Novel Insights into the Development and Function of Cilia Using the Advantages of the Paramecium Cell and Its Many Cilia
Abstract
:1. Introduction
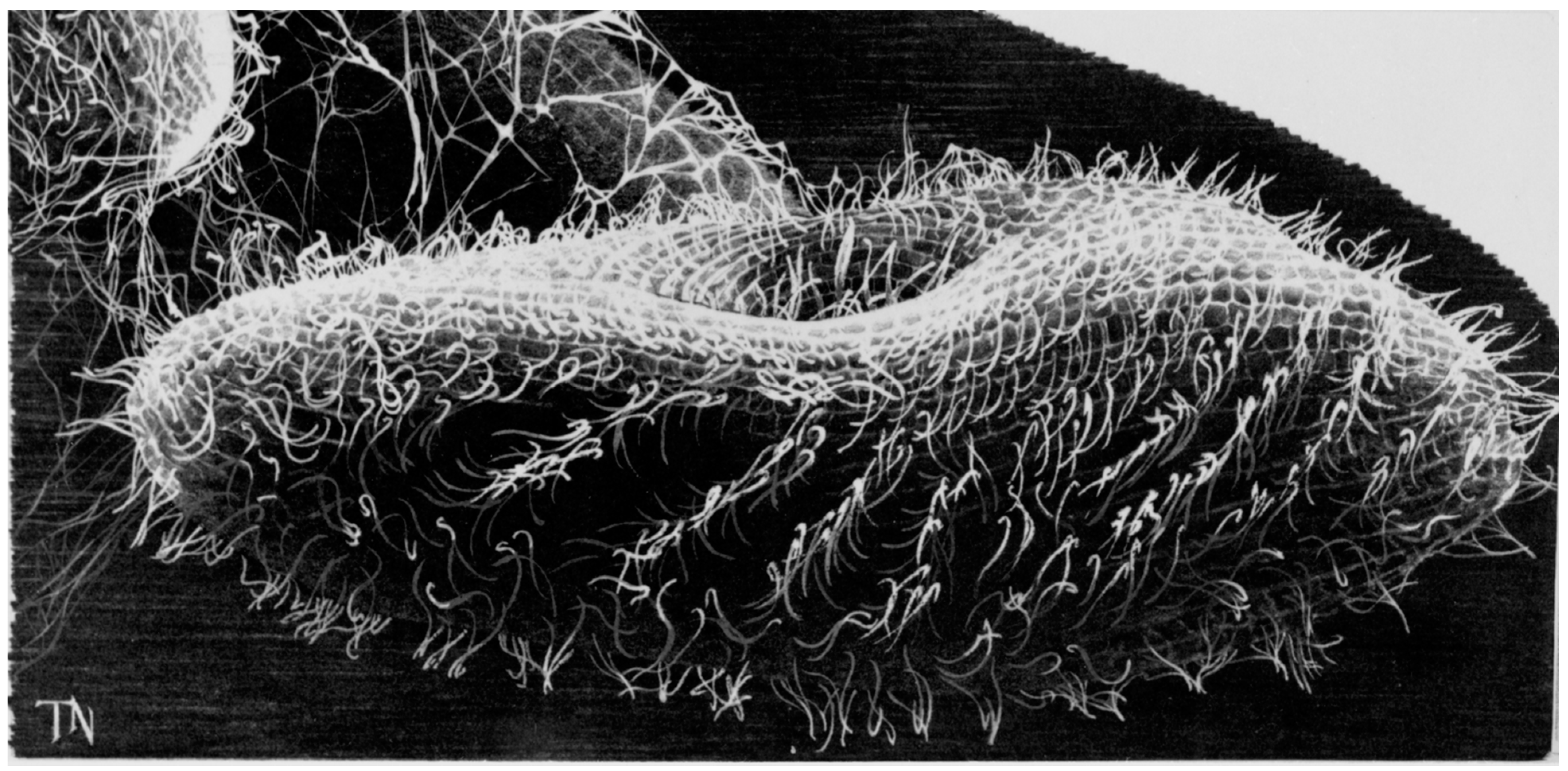
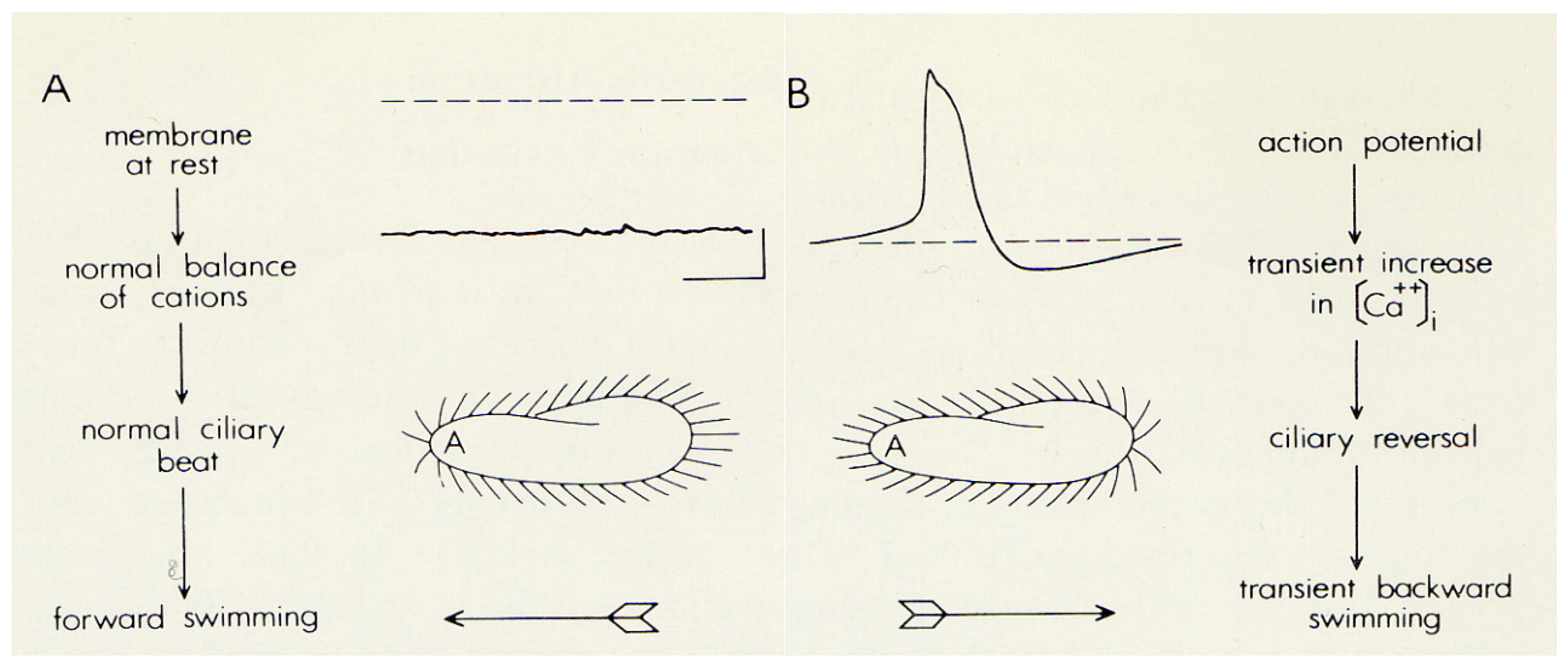
2. The Advantages of having an Excitable Membrane: Paramecium Broadcasts the Activity of Its Ciliary Ion Channels through Its Behavior
3. Cilia are Signal Transduction Compartments
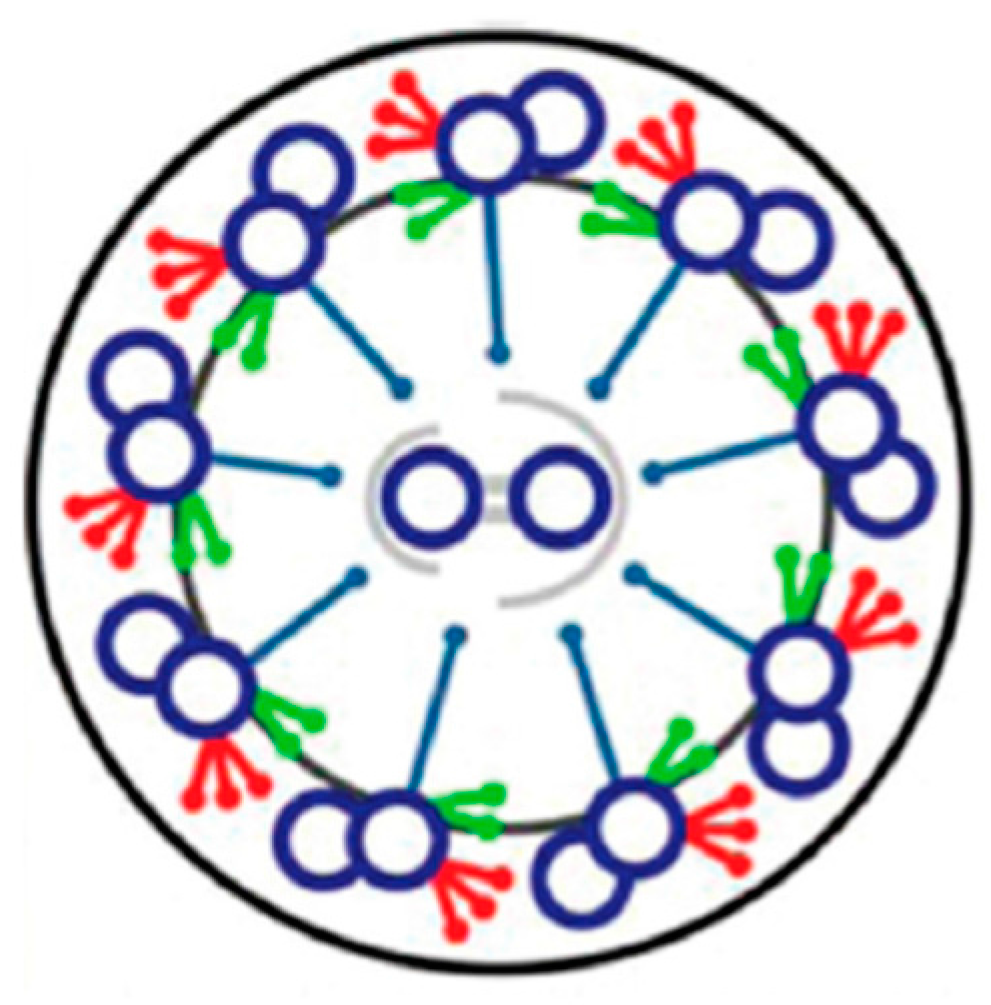
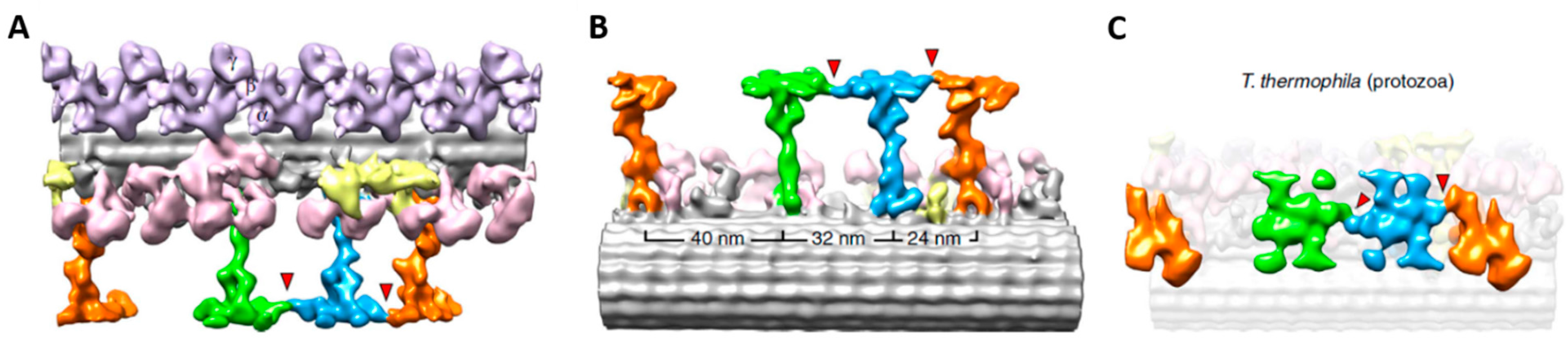
4. Depolarization and Signal Transduction in the Ciliary Compartment
5. Hyperpolarization and Signal Transduction in the Ciliary Compartment
6. The Advantage of Mutants
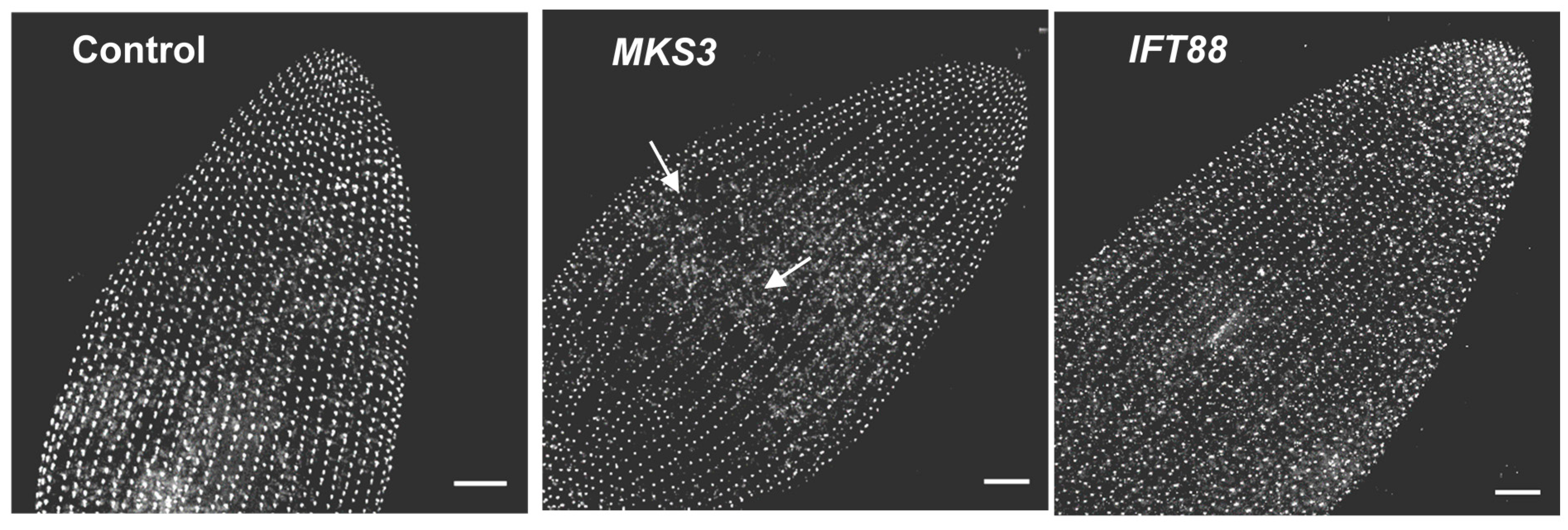
7. Depletion of Ciliary Proteins
