Heparanase: A Multitasking Protein Involved in Extracellular Matrix (ECM) Remodeling and Intracellular Events
Abstract
1. Introduction
Extracellular Matrix, Heparan Sulfate Proteoglycans and Heparanase
2. Heparanase Structure and Activity
2.1. Heparanase Processing and Structure
2.2. Heparanase Enzymatic Activity
2.3. Heparanase Non-Enzymatic Activities
3. Role of Heparanase in Pathological Conditions
3.1. Heparanase and Cancer Motility, Invasion and Metastasis
3.2. Heparanase and Angiogenesis
3.3. Heparanase and Coagulation
3.4. Heparanase and Inflammation
3.5. Heparanase and Fibrosis
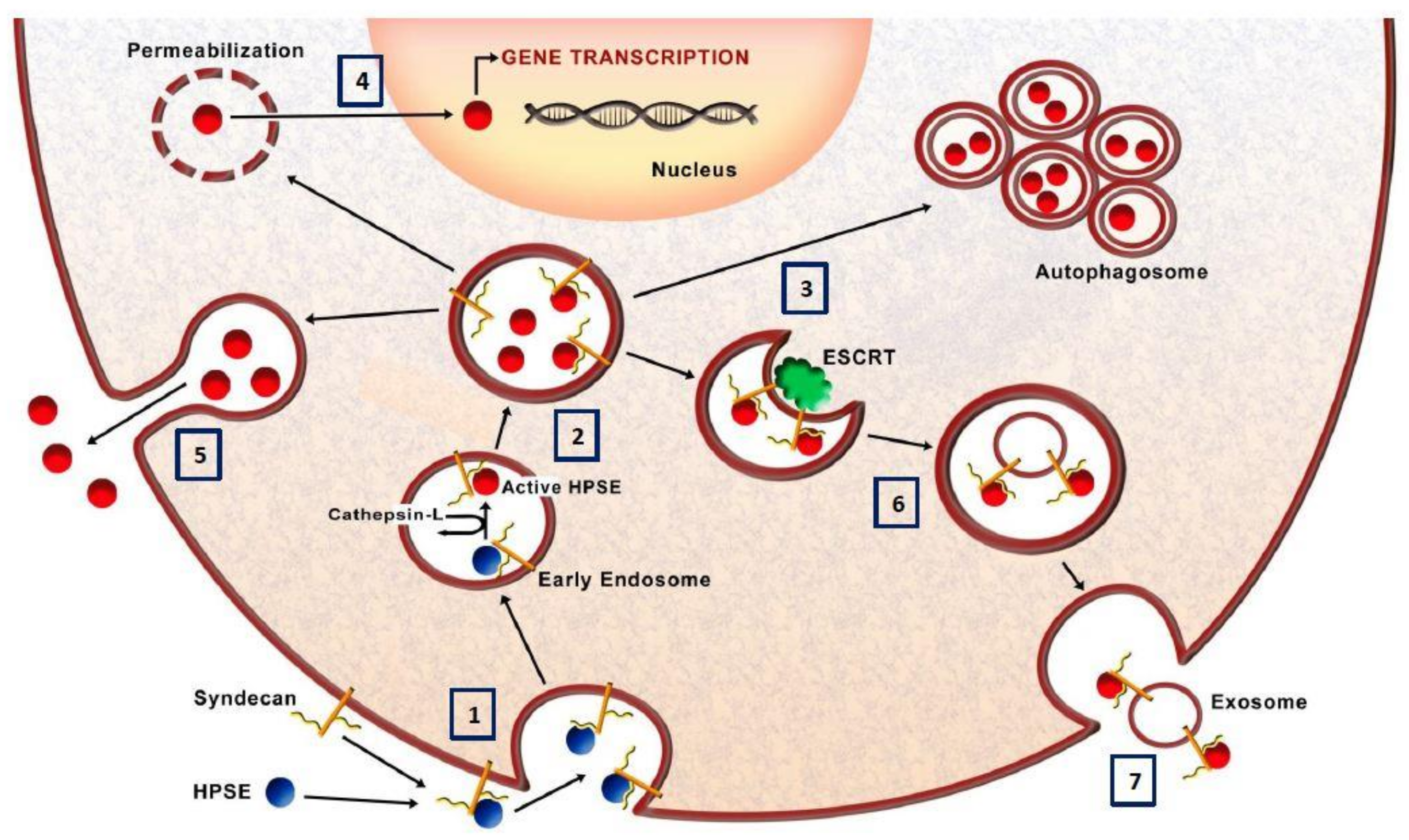
3.6. Heparanase and Autophagy
3.7. Heparanase and Exosome Production
3.8. Heparanase Nuclear Activity
3.9. Heparanase in Viral Pathogenesis
4. Heparanase Inhibition as Pharmacological Strategy
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ramani, V.C.; Purushothaman, A.; Stewart, M.D.; Thompson, C.A.; Vlodavsky, I.; Au, J.L.; Sanderson, R.D. The heparanase/syndecan-1 axis in cancer: Mechanisms and therapies. FEBS J. 2013, 280, 2294–2306. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Singh, P.; Boyango, I.; Gutter-Kapon, L.; Elkin, M.; Sanderson, R.D.; Ilan, N. Heparanase: From basic research to therapeutic applications in cancer and inflammation. Drug Resist. Updat. 2016, 29, 54–75. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, R.D.; Elkin, M.; Rapraeger, A.C.; Ilan, N.; Vlodavsky, I. Heparanase regulation of cancer, autophagy and inflammation: New mechanisms and targets for therapy. FEBS J. 2017, 284, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Meirovitz, A.; Goldberg, R.; Binder, A.; Rubinstein, A.M.; Hermano, E.; Elkin, M. Heparanase in inflammation and inflammation-associated cancer. FEBS J. 2013, 280, 2307–2319. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, R.D.; Bandari, S.K.; Vlodavsky, I. Proteases and glycosidases on the surface of exosomes: Newly discovered mechanisms for extracellular remodeling. Matrix Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Lv, Q.; Zeng, J.; He, L. The advancements of heparanase in fibrosis. Int. J. Mol. Epidemiol. Genet. 2016, 7, 137–140. [Google Scholar] [PubMed]
- Iozzo, R.V.; Schaefer, L. Proteoglycan form and function: A comprehensive nomenclature of proteoglycans. Matrix Biol. 2015, 42, 11–55. [Google Scholar] [CrossRef] [PubMed]
- Iozzo, R.V. Basement membrane proteoglycans: From cellar to ceiling. Nat. Rev. Mol. Cell Biol. 2005, 6, 646–656. [Google Scholar] [CrossRef] [PubMed]
