despite infrequent cellular fusion or differentiation. Mol. Ther. J. Am. Soc. Gene Ther. 2006, 14, 840–850. [Google Scholar] [CrossRef]
- Gnecchi, M.; He, H.; Noiseux, N.; Liang, O.D.; Zhang, L.; Morello, F.; Mu, H.; Melo, L.G.; Pratt, R.E.; Ingwall, J.S.; et al. Evidence supporting paracrine hypothesis for Akt-modified mesenchymal stem cell-mediated cardiac protection and functional improvement. FASEB J. 2006, 20, 661–669. [Google Scholar] [CrossRef]
- Jiang, S.; Haider, H.; Idris, N.M.; Salim, A.; Ashraf, M. Supportive interaction between cell survival signaling and angiocompetent factors enhances donor cell survival and promotes angiomyogenesis for cardiac repair. Circ. Res. 2006, 99, 776–784. [Google Scholar] [CrossRef]
- Chao, J.; Bledsoe, G.; Chao, L. Kallikrein-kinin in stem cell therapy. World J. Stem Cells 2014, 6, 448–457. [Google Scholar] [CrossRef]
- Devetzi, M.; Goulielmaki, M.; Khoury, N.; Spandidos, D.A.; Sotiropoulou, G.; Christodoulou, I.; Zoumpourlis, V. Genetically modified stem cells in treatment of human diseases: Tissue kallikrein (KLK1) based targeted therapy (Review). Int. J. Mol. Med. 2018, 41, 1177–1186. [Google Scholar] [CrossRef]
- Yin, H.; Chao, L.; Chao, J. Nitric oxide mediates cardiac protection of tissue kallikrein by reducing inflammation and ventricular remodeling after myocardial ischemia/reperfusion. Life Sci. 2008, 82, 156–165. [Google Scholar] [CrossRef]
- Westermann, D.; Schultheiss, H.P.; Tschope, C. New perspective on the tissue kallikrein-kinin system in myocardial infarction: Role of angiogenesis and cardiac regeneration. Int. Immunopharmacol. 2008, 8, 148–154. [Google Scholar] [CrossRef]
- Yao, Y.Y.; Yin, H.; Shen, B.; Smith, R.S., Jr.; Liu, Y.; Gao, L.; Chao, L.; Chao, J. Tissue kallikrein promotes neovascularization and improves cardiac function by the Akt-glycogen synthase kinase-3beta pathway. Cardiovasc. Res. 2008, 80, 354–364. [Google Scholar] [CrossRef]
- Yin, H.; Chao, L.; Chao, J. Kallikrein/kinin protects against myocardial apoptosis after ischemia/reperfusion via Akt-glycogen synthase kinase-3 and Akt-Bad.14-3-3 signaling pathways. J. Biol. Chem. 2005, 280, 8022–8030. [Google Scholar] [CrossRef]
- Schumann, G.B.; Crisman, L.G. Cerebrospinal fluid cytopathology. Clin. Lab. Med. 1985, 5, 275–302. [Google Scholar] [CrossRef]
- Spillmann, F.; Graiani, G.; Van Linthout, S.; Meloni, M.; Campesi, I.; Lagrasta, C.; Westermann, D.; Tschope, C.; Quaini, F.; Emanueli, C.; et al. Regional and global protective effects of tissue kallikrein gene delivery to the peri-infarct myocardium. Regen. Med. 2006, 1, 235–254. [Google Scholar] [CrossRef]
- Gao, L.; Bledsoe, G.; Yin, H.; Shen, B.; Chao, L.; Chao, J. Tissue kallikrein-modified mesenchymal stem cells provide enhanced protection against ischemic cardiac injury after myocardial infarction. Circ. J. 2013, 77, 2134–2144. [Google Scholar] [CrossRef]