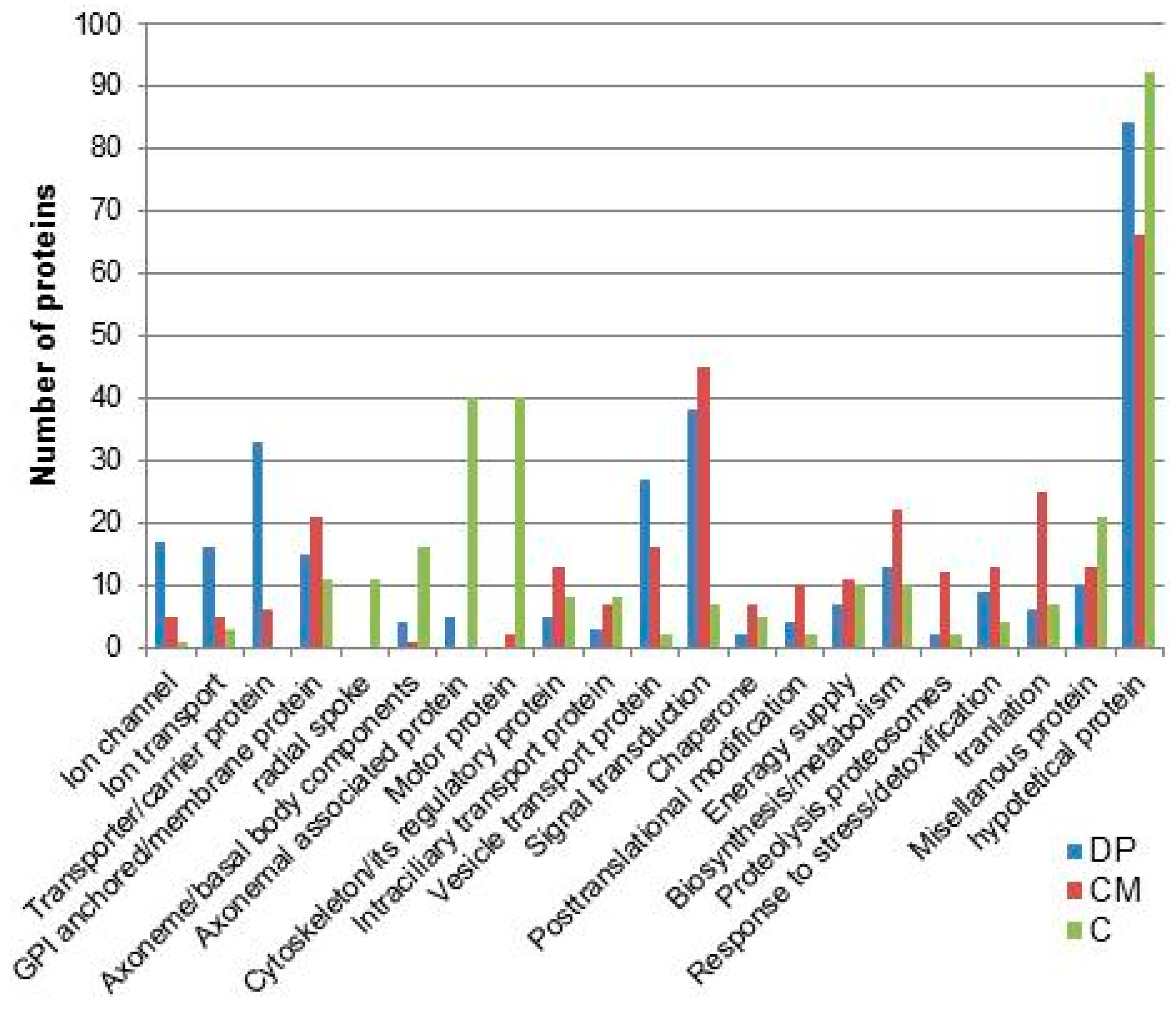
8. Ciliary Membrane Proteomics
9. Summary
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jennings, H.S. Behavior of the Lower Organisms; The Columbia University Press, The MacMillan Company: New York, NY, USA, 1906. [Google Scholar]
- Kleene, S.; Van Houten, J.L. Electrical signaling in motile and primary cilia. BioScience 2014, 64, 1092–1102. [Google Scholar] [CrossRef] [PubMed]
- Kung, C.; Chang, S.Y.; Satow, Y.; Houten, J.V.; Hansma, H. Genetic dissection of behavior in Paramecium. Science 1975, 188, 898–904. [Google Scholar] [PubMed]
- Wright, M.V.; Van Houten, J.L. Characterization of a putative Ca2+-transporting Ca2+-ATPase in the pellicles of Paramecium tetraurelia. Biochim. Biophys. Acta 1990, 1029, 241–251. [Google Scholar] [CrossRef]