- Marneros, A.G.; Olsen, B.R. Physiological role of collagen XVIII and endostatin. FASEB J. 2005, 19, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Rivara, S.; Milazzo, F.M.; Giannini, G. Heparanase: A rainbow pharmacological target associated to multiple pathologies including rare diseases. Future Med. Chem. 2016, 8, 647–680. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Gross-Cohen, M.; Weissmann, M.; Ilan, N.; Sanderson, R.D. Opposing Functions of Heparanase-1 and Heparanase-2 in Cancer Progression. Trends Biochem. Sci. 2018, 43, 18–31. [Google Scholar] [CrossRef] [PubMed]
- Levy-Adam, F.; Feld, S.; Cohen-Kaplan, V.; Shteingauz, A.; Gross, M.; Arvatz, G.; Naroditsky, I.; Ilan, N.; Doweck, I.; Vlodavsky, I. Heparanase 2 interacts with heparan sulfate with high affinity and inhibits heparanase activity. J. Biol. Chem. 2010, 285, 28010–28019. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Jemth, P.; Escobar Galvis, M.L.; Vlodavsky, I.; Horner, A.; Lindahl, U.; Li, J.P. Processing of macromolecular heparin by heparanase. J. Biol. Chem. 2003, 278, 35152–35158. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Viola, C.M.; Brzozowski, A.M.; Davies, G.J. Structural characterization of human heparanase reveals insights into substrate recognition. Nat. Struct. Mol. Biol. 2015, 22, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Hulett, M.D.; Hornby, J.R.; Ohms, S.J.; Zuegg, J.; Freeman, C.; Gready, J.E.; Parish, C.R. Identification of active-site residues of the pro-metastatic endoglycosidase heparanase. Biochemistry 2000, 39, 15659–15667. [Google Scholar] [CrossRef] [PubMed]
- Shafat, I.; Vlodavsky, I.; Ilan, N. Characterization of mechanisms involved in secretion of active heparanase. J. Biol. Chem. 2006, 281, 23804–23811. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Zhou, S. The functions of heparanase in human diseases. Mini Rev. Med. Chem. 2017, 17, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Fux, L.; Ilan, N.; Sanderson, R.D.; Vlodavsky, I. Heparanase: Busy at the cell surface. Trends Biochem. Sci. 2009, 34, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Gingis-Velitski, S.; Zetser, A.; Flugelman, M.Y.; Vlodavsky, I.; Ilan, N. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. J. Biol. Chem. 2004, 279, 23536–23541. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zaken, O.; Gingis-Velitski, S.; Vlodavsky, I.; Ilan, N. Heparanase induces Akt phosphorylation via a lipid raft receptor. Biochem. Biophys. Res. Commun. 2007, 361, 829–834. [Google Scholar] [CrossRef] [PubMed]
- Ilan, N.; Elkin, M.; Vlodavsky, I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int. J. Biochem. Cell. Biol. 2006, 38, 2018–2039. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Beckhove, P.; Lerner, I.; Pisano, C.; Meirovitz, A.; Ilan, N.; Elkin, M. Significance of heparanase in cancer and inflammation. Cancer Microenviron. 2012, 5, 115–132. [Google Scholar] [CrossRef] [PubMed]
- Vreys, V.; David, G. Mammalian heparanase: What is the message? J. Cell. Mol. Med. 2007, 11, 427–452. [Google Scholar] [CrossRef] [PubMed]
