Regulation of TGF-β Superfamily Signaling by SMAD Mono-Ubiquitination
Abstract
:1. Introduction
1.1. Ubiquitin and Ubiquitination

1.2. TGF-β/SMAD and BMP/SMAD Signaling
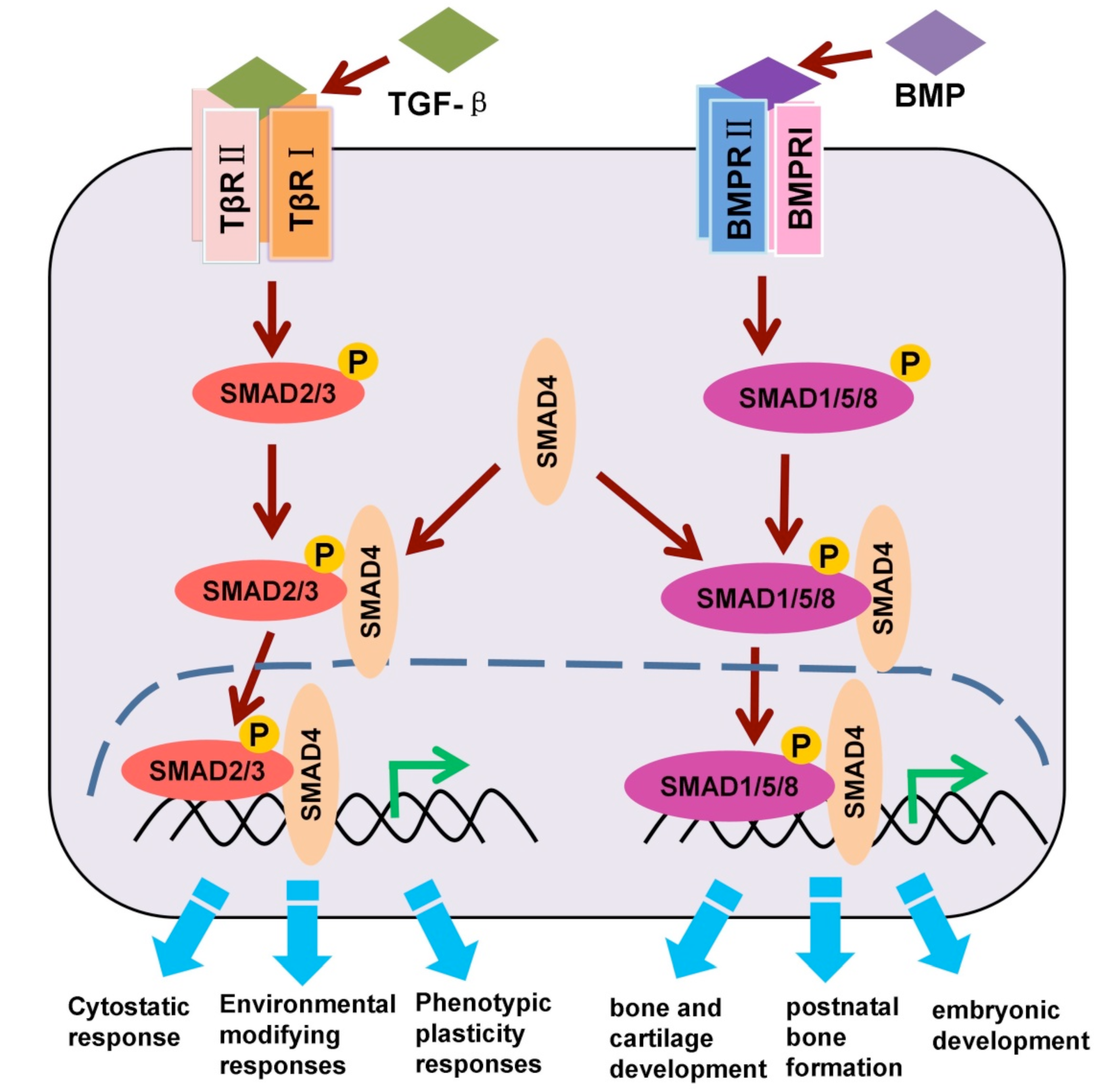
2. Critical Regulation of SMADs by Mono-Ubiquitination
2.1. Mono-Ubiquitination of SMAD3 at Multiple Lysines

2.2. Mono-Ubiquitination of SMAD4
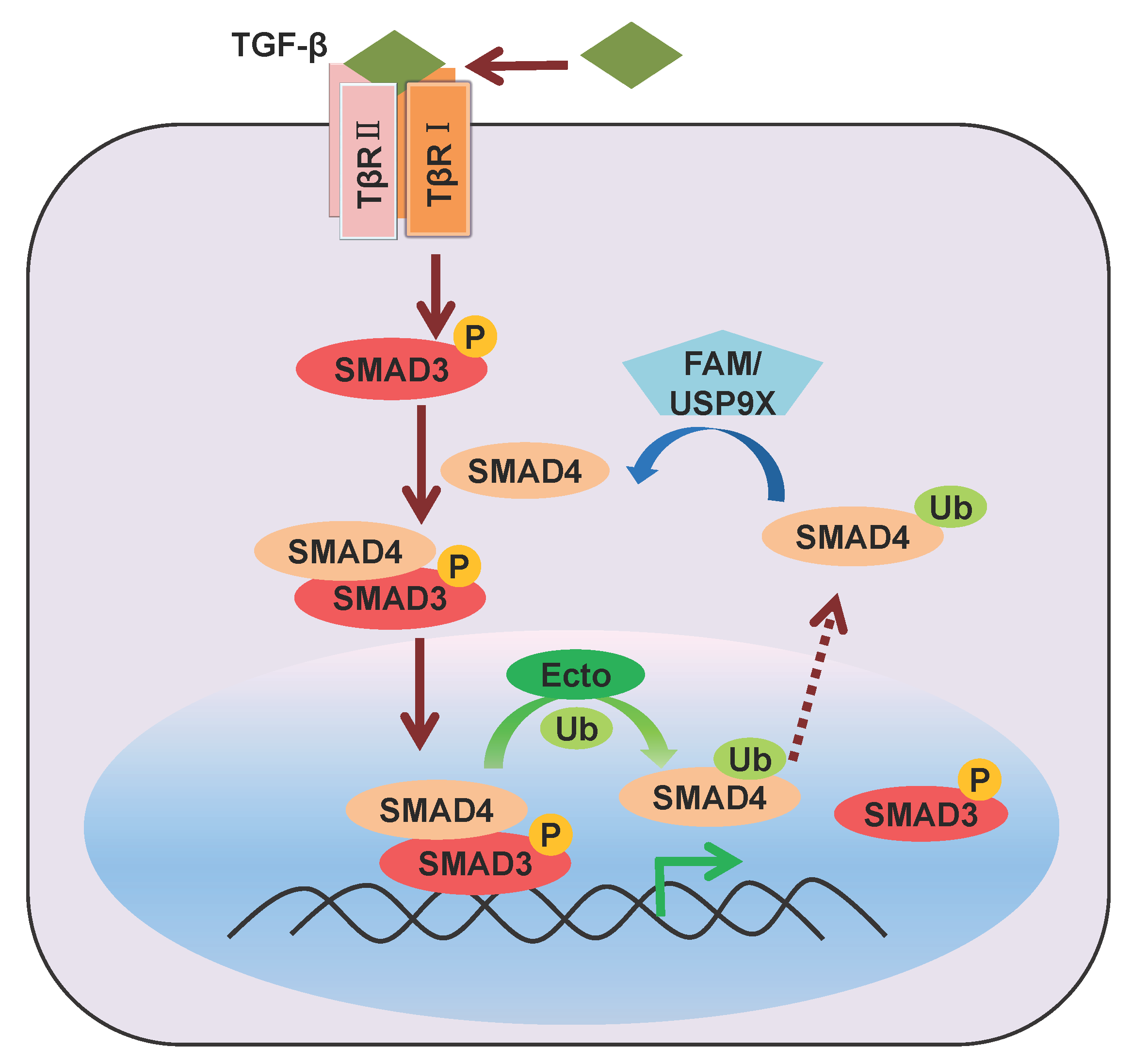
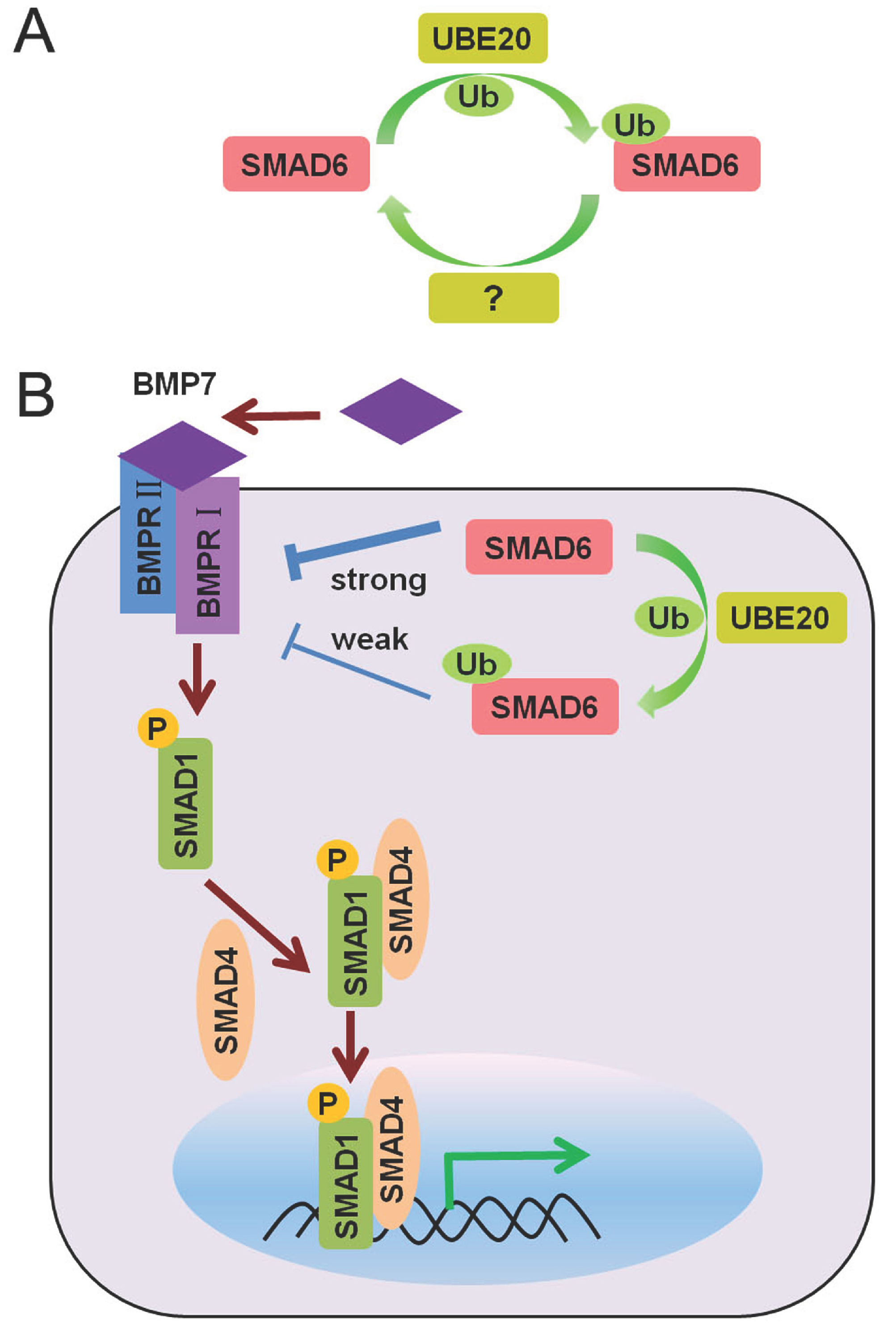
2.3. Mono-Ubiquitination of SMAD6
3. Conclusions and Future Perspectives
Supplementary Materials
Acknowledgments
Conflicts of Interest
References
- Moustakas, A.; Heldin, C.H. The regulation of TGFbeta signal transduction. Development 2009, 136, 3699–3714. [Google Scholar] [PubMed]
- Kang, J.S.; Liu, C.; Derynck, R. New regulatory mechanisms of TGF-beta receptor function. Trends Cell Biol. 2009, 19, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Sasaki, H.; Lowe, L.; Hogan, B.L.; Kuehn, M.R. Nodal is a novel TGF-beta-like gene expressed in the mouse node during gastrulation. Nature 1993, 361, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Schier, A.F.; Shen, M.M. Nodal signalling in vertebrate development. Nature 2000, 403, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Feldman, B.; Gates, M.A.; Egan, E.S.; Dougan, S.T.; Rennebeck, G.; Sirotkin, H.I.; Schier, A.F.; Talbot, W.S. Zebrafish organizer development and germ-layer formation require nodal-related signals. Nature 1998, 395, 181–185. [Google Scholar] [CrossRef] [PubMed]
- Gritsman, K.; Zhang, J.; Cheng, S.; Heckscher, E.; Talbot, W.S.; Schier, A.F. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell 1999, 97, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Plouhinec, J.L.; Zakin, L.; de Robertis, E.M. Systems control of BMP morphogen flow in vertebrate embryos. Curr. Opin. Genet. Dev. 2011, 21, 696–703. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.K.; Kugler, M.C.; Wolters, P.J.; Robillard, L.; Galvez, M.G.; Brumwell, A.N.; Sheppard, D.; Chapman, H.A. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc. Natl. Acad. Sci. USA 2006, 103, 13180–13185. [Google Scholar] [CrossRef] [PubMed]