- Duan, H.F.; Wu, C.T.; Wu, D.L.; Lu, Y.; Liu, H.J.; Ha, X.Q.; Zhang, Q.W.; Wang, H.; Jia, X.X.; Wang, L.S. Treatment of myocardial ischemia with bone marrow-derived mesenchymal stem cells overexpressing hepatocyte growth factor. Mol. Ther. J. Am. Soc. Gene Ther. 2003, 8, 467–474. [Google Scholar] [CrossRef]
- Song, H.; Kwon, K.; Lim, S.; Kang, S.M.; Ko, Y.G.; Xu, Z.; Chung, J.H.; Kim, B.S.; Lee, H.; Joung, B.; et al. Transfection of mesenchymal stem cells with the FGF-2 gene improves their survival under hypoxic conditions. Mol. Cell. 2005, 19, 402–407. [Google Scholar]
- Haider, H.; Jiang, S.; Idris, N.M.; Ashraf, M. IGF-1-overexpressing mesenchymal stem cells accelerate bone marrow stem cell mobilization via paracrine activation of SDF-1alpha/CXCR4 signaling to promote myocardial repair. Circ. Res. 2008, 103, 1300–1308. [Google Scholar] [CrossRef]
- Song, S.W.; Chang, W.; Song, B.W.; Song, H.; Lim, S.; Kim, H.J.; Cha, M.J.; Choi, E.; Im, S.H.; Chang, B.C.; et al. Integrin-linked kinase is required in hypoxic mesenchymal stem cells for strengthening cell adhesion to ischemic myocardium. Stem Cell. 2009, 27, 1358–1365. [Google Scholar] [CrossRef]
- Tang, J.; Wang, J.; Guo, L.; Kong, X.; Yang, J.; Zheng, F.; Zhang, L.; Huang, Y.; et al. Mesenchymal stem cells modified with stromal cell-derived factor 1 alpha improve cardiac remodeling via paracrine activation of hepatocyte growth factor in a rat model of myocardial infarction. Mol. Cell. 2010, 29, 9–19. [Google Scholar] [CrossRef]
- Bao, C.; Guo, J.; Zheng, M.; Chen, Y.; Lin, G.; Hu, M. Enhancement of the survival of engrafted mesenchymal stem cells in the ischemic heart by TNFR gene transfection. Biochemistry and cell biology. Biochem. Cell Biol. 2010, 88, 629–634. [Google Scholar] [CrossRef]
- Tang, Y.L.; Tang, Y.; Zhang, Y.C.; Qian, K.; Shen, L.; Phillips, M.I. Improved graft mesenchymal stem cell survival in ischemic heart with a hypoxia-regulated heme oxygenase-1 vector. J. Am. Coll. Cardiol. 2005, 46, 1339–1350. [Google Scholar] [CrossRef]
- Yao, Y.; Sheng, Z.; Li, Y.; Yan, F.; Fu, C.; Li, Y.; Ma, G.; Liu, N.; Chao, J.; Chao, L. Tissue kallikrein promotes cardiac neovascularization by enhancing endothelial progenitor cell functional capacity. Hum. Gene Ther. 2012, 23, 859–870. [Google Scholar] [CrossRef]
- Yao, Y.; Sheng, Z.; Li, Y.; Fu, C.; Ma, G.; Liu, N.; Chao, J.; Chao, L. Tissue kallikrein-modified human endothelial progenitor cell implantation improves cardiac function via enhanced activation of akt and increased angiogenesis. Lab. Investig. 2013, 93, 577–591. [Google Scholar] [CrossRef]
- Sen, S.; Merchan, J.; Dean, J.; Ii, M.; Gavin, M.; Silver, M.; Tkebuchava, T.; Yoon, Y.S.; Rasko, J.E.; Aikawa, R. Autologous transplantation of endothelial progenitor cells genetically modified by adeno-associated viral vector delivering insulin-like growth factor-1 gene after myocardial infarction. Hum. Gene Ther. 2010, 21, 1327–1334. [Google Scholar] [CrossRef]