- Secchi, M.F.; Masola, V.; Zaza, G.; Lupo, A.; Gambaro, G.; Onisto, M. Recent data concerning heparanase: Focus on fibrosis, inflammation and cancer. Biomol. Concepts 2015, 6, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Cassinelli, G.; Favini, E.; Dal Bo, L.; Tortoreto, M.; De Maglie, M.; Dagrada, G.; Pilotti, S.; Zunino, F.; Zaffaroni, N.; Lanzi, C. Antitumor efficacy of the heparan sulfate mimic roneparstat (SST0001) against sarcoma models involves multi-target inhibition of receptor tyrosine kinases. Oncotarget 2016, 7, 47848–47863. [Google Scholar] [CrossRef] [PubMed]
- Ostapoff, K.T.; Awasthi, N.; Cenik, B.K.; Hinz, S.; Dredge, K.; Schwarz, R.E.; Brekken, R.A. PG545, an angiogenesis and heparanase inhibitor, reduces primary tumor growth and metastasis in experimental pancreatic cancer. Mol. Cancer Ther. 2013, 12, 1190–1201. [Google Scholar] [CrossRef] [PubMed]
- Zcharia, E.; Metzger, S.; Chajek-ShaulL, T.; Aingorn, H.; Elkin, M.; Friedmann, Y.; Weinstein, T.; Li, J.P.; Lindahl, U.; Vlodavsky, I. Transgenic expression of mammalian heparanase uncovers physiological functions of heparan sulfate in tissue morphogenesis, vascularization, and feeding behavior. FASEB J. 2004, 18, 252–263. [Google Scholar] [CrossRef] [PubMed]
- Ellis, L.M.; Staley, C.A.; Liu, W.; Fleming, R.Y.; Parikh, N.U.; Bucana, C.D.; Gallick, G.E. Down-regulation of vascular endothelial growth factor in a human colon carcinoma cell line transfected with an antisense expression vector specific for c-src. J. Biol. Chem. 1998, 273, 1052–1057. [Google Scholar] [CrossRef] [PubMed]
- Nadir, Y.; Brenner, B. Heparanase procoagulant activity in cancer progression. Thromb. Res. 2016, 140 (Suppl. 1), S44–S48. [Google Scholar] [CrossRef]
- Axelsson, J.; Xu, D.; Kang, B.N.; Nussbacher, J.K.; Handel, T.M.; Ley, K.; Sriramarao, P.; Esko, J.D. Inactivation of heparan sulfate 2-O-sulfotransferase accentuates neutrophil infiltration during acute inflammation in mice. Blood 2012, 120, 1742–1751. [Google Scholar] [CrossRef] [PubMed]
- Gotte, M. Syndecans in inflammation. FASEB J. 2003, 17, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Parish, C.R. The role of heparan sulphate in inflammation. Nat. Rev. Immunol. 2003, 6, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Eldor, A.; Haimovitz-Friedman, A.; Matzner, Y.; Ishai-Michaeli, R.; Lider, O.; Naparstek, Y.; Cohen, I.R.; Fuks, Z. Expression of heparanase by platelets and circulating cells of the immune system: Possible involvement in diapedesis and extravasation. Invasion Metastasis 1992, 12, 112–127. [Google Scholar]
- Lerner, I.; Hermano, E.; Zcharia, E.; Rodkin, D.; Bulvik, R.; Doviner, V.; Rubinstein, A.M.; Ishai-Michaeli, R.; Atzmon, R.; Sherman, Y.; et al. Heparanase powers a chronic inflammatory circuit that promotes colitis-associated tumorigenesis in mice. J. Clin. Investig. 2011, 121, 1709–1721. [Google Scholar] [CrossRef] [PubMed]