- Ten Dijke, P.; Egorova, A.D.; Goumans, M.J.; Poelmann, R.E.; Hierck, B.P. TGF-beta signaling in endothelial-to-mesenchymal transition: the role of shear stress and primary cilia. Sci. Signal 2012, 5, pt2. [Google Scholar] [CrossRef] [PubMed]
- Ikushima, H.; Miyazono, K. TGFbeta signalling: a complex web in cancer progression. Nat. Rev. Cancer 2010, 10, 415–424. [Google Scholar] [CrossRef] [PubMed]
- Massague, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef] [PubMed]
- Mu, Y.; Gudey, S.K.; Landstrom, M. Non-Smad signaling pathways. Cell Tissue Res. 2012, 347, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.E. Non-Smad pathways in TGF-beta signaling. Cell Res. 2009, 19, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 2007, 28, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Durocher, D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 2013, 49, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M.; Eddins, M.J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta 2004, 1695, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I. A new ubiquitin chain, a new signal. Nat. Rev. Mol. Cell Biol. 2009, 10, 306. [Google Scholar] [CrossRef] [PubMed]
- Weissman, A.M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001, 2, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, A.L.; Ciechanover, A. Targeting proteins for destruction by the ubiquitin system: implications for human pathobiology. Annu. Rev. Pharmacol. Toxicol. 2009, 49, 73–96. [Google Scholar] [CrossRef]
- Shi, Y.; Massague, J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef] [PubMed]
- Itoh, S.; ten Dijke, P. Negative regulation of TGF-belta receptor/Smad signal transduction. Curr. Opin. Cell Biol. 2007, 19, 176–184. [Google Scholar] [CrossRef] [PubMed]