- Monsanto, M.M.; White, K.S.; Kim, T.; Wang, B.J.; Fisher, K.; Ilves, K.; Khalafalla, F.G.; Casillas, A.; Broughton, K.; Mohsin, S.; et al. Concurrent Isolation of 3 Distinct Cardiac Stem Cell Populations from a Single Human Heart Biopsy. Circ. Res. 2017, 121, 113–124. [Google Scholar] [CrossRef]
- Nadal-Ginard, B.; Ellison, G.M.; Torella, D. The cardiac stem cell compartment is indispensable for myocardial cell homeostasis, repair and regeneration in the adult. Stem Cell Res. 2014, 13, 615–630. [Google Scholar] [CrossRef] [PubMed]
- Messina, E.; De Angelis, L.; Frati, G.; Morrone, S.; Chimenti, S.; Fiordaliso, F.; Salio, M.; Battaglia, M.; Latronico, M.V.; Coletta, M.; et al. Isolation and expansion of adult cardiac stem cells from human and murine heart. Circ. Res. 2004, 95, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Beltrami, A.P.; Barlucchi, L.; Torella, D.; Baker, M.; Limana, F.; Chimenti, S.; Kasahara, H.; Rota, M.; Musso, E.; Urbanek, K.; et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003, 114, 763–776. [Google Scholar] [CrossRef]
- Molgat, A.S.; Tilokee, E.L.; Rafatian, G.; Vulesevic, B.; Ruel, M.; Milne, R.; Suuronen, E.J.; Davis, D.R. Hyperglycemia inhibits cardiac stem cell-mediated cardiac repair and angiogenic capacity. Circulation 2014, 130 (Suppl 1), S70–S76. [Google Scholar] [CrossRef]
- Hsiao, L.C.; Perbellini, F.; Gomes, R.S.; Tan, J.J.; Vieira, S.; Faggian, G.; Clarke, K.; Carr, C.A. Murine cardiosphere-derived cells are impaired by age but not by cardiac dystrophic dysfunction. Stem Cells Dev. 2014, 23, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Olivieri, F.; Recchioni, R.; Marcheselli, F.; Abbatecola, A.M.; Santini, G.; Borghetti, G.; Antonicelli, R.; Procopio, A.D. Cellular senescence in cardiovascular diseases: Potential age-related mechanisms and implications for treatment. Curr. Pharm. Des. 2013, 19, 1710–1719. [Google Scholar] [PubMed]
- Jackson, R.; Tilokee, E.L.; Latham, N.; Mount, S.; Rafatian, G.; Strydhorst, J.; Ye, B.; Boodhwani, M.; Chan, V.; Ruel, M.; et al. Paracrine Engineering of Human Cardiac Stem Cells with Insulin-Like Growth Factor 1 Enhances Myocardial Repair. J. Am. Heart Assoc. 2015, 4, e002104. [Google Scholar] [CrossRef]
- Muraski, J.A.; Rota, M.; Misao, Y.; Fransioli, J.; Cottage, C.; Gude, N.; Esposito, G.; Delucchi, F.; Arcarese, M.; Alvarez, R.; et al. Pim-1 regulates cardiomyocyte survival downstream of Akt. Nat. Med. 2007, 13, 1467–1475. [Google Scholar] [CrossRef]
- Mohsin, S.; Khan, M.; Nguyen, J.; Alkatib, M.; Siddiqi, S.; Hariharan, N.; Wallach, K.; Monsanto, M.; Gude, N.; Dembitsky, W.; et al. Rejuvenation of human cardiac progenitor cells with Pim-1 kinase. Circ. Res. 2013, 113, 1169–1179. [Google Scholar] [CrossRef]
- Mohsin, S.; Khan, M.; Toko, H.; Bailey, B.; Cottage, C.T.; Wallach, K.; Nag, D.; Lee, A.; Siddiqi, S.; Lan, F.; et al. Human cardiac progenitor cells engineered with Pim-I kinase enhance myocardial repair. J. Am. Coll. Cardiol. 2012, 60, 1278–1287. [Google Scholar] [CrossRef]