- Li, R.W.; Freeman, C.; Yu, D.; Hindmarsh, E.J.; Tymms, K.E.; Parish, C.R.; Smith, P.N. Dramatic regulation of heparanase activity and angiogenesis gene expression in synovium from patients with rheumatoid arthritis. Arthr. Rheum. 2008, 58, 1590–1600. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, E.P.; Yang, Y.; Janssen, W.J.; Gandjeva, A.; Perez, M.J.; Barthel, L.; Zemans, R.L.; Bowman, J.C.; Koyanagi, D.E.; Yunt, Z.X.; et al. The pulmonary endothelial glycocalyx regulates neutrophil adhesion and lung injury during experimental sepsis. Nat. Med. 2012, 18, 1217–1223. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, L.; Chen, S. Endogenous toll-like receptor ligands and their biological significance. J. Cell. Mol. Med. 2010, 14, 2592–2603. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.B.; Brunn, G.J.; Kodaira, Y.; Platt, J.L. Receptor-mediated monitoring of tissue well-being via detection of soluble heparan sulfate by Toll-like receptor 4. J. Immunol. 2002, 168, 5233–5239. [Google Scholar] [CrossRef] [PubMed]
- Brunn, G.J.; Bungum, M.K.; Johnson, G.B.; Platt, J.L. Conditional signaling by Toll-like receptor 4. FASEB J. 2005, 19, 872–874. [Google Scholar] [CrossRef] [PubMed]
- Masola, V.; Zaza, G.; Bellin, G.; Dall’Olmo, L.; Granata, S.; Vischini, G.; Secchi, M.F.; Lupo, A.; Gambaro, G.; Onisto, M. Heparanase regulates the M1 polarization of renal macrophages and their crosstalk with renal epithelial tubular cells after ischemia/reperfusion injury. FASEB J. 2017, 32, 742–756. [Google Scholar] [CrossRef] [PubMed]
- Masola, V.; Zaza, G.; Onisto, M.; Lupo, A.; Gambaro, G. Impact of heparanase on renal fibrosis. J. Transl. Med. 2015, 13, 181. [Google Scholar] [CrossRef] [PubMed]
- Secchi, M.F.; Crescenzi, M.; Masola, V.; Russo, F.P.; Floreani, A.; Onisto, M. Heparanase and macrophage interplay in the onset of liver fibrosis. Sci. Rep. 2017, 7, 14956. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Sun, F.; Wang, Y.; Zhu, J.; Fang, J.; Zhang, S.; Yu, Q.; Gong, Q.; Ren, B.; Xiang, X.; et al. HMGB1 exacerbates bronchiolitis obliterans syndrome via RAGE/NF-κB/HPSE signaling to enhance latent TGF-β release from ECM. Am. J. Transl. Res. 2016, 8, 1971–1984. [Google Scholar] [PubMed]
- Masola, V.; Gambaro, G.; Tibaldi, E.; Onisto, M.; Abaterusso, C.; Lupo, A. Regulation of heparanase by albumin and advanced glycation end products in proximal tubular cells. Biochim. Biophys. Acta. 2011, 1813, 1475–1482. [Google Scholar] [CrossRef] [PubMed]