- Adoutte, A.; Ramanathan, R.; Lewis, R.M.; Dute, R.R.; Ling, K.Y.; Kung, C.; Nelson, D.L. Biochemical studies of the excitable membrane of Paramecium tetraurelia. III. Proteins of cilia and ciliary membranes. J. Cell Biol. 1980, 84, 717–738. [Google Scholar] [CrossRef] [PubMed]
- Fliegauf, M.; Benzing, T.; Omran, H. Mechanisms of disease - when cilia go bad: Cilia defects and ciliopathies. Nat. Rev. Mol. Cell. Biol. 2007, 8, 880–893. [Google Scholar] [CrossRef] [PubMed]
- Weeraratne, S.D. GPI receptors in folate chemosensory transduction in Paramecium tetraurelia. Ph.D. Thesis, University of Vermont, Burlington, VT, USA, 2007. [Google Scholar]
- Jacobs, C.L. NMDA receptor associated protein in Paramecium and it involvement in glutamate chemoresponse. MS thesis, University of Vermont, Burlington, VT, USA, 2007. [Google Scholar]
- Valentine, M.S.; Rajendran, A.; Yano, J.; Weeraratne, S.D.; Beisson, J.; Cohen, J.; Koll, F.; Van Houten, J. Paramecium BBS genes are key to presence of channels in cilia. Cilia 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Machemer, H.; Ogura, A. Ionic conductances of membranes in ciliated and deciliated Paramecium. J. Physiol. 1979, 296, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Fleischman, J. Old lab animal model systems gain new respect - Paramecium redux. ASCB Post. 2014. Available online: http://www.ascb.org/old-lab-animal-model-systems-gain-new-respect-paramecium-redux/ (accessed on 2 July 2015).