- Bhute, V.J.; Bao, X.; Palecek, S.P. Advances in Applications of Metabolomics in Pluripotent Stem Cell Research. Curr. Opin. Chem. Eng. 2017, 15, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Ezashi, T.; Das, P.; Roberts, R.M. Low O2 tensions and the prevention of differentiation of hES cells. Proc. Natl. Acad. Sci. USA 2005, 102, 4783–4788. [Google Scholar] [CrossRef] [PubMed]
- Mohyeldin, A.; Garzon-Muvdi, T.; Quinones-Hinojosa, A. Oxygen in stem cell biology: A critical component of the stem cell niche. Cell Stem Cell 2010, 7, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.M.; Kwon, S.; Pak, Y.K.; Seol, H.W.; Choi, Y.M.; Park, D.J.; Park, K.S.; Lee, H.K. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem. Biophys. Res. Commun. 2006, 348, 1472–1478. [Google Scholar] [CrossRef] [PubMed]
- Facucho-Oliveira, J.M.; Alderson, J.; Spikings, E.C.; Egginton, S.; St John, J.C. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J. Cell Sci. 2007, 120, 4025–4034. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, T. Metabolic regulation of mesenchymal stem cell in expansion and therapeutic application. Biotechnol. Prog. 2015, 31, 468–481. [Google Scholar] [CrossRef]
- Zhang, J.; Nuebel, E.; Daley, G.Q.; Koehler, C.M.; Teitell, M.A. Metabolic regulation in pluripotent stem cells during reprogramming and self-renewal. Cell Stem Cell 2012, 11, 589–595. [Google Scholar] [CrossRef]
- Kuppusamy, K.T.; Jones, D.C.; Sperber, H.; Madan, A.; Fischer, K.A.; Rodriguez, M.L.; Pabon, L.; Zhu, W.Z.; Tulloch, N.L.; Yang, X.; et al. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc. Natl. Acad. Sci. USA 2015, 112, E2785–E2794. [Google Scholar] [CrossRef]
- Lener, T.; Gimona, M.; Aigner, L.; Borger, V.; Buzas, E.; Camussi, G.; Chaput, N.; Chatterjee, D.; Court, F.A.; Del Portillo, H.A.; et al. Applying extracellular vesicles based therapeutics in clinical trials—An ISEV position paper. J. Extracell. Vesicles 2015, 4, 30087. [Google Scholar] [CrossRef]
- Ratajczak, M.Z.; Ratajczak, J. Extracellular Microvesicles as Game Changers in Better Understanding the Complexity of Cellular Interactions-From Bench to Clinical Applications. Am. J. Med. Sci. 2017, 354, 449–452. [Google Scholar] [CrossRef]
- Li, M.; Zeringer, E.; Barta, T.; Schageman, J.; Cheng, A.; Vlassov, A.V. Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Phil. Trans. R. Soc. B 2014, 369, 20130502. [Google Scholar] [CrossRef] [PubMed]
- Iaconetti, C.; Sorrentino, S.; De Rosa, S.; Indolfi, C. Exosomal miRNAs in Heart Disease. Physiology 2016, 31, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Arslan, F.; Lai, R.C.; Smeets, M.B.; Akeroyd, L.; Choo, A.; Aguor, E.N.; Timmers, L.; van Rijen, H.V.; Doevendans, P.A.; Pasterkamp, G.; et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013, 10, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Adamiak, M.; Sahoo, S. Exosomes in Myocardial Repair: Advances and Challenges in the Development of Next-Generation Therapeutics. Mol. Ther. J. Am. Soc. Gene Ther. 2018, 26, 1635–1643. [Google Scholar] [CrossRef]