- Masola, V.; Zaza, G.; Gambaro, G.; Onisto, M.; Bellin, G.; Vischini, G.; Khamaysi, I.; Hassan, A.; Hamoud, S.; Nativ, O.; et al. Heparanase: A Potential New Factor Involved in the Renal Epithelial Mesenchymal Transition (EMT) Induced by Ischemia/Reperfusion (I/R) Injury. PLoS ONE 2016, 11, e0160074. [Google Scholar] [CrossRef] [PubMed]
- Gil, N.; Goldberg, R.; Neuman, T.; Garsen, M.; Zcharia, E.; Rubinstein, A.M.; van Kuppevelt, T.; Meirovitz, A.; Pisano, C.; Li, J.P.; et al. Heparanase is essential for the development of diabetic nephropathy in mice. Diabetes 2012, 61, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Masola, V.; Gambaro, G.; Tibaldi, E.; Brunati, A.M.; Gastaldello, A.; D’Angelo, A.; Onisto, M.; Lupo, A. Heparanase and syndecan-1 interplay orchestrates fibroblast growth factor-2-induced epithelial-mesenchymal transition in renal tubular cells. J. Biol. Chem. 2012, 287, 1478–1488. [Google Scholar] [CrossRef] [PubMed]
- Masola, V.; Zaza, G.; Secchi, M.F.; Gambaro, G.; Lupo, A.; Onisto, M. Heparanse is a key player in the renal fibrosis by regulating TGF-b expression and activity. BBA Mol. Cell Res. 2014, 1843, 2122–2128. [Google Scholar]
- Ikeguchi, M.; Hirooka, Y.; Kaibara, N. Heparanase gene expression and its correlation with spontaneous apoptosis in hepatocytes of cirrhotic liver and carcinoma. Eur. J. Cancer 2003, 39, 86–90. [Google Scholar] [CrossRef]
- Xiao, Y.; Kleeff, J.; Shi, X.; Büchler, M.W.; Friess, H. Heparanase expression in hepatocellular carcinoma and the cirrhotic liver. Hepatol. Res. 2003, 26, 192–198. [Google Scholar] [CrossRef]
- Goldshmidt, O.; Yeikilis, R.; Mawasi, N.; Paizi, M.; Gan, N.; Ilan, N.; Pappo, O.; Vlodavsky, I.; Spira, G. Heparanase expression during normal liver development and following partial hepatectomy. J. Pathol. 2004, 203, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Ohayon, O.; Mawasi, N.; Pevzner, A.; Tryvitz, A.; Gildor, T.; Pines, M.; Rojkind, M.; Paizi, M.; Spira, G. Halofuginone upregulates the expression of heparanase in thioacetamide-induced liver fibrosis in rats. Lab Investig. 2008, 88, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Shteingauz, A.; Boyango, I.; Naroditsky, I.; Hammond, E.; Gruber, M.; Doweck, I.; Ilan, N.; Vlodavsky, I. Heparanase enhances tumor growth and chemoresistance by promoting autophagy. Cancer Res. 2015, 75, 3946–3957. [Google Scholar] [CrossRef] [PubMed]
- Baietti, M.F.; Zhang, Z.; Mortier, E.; Melchior, A.; Degeest, G.; Geeraerts, A.; Ivarsson, Y.; Depoortere, F.; Coomans, C.; Vermeiren, E.; et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 2012, 14, 677–685. [Google Scholar] [CrossRef] [PubMed]
- Roucourt, B.; Meeussen, S.; Bao, J.; Zimmermann, P.; David, G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015, 25, 412–428. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.A.; Purushothaman, A.; Ramani, V.C.; Vlodavsky, I.; Sanderson, R.D. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J. Biol. Chem. 2013, 288, 10093–10099. [Google Scholar] [CrossRef] [PubMed]