- Beisson, J.; Bétermier, M.; Bré, M.-H.; Cohen, J.; Duharcourt, S.; Duret, L.; Kung, C.; Malinsky, S.; Meyer, E.; Preer, J.R.; et al. Paramecium tetraurelia: The renaissance of an early unicellular model. Cold Spring Harb. Protoc. 2010, 2010. [Google Scholar] [CrossRef] [PubMed]
- Machemer, H. Electrophysiology. In Paramecium; Gortz, H.-D., Ed.; Springer-Verlag: Berlin, Germany, 1988; pp. 186–215. [Google Scholar]
- Machemer, H. Motor control of cilia. In Paramecium; Gortz, H.-D., Ed.; Springer-Verlag: Berlin, Germany, 1988; pp. 216–235. [Google Scholar]
- Brehm, P.; Eckert, R. An electrophysiological study of the regulation of ciliary beating frequency in Paramecium. J. Physiol. 1978, 283, 557–568. [Google Scholar] [CrossRef] [PubMed]
- Naitoh, Y.; Kaneko, H. Reactivated triton-extraced models of Paramecium: Modifications of ciliary movement by calcium ions. Science 1972, 176, 523–524. [Google Scholar] [CrossRef]
- Preston, R.R.; Kink, J.A.; Hinrichsen, R.D.; Saimi, Y.; Kung, C. Calmodulin mutants and Ca2+-dependent channels in Paramecium. Annu. Rev. Physiol. 1991, 53, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Kung, C.; Saimi, Y. Ca2+ channels of Paramecium: A multidisciplinary study. In Current Topics in Membranes and Transport; Academic Press: San Diego, CA, USA, 1985; Volume 23, pp. 45–66. [Google Scholar]
- Dunlap, K. Localization of calcium channels in Paramecium caudatum. J. Physiol. 1977, 271, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Van Houten, J.; Van Houten, J. Computer analysis of Paramecium chemokinesis behavior. J. Theor. Biol. 1982, 98, 4453–4468. [Google Scholar]
- Eckert, R. Bioelectric control of ciliary activity. Science 1972, 176, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Plattner, H. Calcium signalling in the ciliated protozoan model, Paramecium: Strict signal localisation by epigenetically controlled positioning of different Ca2+-channels. Cell Calcium 2015, 57, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Preston, R.; Saimi, Y. Calcium ions and the regulation of motility in Paramecium. In Ciliary and Flagellar Membranes; Bloodgood, R.A., Ed.; Plenum Publishing Corp.: New York, NY, USA, 1990; pp. 173–200. [Google Scholar]
- Lieberman, S.J.; Hamasaki, T.; Satir, P. Ultrastructure and motion analysis of permeabilized Paramecium capable of motility and regulation of motility. Cell. Motil. Cytoskel. 1988, 9, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Van Houten, J. Chemosensory transduction in Paramecium. Eur. J. Protistol. 1998, 34, 301–307. [Google Scholar] [CrossRef]
- Satir, P.; Heuser, T.; Sale, W. A structural basis for how motile cilia beat. BioScience 2014, 64, 1073–1083. [Google Scholar] [CrossRef]
- Porter, M.; Sale, W. The 9+2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J. Cell Biol. 2000, 151, F37–F42. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Yin, W.; Smith, M.C.; Song, K.; Leigh, M.W.; Zariwala, M.A.; Knowles, M.R.; Ostrowski, L.E.; Nicastro, D. Cryo-electron tomography reveals ciliary defects underlying human RSPH1 primary ciliary dyskinesia. Nat. Commun. 2014, 5, 5727. [Google Scholar] [CrossRef] [PubMed]
- Wirschell, M.; Yamamoto, R.; Alford, L.M.; Gokhale, A.; Gaillard, A.R.; Sale, W. Regulation of ciliary motility: Conserved protein kinases and phosphatases are targeted and anchored in the ciliary axoneme. Arch. Biochem. Biophys. 2011, 510, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, L.; Schroder, J.; Satir, P.; Christensen, S. The ciliary cytoskeleton. Compr. Physiol. 2012, 2, 779–803. [Google Scholar] [PubMed]
- DeCaen, P.G.; Delling, M.; Vien, T.N.; Clapham, D.E. Direct recording and molecular identification of the calcium channel of primary cilia. Nature 2013, 504, 315–318. [Google Scholar] [CrossRef] [PubMed]
- Delling, M.; DeCaen, P.G.; Doerner, J.F.; Febvay, S.; Clapham, D.E. Primary cilia are specialized calcium signalling organelles. Nature 2013, 504, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Delmas, P.; Padilla, F.; Osorio, N.; Coste, A.; Baasner, A.; Raoux, M.; Crest, M. Polycystins, calcium signaling and human diseases. Biochem. Biophys. Res. Commun. 2004, 322, 1374–1383. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Mohieldin, A.M.; Muntean, B.S.; Green, J.A.; Shah, J.V.; Mykytyn, K.; Nauli, S.M. Cilioplasm is a cellular compartment for calcium signaling in response to mechanical and chemical stimuli. Cell. Mol. Life Sci. 2014, 71, 2165–2178. [Google Scholar] [CrossRef] [PubMed]