- Davidson, S.M.; Yellon, D.M. Exosomes and cardioprotection—A critical analysis. Mol. Asp. Med. 2018, 60, 104–114. [Google Scholar] [CrossRef]
- Gartz, M.; Strande, J.L. Examining the Paracrine Effects of Exosomes in Cardiovascular Disease and Repair. J. Am. Heart Assoc. 2018, 7, e007954. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, X.; Cao, W.; Ma, J.; Sun, L.; Qian, H.; Zhu, W.; Xu, W. Exosomes Derived from Human Umbilical Cord Mesenchymal Stem Cells Relieve Acute Myocardial Ischemic Injury. Stem Cells Int. 2015, 2015, 761643. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.C.; Arslan, F.; Lee, M.M.; Sze, N.S.; Choo, A.; Chen, T.S.; Salto-Tellez, M.; Timmers, L.; Lee, C.N.; El Oakley, R.M.; et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010, 4, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Bian, S.; Zhang, L.; Duan, L.; Wang, X.; Min, Y.; Yu, H. Extracellular vesicles derived from human bone marrow mesenchymal stem cells promote angiogenesis in a rat myocardial infarction model. J. Mol. Med. 2014, 92, 387–397. [Google Scholar] [CrossRef]
- Cui, X.; He, Z.; Liang, Z.; Chen, Z.; Wang, H.; Zhang, J. Exosomes from Adipose-derived Mesenchymal Stem Cells Protect the Myocardium Against Ischemia/Reperfusion Injury Through Wnt/beta-Catenin Signaling Pathway. J. Cardiovasc. Pharmacol. 2017, 70, 225–231. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, J.; Yan, W.; Li, Y.; Shen, Z.; Asahara, T. Pretreatment of Cardiac Stem Cells with Exosomes Derived from Mesenchymal Stem Cells Enhances Myocardial Repair. J. Am. Heart Assoc. 2016, 5, e002856. [Google Scholar] [CrossRef] [PubMed]
- Gallet, R.; Dawkins, J.; Valle, J.; Simsolo, E.; de Couto, G.; Middleton, R.; Tseliou, E.; Luthringer, D.; Kreke, M.; Smith, R.R.; et al. Exosomes secreted by cardiosphere-derived cells reduce scarring, attenuate adverse remodelling, and improve function in acute and chronic porcine myocardial infarction. Eur. Heart J. 2017, 38, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Schey, K.L.; Luther, J.M.; Rose, K.L. Proteomics characterization of exosome cargo. Methods 2015, 87, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Choi, D.Y.; Yun, S.J.; Choi, S.M.; Kang, J.W.; Jung, J.W.; Hwang, D.; Kim, K.P.; Kim, D.W. Proteomic analysis of microvesicles derived from human mesenchymal stem cells. J. Proteome Res. 2012, 11, 839–849. [Google Scholar] [CrossRef] [PubMed]
- Lai, R.C.; Tan, S.S.; Teh, B.J.; Sze, S.K.; Arslan, F.; de Kleijn, D.P.; Choo, A.; Lim, S.K. Proteolytic Potential of the MSC Exosome Proteome: Implications for an Exosome-Mediated Delivery of Therapeutic Proteasome. Int. J. Proteom. 2012, 2012, 971907. [Google Scholar] [CrossRef] [PubMed]