- Kavsak, P.; Rasmussen, R.K.; Causing, C.G.; Bonni, S.; Zhu, H.; Thomsen, G.H.; Wrana, J.L. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 2000, 6, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, C.; Murakami, G.; Fukuchi, M.; Shimanuki, T.; Shikauchi, Y.; Imamura, T.; Miyazono, K. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J. Biol. Chem. 2002, 277, 39919–39925. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Kavsak, P.; Abdollah, S.; Wrana, J.L.; Thomsen, G.H. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 1999, 400, 687–693. [Google Scholar]
- Zhang, Y.; Chang, C.; Gehling, D.J.; Hemmati-Brivanlou, A.; Derynck, R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA 2001, 98, 974–979. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xin, H.; Xu, X.; Huang, M.; Zhang, X.; Chen, Y.; Zhang, S.; Fu, X.Y.; Chang, Z. CHIP mediates degradation of Smad proteins and potentially regulates Smad-induced transcription. Mol. Cell Biol. 2004, 24, 856–864. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liang, M.; Feng, X.H. Smurf2 is a ubiquitin E3 ligase mediating proteasome-dependent degradation of Smad2 in transforming growth factor-beta signaling. J. Biol. Chem. 2000, 275, 36818–36822. [Google Scholar] [CrossRef] [PubMed]
- Kuratomi, G.; Komuro, A.; Goto, K.; Shinozaki, M.; Miyazawa, K.; Miyazono, K.; Imamura, T. NEDD4-2 (neural precursor cell expressed, developmentally down-regulated 4-2) negatively regulates TGF-beta (transforming growth factor-beta) signalling by inducing ubiquitin-mediated degradation of Smad2 and TGF-beta type I receptor. Biochem. J. 2005, 386, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.R.; Lallemand, F.; Ferrand, N.; Pessah, M.; L’Hoste, S.; Camonis, J.; Atfi, A. The novel E3 ubiquitin ligase Tiul1 associates with TGIF to target Smad2 for degradation. EMBO J. 2004, 23, 3780–3792. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Xu, X.; Li, L.; Ning, H.; Rong, Y.; Shang, Y.; Wang, Y.; Fu, X.Y.; Chang, Z. CHIP controls the sensitivity of transforming growth factor-beta signaling by modulating the basal level of Smad3 through ubiquitin-mediated degradation. J. Biol. Chem. 2005, 280, 20842–20850. [Google Scholar] [CrossRef] [PubMed]