- Brehm, P.; Dunlap, K.; Eckert, R. Calcium-dependent repolarization in Paramecium. J. Physiol. 1978, 274, 639–654. [Google Scholar] [CrossRef] [PubMed]
- Satow, Y.; Kung, C. Ca-induced K+-outward current in Paramecium tetraurelia. J. Exp. Biol. 1980, 88, 293–303. [Google Scholar] [PubMed]
- Husser, M.; Hardt, M.; Blanchard, M.-P.; Hentschel, J.; Klauke, N.; Plattner, H. One-way calcium spill-over during signal transduction in Paramecium cells: From cortex into cilia but not the reverse. Cell Calcium 2004, 36, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Valentine, M.; (University of Vermont: Burlington, VT, USA). Personal communication, 2015.
- Gerdes, J.M.; Davis, E.E.; Katsanis, N. The vertebrate primary cilium in development, homeostasis, and disease. Cell 2009, 137, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Berbari, N.; O'Connor, A.; Haycraft, C.; Yoder, B. The primary cilium is a complex signaling center. Curr. Biol. 2009, 19, R526–R535. [Google Scholar] [CrossRef] [PubMed]
- Haynes, W.J.; Kung, C.; Saimi, Y.; Preston, R.R. An exchanger-like protein underlies the large Mg2+ current in Paramecium. Proc. Natl. Acad. Sci. USA 2002, 99, 15717–15722. [Google Scholar] [CrossRef] [PubMed]
- Preston, R.R.; Kung, C. Isolation and characterization of Paramecium mutants defective in their response to magnesium. Genetics 1994, 137, 759–769. [Google Scholar] [PubMed]
- Schultz, J.; Polh, T.; Klumpp, S. Voltage-gated Ca2+ entry into Paramecium linked to intraciliary increase in cyclic GMP. Nature 1986, 322, 271–273. [Google Scholar] [CrossRef]
- Noguchi, M.; Kurahashi, S.; Kamachi, H.; Inoue, H. Control of the ciliary beat by cyclic nucleotides in intact cortical sheets from Paramecium. Zool. Sci. 2004, 21, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Bonini, N.; Nelson, D. Differential regulation of Paramecium ciliary motility by cAMP and cGMP. J. Cell Biol. 1988, 106, 1615–1623. [Google Scholar] [CrossRef] [PubMed]
- Machemer, H. Frequency and directional responses of cilia to membrane potential changes in Paramecium. J. Comp. Physiol. 1974, 92, 293–316. [Google Scholar] [CrossRef]
- Machemer, H. Cellular behavior modulated by ions: Electrophysiological implications. J. Protozool. 1989, 36, 463–487. [Google Scholar] [CrossRef]
- Van Houten, J.L. Membrane potential changes during chemokinesis in Paramecium. Science 1979, 204, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Preston, R.; Van Houten, J. Chemoreception in Paramecium: Acetate - and folate-induced membrane hyperpolarization. J. Comp. Physiol. 1987, 160, 525–536. [Google Scholar] [CrossRef]
- Yang, W.; Braun, C.; Plattner, H.; Purvee, J.; Van Houten, J.L. Cyclic nucletotides in gluamate chemosensory signal transduction of Paramecium. J. Cell Sci. 1997, 110, 1567–1572. [Google Scholar]
- Schultz, J.E.; Klumpp, S.; Benz, R.; Schurhoff-Goeters, W.J.; Schmid, A. Regulation of adenylyl cyclase from Paramecium by an intrinsic potassium conductance. Science 1992, 255, 600–603. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.H.; Vishnyakov, A.; Hambach, K.; Schultz, A.; Schultz, J.E.; Linder, J.U. Adenylyl cyclases from Plasmodium, Paramecium and Tetrahymena are novel ion channel/enzyme fusion proteins. Cell. Signal. 2004, 16, 115–125. [Google Scholar] [CrossRef]
- Klumpp, S.; Gierlich, D.; Schultz, J.E. Adenylate cyclase and guanylate cyclase in the excitable ciliary membrane from Paramecium: Separation and regulation. FEBS Lett. 1984, 171, 95–99. [Google Scholar] [CrossRef]
- Kutomi, O.; Hori, M.; Ishida, M.; Tominaga, T.; Kamachi, H.; Koll, F.; Cohen, J.; Yamada, N.; Noguchi, M. Outer dynein arm light chain 1 is essential for controlling the ciliary response to cyclic AMP in Paramecium tetraurelia. Eukaryot. Cell 2012, 11, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki, T.; Barkalow, K.; Richmond, J.; Satir, P. cAMP-stimulated phosphorylation of an axonemal polypeptide that copurifies with the 22S dynein arm regulates microtubule translocation velocity and swimming speed in Paramecium. Proc. Natl. Acad. Sci. USA 1991, 88, 7918–7922. [Google Scholar] [CrossRef] [PubMed]