- Vallabhaneni, K.C.; Penfornis, P.; Dhule, S.; Guillonneau, F.; Adams, K.V.; Mo, Y.Y.; Xu, R.; Liu, Y.; Watabe, K.; Vemuri, M.C.; et al. Extracellular vesicles from bone marrow mesenchymal stem/stromal cells transport tumor regulatory microRNA, proteins, and metabolites. Oncotarget 2015, 6, 4953–4967. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, S.; Klychko, E.; Thorne, T.; Misener, S.; Schultz, K.M.; Millay, M.; Ito, A.; Liu, T.; Kamide, C.; Agrawal, H.; et al. Exosomes from human CD34(+) stem cells mediate their proangiogenic paracrine activity. Circ. Res. 2011, 109, 724–728. [Google Scholar] [CrossRef]
- Mocharla, P.; Briand, S.; Giannotti, G.; Dorries, C.; Jakob, P.; Paneni, F.; Luscher, T.; Landmesser, U. AngiomiR-126 expression and secretion from circulating CD34(+) and CD14(+) PBMCs: Role for proangiogenic effects and alterations in type 2 diabetics. Blood 2013, 121, 226–236. [Google Scholar] [CrossRef]
- Gray, W.D.; French, K.M.; Ghosh-Choudhary, S.; Maxwell, J.T.; Brown, M.E.; Platt, M.O.; Searles, C.D.; Davis, M.E. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ. Res. 2015, 116, 255–263. [Google Scholar] [CrossRef]
- Namazi, H.; Mohit, E.; Namazi, I.; Rajabi, S.; Samadian, A.; Hajizadeh-Saffar, E.; Aghdami, N.; Baharvand, H. Exosomes secreted by hypoxic cardiosphere-derived cells enhance tube formation and increase pro-angiogenic miRNA. J. Cell. Biochem. 2018, 119, 4150–4160. [Google Scholar] [CrossRef]
- Meng, Y.; Eirin, A.; Zhu, X.Y.; Tang, H.; Chanana, P.; Lerman, A.; Van Wijnen, A.J.; Lerman, L.O. The metabolic syndrome alters the miRNA signature of porcine adipose tissue-derived mesenchymal stem cells. Cytom. Part A 2018, 93, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Henry, V.J.; Bandrowski, A.E.; Pepin, A.S.; Gonzalez, B.J.; Desfeux, A. OMICtools: An informative directory for multi-omic data analysis. Database J. Biol. Databases Curation 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Torre, D.; Krawczuk, P.; Jagodnik, K.M.; Lachmann, A.; Wang, Z.; Wang, L.; Kuleshov, M.V.; Ma’ayan, A. Datasets2Tools, repository and search engine for bioinformatics datasets, tools and canned analyses. Sci. Data 2018, 5, 180023. [Google Scholar] [CrossRef] [PubMed]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef] [PubMed]
- Vilne, B.; Schunkert, H. Integrating Genes Affecting Coronary Artery Disease in Functional Networks by Multi-OMICs Approach. Front. Cardiovasc. Med. 2018, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wu, H.; Wu, S.; Wang, C. Single-Cell Sequencing Technologies for Cardiac Stem Cell Studies. Stem Cells Dev. 2017, 26, 1540–1551. [Google Scholar] [CrossRef] [PubMed]
| SC Type | Abnormality Type | % of Abnormal Cell Lines | Passage Number | Affected Gene | Encoded Protein | Protein Role | Ref. |
|---|---|---|---|---|---|---|---|
| ESCs, iPSCs | Trisomy 12 | 12–20 | 14 | NANOG | Homeobox protein NANOG | Pluripotency | [7,8,9,10] |
| GDF3 | Growth differentiation factor-3 | Pluripotency | |||||
| Trisomy 8 | 9–20 | 19–26 | PTP4A3 | Protein tyrosine phosphatase type IVA, member 3 | Cell proliferation | [7,8,11] | |
| NDRG1 | N-myc downstream regulated 1 | Cell growth | |||||
| Trisomy X | 1–5 | 5–8 | FAM58A | Family with sequence similarity 58, member A | Cell proliferation | [7,8,11] | |