- Nobuhisa, T.; Naomoto, Y.; Okawa, T.; Takaoka, M.; Gunduz, M.; Motoki, T.; Nagatsuka, H.; Tsujigiwa, H.; Shirakawa, Y.; Yamatsuji, T.; et al. Translocation of heparanase into nucleus results in cell differentiation. Cancer Sci. 2007, 98, 535–540. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, A.; Hurst, D.R.; Pisano, C.; Mizumoto, S.; Sugahara, K.; Sanderson, R.D. Heparanase-mediated loss of nuclear syndecan-1 enhances histone acetyltransferase (HAT) activity to promote expression of genes that drive an aggressive tumor phenotype. J. Biol. Chem. 2011, 286, 30377–30383. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gorzelanny, C.; Bauer, A.T.; Halter, N.; Komljenovic, D.; Bäuerle, T.; Borsig, L.; Roblek, M.; Schneider, S.W. Nuclear heparanase-1 activity suppresses melanoma progression via its DNA-binding affinity. Oncogene 2015, 34, 5832–5842. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Y.; Zhang, D.; Puthanveetil, P.; Johnson, J.D.; Rodrigues, B. Fatty acid-induced nuclear translocation of heparanase uncouples glucose metabolism in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 406–414. [Google Scholar] [CrossRef] [PubMed]
- Schubert, S.Y.; Ilan, N.; Shushy, M.; Ben-Izhak, O.; Vlodavsky, I.; Goldshmidt, O. Human heparanase nuclear localization and enzymatic activity. Lab Investig. 2004, 8, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Lanzi, C.; Zaffaroni, N.; Cassinelli, G. Targeting Heparan Sulfate Proteoglycans and their Modifying Enzymes to Enhance Anticancer Chemotherapy Efficacy and Overcome Drug Resistance. Curr. Med. Chem. 2017, 24, 2860–2886. [Google Scholar] [CrossRef] [PubMed]
- Bandari, S.K.; Purushothaman, A.; Ramani, V.C.; Brinkley, G.J.; Chandrashekar, D.S.; Varambally, S.; Mobley, J.A.; Zhang, Y.; Brown, E.E.; Vlodavsky, I.; et al. Chemotherapy induces secretion of exosomes loaded with heparanase that degrades extracellular matrix and impacts tumor and host cell behavior. Matrix Biol. 2017, 65, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Agelidis, A.M.; Hadigal, S.R.; Jaishankar, D.; Shukla, D. Viral Activation of Heparanase Drives Pathogenesis of Herpes Simplex Virus-1. Cell Rep. 2017, 20, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Surviladze, Z.; Sterkand, R.T.; Ozbun, M.A. Interaction of human papillomavirus type 16 particles with heparan sulfate and syndecan-1 molecules in the keratinocyte extracellular matrix plays an active role in infection. J. Gen. Virol. 2015, 96, 2232–2241. [Google Scholar] [CrossRef] [PubMed]