- Bonini, N.M.; Gustin, M.C.; Nelson, D.L. Regulation of ciliary motility by membrane potential in Paramecium: A role for cyclic AMP. Cell. Motil. Cytoskel. 1986, 6, 256–272. [Google Scholar] [CrossRef] [PubMed]
- Preston, R.R.; Saimi, Y.; Kung, C. Evidence for two K+ currents activated upon hyperpolarization of Paramecium tetraurelia. J. Membr. Biol. 1990, 115, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.; Rajendran, A.; Valentine, M.S.; Saha, M.; Ballif, B.A.; Van Houten, J.L. Proteomic analysis of the cilia membrane of Paramecium tetraurelia. J. Proteomics 2013, 78, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, M.; Sasaki, J.Y.; Kamachi, H.; Inoue, H. Protein phosphatase 2C is involved in the cAMP-dependent ciliary control in Paramecium caudatum. Cell. Motil. Cytoskel. 2003, 54, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Bonini, N.; Nelson, D. Phosphoproteins associated with cyclic nucleotide stimulation of ciliary motility in Paramecium. J. Cell Sci. 1990, 95, 219–230. [Google Scholar] [PubMed]
- Preston, R. Genetic dissection of Ca2+-dependent ion channel function in Paramecium. BioEssays 1990, 12, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Saimi, Y.; Kung, C. Behavioral genetics of Paramecium. Annu. Rev. Genet. 1987, 21, 47–65. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M. Behavioral mutants in Paramecium caudatum. Genetics 1979, 91, 393–408. [Google Scholar] [PubMed]
- Oertel, D.; Schein, S.; Kung, C. Separation of membrane currents using a Paramecium mutant. Nat. Neurosci. 1977, 268, 120–124. [Google Scholar] [CrossRef]
- Satow, Y.; Kung, C. Membrane currents of pawn mutants of the pwA group in Paramecium tetraurelia. J. Exp. Biol. 1980, 84, 57–71. [Google Scholar] [PubMed]
- Gonda, K.; Yoshida, A.; Oami, K.; Takahashi, M. Centrin is essential for the activity of the ciliary reversal-coupled voltage-gated Ca2+ channels. Biochem. Biophys. Res. Commun. 2004, 323, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Lodh, S. Characterization of pwA and pwB Proteins in Paramecium. Ph.D. Thesis, University of Vermont, Burlington, VT, USA, 2012. [Google Scholar]
- Kung, C.; Preston, R.R.; Maley, M.E.; Ling, K.Y.; Kanabrocki, J.A.; Seavey, B.R.; Saimi, Y. In vivo Paramecium mutants show that calmodulin orchestrates membrane responses to stimuli. Cell Calcium 1992, 13, 413–425. [Google Scholar] [CrossRef]
- Kink, J.A.; Maley, M.E.; Preston, R.R.; Ling, K.Y.; Wallen-Friedman, M.A.; Saimi, Y.; Kung, C. Mutations in Paramecium calmodulin indicate functional differences between the C-terminal and N-terminal lobes in vivo. Cell 1990, 62, 165–174. [Google Scholar] [CrossRef]
- Preston, R.R.; Saimi, Y.; Martinac, B.; Kung, C. Genetic analysis of ion channels of prokaryotes and lower eukaryotes. Curr. Opin. Genet. Dev. 1992, 2, 780–784. [Google Scholar] [CrossRef]
- Reiter, J.F.; Blacque, O.E.; Leroux, M.R. The base of the cilium: Roles for transition fibers and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 2012, 13, 608–618. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeister, H.; Babinger, K.; Gurster, S.; Cedzich, A.; Meese, C.; Schadendorf, K.; Osten, L.; de Vries, U.; Rascle, A.; Witzgall, R. Polycystin-2 takes different routes to the somatic and ciliary plasma membrane. J. Cell Biol. 2011, 192, 631–645. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; White, S.R.; Shida, T.; Schulz, S.; Aguiar, M.; Gygi, S.P.; Bazan, J.F.; Nachury, M.V. The conserved Bardet-Biedl Syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell 2010, 141, 1208–1219. [Google Scholar] [CrossRef] [PubMed]
- Klinger, M.; Wang, W.; Kuhns, S.; Barenz, F.; Drager-Meurer, S.; Pereira, G.; Gruss, O.J. The novel centriolar satellite protein SSX2IP targets CEP290 to the ciliary transition zone. Mol. Biol. Cell 2014, 25, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Dolphin, A.C. Calcium channel auxiliary α2δ and β subunits: Trafficking and one step beyond. Nat. Rev. Neurosci. 2012, 13, 542–555. [Google Scholar] [CrossRef] [PubMed]
- Picariello, T.; Valentine, M.S.; Yano, J.; Van Houten, J. Reduction of meckelin leads to general loss of cilia, ciliary microtubule misalignment and distorted cell surface organization. Cilia 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Galvani, A.; Sperling, L. RNA interference by feeding in Paramecium. Trends Genet. 2002, 18, 11–12. [Google Scholar] [CrossRef]