| CNVs (20q11.21) | 24–80 | 24–76 | ID1 | Inhibitor of DNA binding 1 | Cell growth | [7,11,12,13,14] | |
| BCL2L1 | B-cell lymphoma-extra large | Anti-apoptotic | |||||
| PDRG1 | p53 and DNA damage-regulated protein 1 | Cell survival | |||||
| TPX2 | Targeting protein for Xklp2 | Cell cycle | |||||
| KIF3B | Kinesin Family Member 3B | Cell cycle | |||||
| MSCs | Trisomy 8 | 4 | 7 | PTP4A3 | Protein tyrosine phosphatase type IVA, member 3 | Cell proliferation | [10,15] |
| CSCs | Trisomy 8 (normoxia vs hypoxia) | 31 vs 0 | 1 vs 6 | PTP4A3 | Protein tyrosine phosphatase type IVA, member 3 | Cell proliferation | [16,17] |
| ADSCs | Trisomy 8 | 8–12 | 2 | PTP4A3 | Protein tyrosine phosphatase type IVA, member 3 | Cell proliferation | [18] |
| SC | Transcription Factor | Active State | Repressed State | Ref. |
|---|---|---|---|---|
| ESCs | OCT4 | Hypomethylated | Hypermethylated | [40,41] |
| NANOG | Hypomethylated | Hypermethylated | ||
| MSCs | OCT4 | Hypermethylated | [42] | |
| NANOG | Hypomethylated | |||
| SOX4 | Hypomethylated |
| Transcript | Source | Effect | Target Molecule/Pathway | Ref. |
|---|---|---|---|---|
| miR-21 | MSC-Exos | ↓ apoptosis | ↓ inhibitors of pro-survival PI3K/Akt pathway | [60,67,82,83] |
| CSCs, CSC-Exos | ↓ apoptosis | ↓ CASP3, CASP8AP2, BAX, PDCD4, FASL, BCL2L11, FOXO3, AK2 | ||
| ↑ proliferation and migration of CSCs | ↓ PTEN, PDCD4 | |||
| iPSC-Exos | ↓ apoptosis | ↓ CASP3/7 | ||
| miR-22 | MSCs, MSC-Exos | ↓ apoptosis; reduces cardiac fibrosis | ↓ MECP2 | [60,84] |
| CSCs | ↑ commitment to SMCs | ↓ EVI1 | ||
| miR-24 | CPCs | ↓ apoptosis | ↓ CASP3, CASP8AP2, BAX, PDCD4, FASL, BCL2L11, FOXO3, AK2 | [67] |
| miR-30b | MSC-Exos | ↑ angiogenesis | ↓ endothelial Dll4 | [85] |
| miR-30c | MSCs-Exos | ↑ angiogenesis | ↓ endothelial Dll4 | [85] |
| miR-126 | MSCs, EPCs EPC-Exos | ↑ migration and survival of MSCs and EPCs; ↑ angiogenesis | ↓ inhibitors of pro-survival PI3K/Akt pathway; ↑ Dll4 expression | [57,64,65,66,86] |
| CDCs, CDC-Exos | ↑ cardioprotection | ↓ PKCδ expression | ||
| miR-146a | CDC-Exos CPC-Exos | ↑ cardioprotection, ↓ fibrosis | ↓ IRAK1 and TRAF6 | [87,88] |
| miR-181b | CDC-Exos | ↑ cardioprotection | ↓ PKCδ and MAP4K4 | [86] |
| miR-199a | CSCs | ↑ cardiomyocyte proliferation; ↓ apoptosis | ↓ P53 activity | [75,89] |
| miR-208 | MSCs | ↑cell proliferation and clonogenicity | ↓ AIMP3/p18 and senescence markers | [90] |
| miR-210 | MSCs | ↓ apoptosis | ↓ CASP8AP2 | [68,91] |
| CDCs, CDC-Exos | ↑ angiogenesis | ↓ EFNA3 | ||
| iPSCs-Exos | ↓ apoptosis | ↓ CASP3/7 | ||
| miR-221 | CPCs | ↓ apoptosis | ↓ CASP3, CASP8AP2, BAX, PDCD4, FASL, BCL2L11, FOXO3, AK2 | [67] |
| miR-291 | ESC-Exos | ↑ CPC survival and proliferation | ↓ P53 activity | [92] |
| miR-294 | ESC-Exos | ↑ CPC survival and proliferation | ↓ P53 activity | [92] |
| miR-295 | ESC-Exos | ↑ CPC survival and proliferation | ↓ P53 activity | [92] |
| let-7f | MSC-Exos | ↑ angiogenesis | ↓ endothelial THBS1 | [85,93] |