- Hirshoren, N.; Bulvik, R.; Neuman, T.; Rubinstein, A.M.; Meirovitz, A.; Elkin, M. Induction of heparanase by HPV E6 oncogene in head and neck squamous cell carcinoma. J. Cell. Mol. Med. 2014, 18, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Thakkar, N.; Yadavalli, T.; Jaishankar, D.; Shukla, D. Emerging Roles of Heparanase in Viral Pathogenesis. Pathogens 2017, 6, 43. [Google Scholar] [CrossRef] [PubMed]
- Weissmann, M.; Arvatz, G.; Horowitz, N.; Feld, S.; Naroditsky, I.; Zhang, Y.; Ng, M.; Hammond, E.; Nevo, E.; Vlodavsky, I.; et al. Heparanase-neutralizing antibodies attenuate lymphoma tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Ferro, V.; Liu, L.; Johnstone, K.D.; Wimmer, N.; Karoli, T.; Handley, P.; Rowley, J.; Dredge, K.; Li, C.P.; Hammond, E.; et al. Discovery of PG545: A highly potent and simultaneous inhibitor of angiogenesis, tumor growth, and metastasis. J. Med. Chem. 2012, 55, 3804–3813. [Google Scholar] [CrossRef] [PubMed]
- Winterhoff, B.; Freyer, L.; Hammond, E.; Giri, S.; Mondal, S.; Roy, D.; Teoman, A.; Mullany, S.; Hoffmann, R.; von Bismarck, A.; et al. PG545 enhances anti-cancer activity of chemotherapy in ovarian models and increases surrogate biomarkers such as VEGF in preclinical and clinical plasma samples. Eur. J. Cancer 2015, 51, 879–892. [Google Scholar] [CrossRef] [PubMed]
- Naggi, A.; Casu, B.; Perez, M.; Torri, G.; Cassinelli, G.; Penco, S.; Pisano, C.; Giannini, G.; Ishai-Michaeli, R.; Vlodavsky, I. Modulation of the heparanase-inhibiting activity of heparin through selective desulfation, graded N-acetylation, and glycol splitting. J. Biol. Chem. 2005, 280, 12103–12113. [Google Scholar] [CrossRef] [PubMed]
- Galli, M.; Chatterjee, M.; Grasso, M.; Specchia, G.; Magen, H.; Einsele, H.; Celeghini, I.; Barbieri, P.; Paoletti, D.; Pace, S.; et al. Phase I study of the heparanase inhibitor Roneparstat: An innovative approach for multiple myeloma therapy. Haematologica 2018, 103, e469–e472. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masola, V.; Bellin, G.; Gambaro, G.; Onisto, M. Heparanase: A Multitasking Protein Involved in Extracellular Matrix (ECM) Remodeling and Intracellular Events. Cells 2018, 7, 236. https://doi.org/10.3390/cells7120236
Masola V, Bellin G, Gambaro G, Onisto M. Heparanase: A Multitasking Protein Involved in Extracellular Matrix (ECM) Remodeling and Intracellular Events. Cells. 2018; 7(12):236. https://doi.org/10.3390/cells7120236
Chicago/Turabian StyleMasola, Valentina, Gloria Bellin, Giovanni Gambaro, and Maurizio Onisto. 2018. "Heparanase: A Multitasking Protein Involved in Extracellular Matrix (ECM) Remodeling and Intracellular Events" Cells 7, no. 12: 236. https://doi.org/10.3390/cells7120236
APA StyleMasola, V., Bellin, G., Gambaro, G., & Onisto, M. (2018). Heparanase: A Multitasking Protein Involved in Extracellular Matrix (ECM) Remodeling and Intracellular Events. Cells, 7(12), 236. https://doi.org/10.3390/cells7120236