- Aury, J.-M.; Jaillon, O.; Duret, L.; Noel, B.; Jubin, C.; Porcel, B.; Segurens, B.; Daubin, V.; Anthouard, V.; Aiach, N.; et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 2006, 444, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz, O.; Malinowska, A.; Klotz, C.; Sperling, L.; Dadlez, M.; Koll, F.; Cohen, J. Cildb: A knowledgebase for centrosomes and cilia. Database 2009, 2009, bap022. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz, O.; Cohen, J.; Tassin, A.-M.; Koll, F. Remodeling Cildb, a popular database for cilia and links for ciliopathies. Cilia 2014, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D. Preparation of cilia and subciliary fractions from Paramecium. In Methods in Cell Biology; Academic Press, Inc.: San Diego, CA, USA, 1995; Volume 47, pp. 17–24. [Google Scholar]
- Arnaiz, O.; Cain, S.; Cohen, J.; Sperling, L. ParameciumDB: A community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 2007, 35, D439–D444. [Google Scholar] [CrossRef] [PubMed]
- Arnaiz, O.; Sperling, L. ParameciumDB in 2011: New tools and new data for functional and comparative genomics of the model ciliate Paramecium tetraurelia. Nucleic Acids Res. 2011, 39, D632–D636. [Google Scholar] [CrossRef] [PubMed]
- Bloodgood, R.A. The future of ciliary and flagellar membrane research. Mol. Biol. Cell. 2012, 23, 2407–2411. [Google Scholar] [CrossRef] [PubMed]
- Mathias, R.A.; Chen, Y.-S.; Kapp, E.A.; Greening, D.W.; Mathivanan, S.; Simpson, R.J. Triton X-114 phase separation in the isolation and purification of mouse liver microsomal membrane proteins. Methods 2011, 54, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Paquette, C.A.; Rakochy, V.; Bush, A.; van Houten, J.L. Glycophosphatidylinositol-anchored proteins in Paramecium tetraurelia: Possible role in chemoresponse. J. Exp. Biol. 2001, 204, 2899–2910. [Google Scholar] [PubMed]
- Yano, J.; Rachochy, V.; van Houten, J.L. Glycosyl phosphatidylinositol-anchored proteins in chemosensory signaling: Antisense manipulation of Paramecium tetraurelia PIG-A gene expression. Eukaryot. Cell 2003, 2, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Capdeville, Y.; Benwakrim, A. The major ciliary membrane proteins in Paramecium primaurelia are all glycosylphosphatidylinositol-anchored proteins. Eur. J. Cell Biol. 1996, 70, 339–346. [Google Scholar] [PubMed]
- Yano, J.; (University of Vermont: Burlington, VT, USA). Personal communication, 2015.
- Rhoads, D.E.; Kaneshiro, E.S. Characterizations of phospholipids from Paramecium tetraurelia cells and cilia. J. Protozool. 1979, 26, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Sepúlveda, M.R.; Hidalgo-Sánchez, M.; Mata, A.M. Localization of endoplasmic reticulum and plasma membrane Ca2+-ATPases in subcellular fractions and sections of pig cerebellum. Eur. J. Neurosci. 2004, 19, 542–551. [Google Scholar] [CrossRef] [PubMed]
- Pani, B.; Singh, B.B. Lipid rafts/caveolae as microdomains of calcium signaling. Cell. Calcium 2009, 45, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Tyler, K.M.; Fridberg, A.; Toriello, K.M.; Olson, C.L.; Cieslak, J.A.; Hazlett, T.L.; Engman, D.M. Flagellar membrane localization via association with lipid rafts. J. Cell Sci. 2009, 122, 859–866. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yano, J.; Valentine, M.S.; Van Houten, J.L. Novel Insights into the Development and Function of Cilia Using the Advantages of the Paramecium Cell and Its Many Cilia. Cells 2015, 4, 297-314. https://doi.org/10.3390/cells4030297
Yano J, Valentine MS, Van Houten JL. Novel Insights into the Development and Function of Cilia Using the Advantages of the Paramecium Cell and Its Many Cilia. Cells. 2015; 4(3):297-314. https://doi.org/10.3390/cells4030297
Chicago/Turabian StyleYano, Junji, Megan S. Valentine, and Judith L. Van Houten. 2015. "Novel Insights into the Development and Function of Cilia Using the Advantages of the Paramecium Cell and Its Many Cilia" Cells 4, no. 3: 297-314. https://doi.org/10.3390/cells4030297
APA StyleYano, J., Valentine, M. S., & Van Houten, J. L. (2015). Novel Insights into the Development and Function of Cilia Using the Advantages of the Paramecium Cell and Its Many Cilia. Cells, 4(3), 297-314. https://doi.org/10.3390/cells4030297