- Koinuma, D.; Shinozaki, M.; Komuro, A.; Goto, K.; Saitoh, M.; Hanyu, A.; Ebina, M.; Nukiwa, T.; Miyazawa, K.; Imamura, T.; et al. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. EMBO J. 2003, 22, 6458–6470. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Rui, H.; Wang, J.; Lin, S.; He, Y.; Chen, M.; Li, Q.; Ye, Z.; Zhang, S.; Chan, S.C.; et al. Axin is a scaffold protein in TGF-beta signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 2006, 25, 1646–1658. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, H.; Zhou, F.; Schimmel, J.; Pardo, C.G.; Zhang, T.; Barakat, T.S.; Sheppard, K.A.; Mickanin, C.; Porter, J.A.; et al. RNF12 controls embryonic stem cell fate and morphogenesis in zebrafish embryos by targeting Smad7 for degradation. Mol. Cell 2012, 46, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.Y.; Yamashita, M.; Coussens, N.P.; Tang, Y.; Wang, X.; Li, C.; Deng, C.X.; Cheng, S.Y.; Zhang, Y.E. Ablation of Smurf2 reveals an inhibition in TGF-beta signalling through multiple mono-ubiquitination of Smad3. EMBO J. 2011, 30, 4777–4789. [Google Scholar] [CrossRef] [PubMed]
- Inui, M.; Manfrin, A.; Mamidi, A.; Martello, G.; Morsut, L.; Soligo, S.; Enzo, E.; Moro, S.; Polo, S.; Dupont, S.; et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat. Cell Biol. 2011, 13, 1368–1375. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Suzuki, H.; Kato, M. Roles of mono-ubiquitinated Smad4 in the formation of Smad transcriptional complexes. Biochem. Biophys. Res. Commun. 2008, 376, 288–292. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Zacchigna, L.; Cordenonsi, M.; Soligo, S.; Adorno, M.; Rugge, M.; Piccolo, S. Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell 2005, 121, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Mamidi, A.; Cordenonsi, M.; Montagner, M.; Zacchigna, L.; Adorno, M.; Martello, G.; Stinchfield, M.J.; Soligo, S.; Morsut, L.; et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell 2009, 136, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, J.; Bauer, A.; Zhang, L.; Selinger, D.W.; Lu, C.X.; Ten Dijke, P. Fine-tuning BMP7 signalling in adipogenesis by UBE2O/E2-230K-mediated monoubiquitination of SMAD6. EMBO J. 2013, 32, 996–1007. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Brooks, C.L.; Wu-Baer, F.; Chen, D.; Baer, R.; Gu, W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 2003, 302, 1972–1975. [Google Scholar] [CrossRef] [PubMed]
- Trotman, L.C.; Wang, X.; Alimonti, A.; Chen, Z.; Teruya-Feldstein, J.; Yang, H.; Pavletich, N.P.; Carver, B.S.; Cordon-Cardo, C.; Erdjument-Bromage, H.; et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 2007, 128, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.J.; Clifton-Bligh, R.J.; Marsh, D.J. Ubiquitination and cancer: Histone H2B monoubiquitination - roles to play in human malignancy. Endocr. Relat. Cancer 2014. [Google Scholar]
- Eichhorn, P.J.; Rodon, L.; Gonzalez-Junca, A.; Dirac, A.; Gili, M.; Martinez-Saez, E.; Aura, C.; Barba, I.; Peg, V.; Prat, A.; et al. USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat. Med. 2012, 18, 429–435. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, F.; Zhang, Z.; Van Dam, H.; Zhang, L.; Zhou, F. Regulation of TGF-β Superfamily Signaling by SMAD Mono-Ubiquitination. Cells 2014, 3, 981-993. https://doi.org/10.3390/cells3040981
Xie F, Zhang Z, Van Dam H, Zhang L, Zhou F. Regulation of TGF-β Superfamily Signaling by SMAD Mono-Ubiquitination. Cells. 2014; 3(4):981-993. https://doi.org/10.3390/cells3040981
Chicago/Turabian StyleXie, Feng, Zhengkui Zhang, Hans Van Dam, Long Zhang, and Fangfang Zhou. 2014. "Regulation of TGF-β Superfamily Signaling by SMAD Mono-Ubiquitination" Cells 3, no. 4: 981-993. https://doi.org/10.3390/cells3040981
APA StyleXie, F., Zhang, Z., Van Dam, H., Zhang, L., & Zhou, F. (2014). Regulation of TGF-β Superfamily Signaling by SMAD Mono-Ubiquitination. Cells, 3(4), 981-993. https://doi.org/10.3390/cells3040981