| Braveheart | ESCs MSCs | ↑ commitment toward the cardiovascular lineage | Activates MESP1, GATA4, HAND1, HAND2, NKX2.5, TBX5, SNAI, TWIST | [79,94] |
| ↑ epigenetic activation of cardiac genes | Binds SUZ12 | |||
| CARMEN | CPCs | ↑ cardiac specification and differentiation of CPCs | Interacts with chromatin remodeling complexes (PRC2) | [80] |
| LincRNA-RoR | iPSCs | ↑ reprogramming | Suppresses P53 pathways | [81,95] |
| ESCs | ↑ self-renewal of human ESCs | Captures miRNAs targeting OCT4, SOX2, NANOG |
| Factor | Source | Effect | Signaling Pathway | Ref. |
|---|---|---|---|---|
| FGF-2 | MSCs | ↑ MSC proliferation; ↑ angiogenesis; ↓ apoptosis | ERK1/2, PI3K-Akt pathways | [101,102,103] |
| TGF-β | MSCs | ↑ MSC proliferation; angiogenesis; ↓ apoptosis | SMAD, PI3K/Akt, MAPK pathways | [101,102,103] |
| VEGF | MSCs, EPCs | ↑ angiogenesis; ↓ apoptosis | PI3K/Akt, MAPK pathways | [101,102,103,104,105,106] |
| HGF | MSCs, CSCs | ↑ angiogenesis; ↓ apoptosis | ERK1/2, p38 MAPKs, PI3K/Akt, NOTCH pathways | [101,102,103] |
| IGF-1 | MSCs, EPCs, CSCs | ↑ angiogenesis; ↓ apoptosis | ERK1/2, PI3K-Akt pathways | [101,102,103,105] |
| Ang-1 | MSCs | ↑ angiogenesis | Tie-2 pathway | [101,102,103] |
| SDF-1 | MSCs, EPCs | ↑ mobilization and homing of BM-MSCs and EPCs; ↑ angiogenesis; ↑ migration and differentiation of CSCs; ↓ apoptosis | ERK1/2, PI3K-Akt pathways | [101,102,103,105] |
| IL-6 | MSCs | ↑ MSC proliferation and “stemness”; ↑ endothelial differentiation of CSCs; ↑ angiogenesis | ERK1/2, JAK-STAT pathway | [101,102,103] |
| Metabolite/ Metabolic Pathway | Effect | Ref. |
|---|---|---|
| SAM | Promotes pluripotency of ESCs and iPSCs | [151] |
| Hypoxia | Promotes pluripotency of ESCs and iPSCs; Promotes undifferentiated state of MSCs and expression of anti-apoptotic and angiogenic factors | [96,152,153] |
| Glycolysis | Promotes pluripotency of ESCs and iPSCs Promotes undifferentiated state of MSCs | [96,154,155,156] |
| Oxidative phosphorylation | Promotes cardiac differentiation of ESCs Promotes differentiation of MSCs | [96,156] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Micheu, M.M.; Scarlatescu, A.I.; Scafa-Udriste, A.; Dorobantu, M. The Winding Road of Cardiac Regeneration—Stem Cell Omics in the Spotlight. Cells 2018, 7, 255. https://doi.org/10.3390/cells7120255
Micheu MM, Scarlatescu AI, Scafa-Udriste A, Dorobantu M. The Winding Road of Cardiac Regeneration—Stem Cell Omics in the Spotlight. Cells. 2018; 7(12):255. https://doi.org/10.3390/cells7120255
Chicago/Turabian StyleMicheu, Miruna Mihaela, Alina Ioana Scarlatescu, Alexandru Scafa-Udriste, and Maria Dorobantu. 2018. "The Winding Road of Cardiac Regeneration—Stem Cell Omics in the Spotlight" Cells 7, no. 12: 255. https://doi.org/10.3390/cells7120255
APA StyleMicheu, M. M., Scarlatescu, A. I., Scafa-Udriste, A., & Dorobantu, M. (2018). The Winding Road of Cardiac Regeneration—Stem Cell Omics in the Spotlight. Cells, 7(12), 255. https://doi.org/10.3390/cells7120255