Correction: Zhou et al. Notoginsenoside R1 Ameliorates Diabetic Retinopathy through PINK1-Dependent Activation of Mitophagy. Cells 2019, 8, 213
Reference
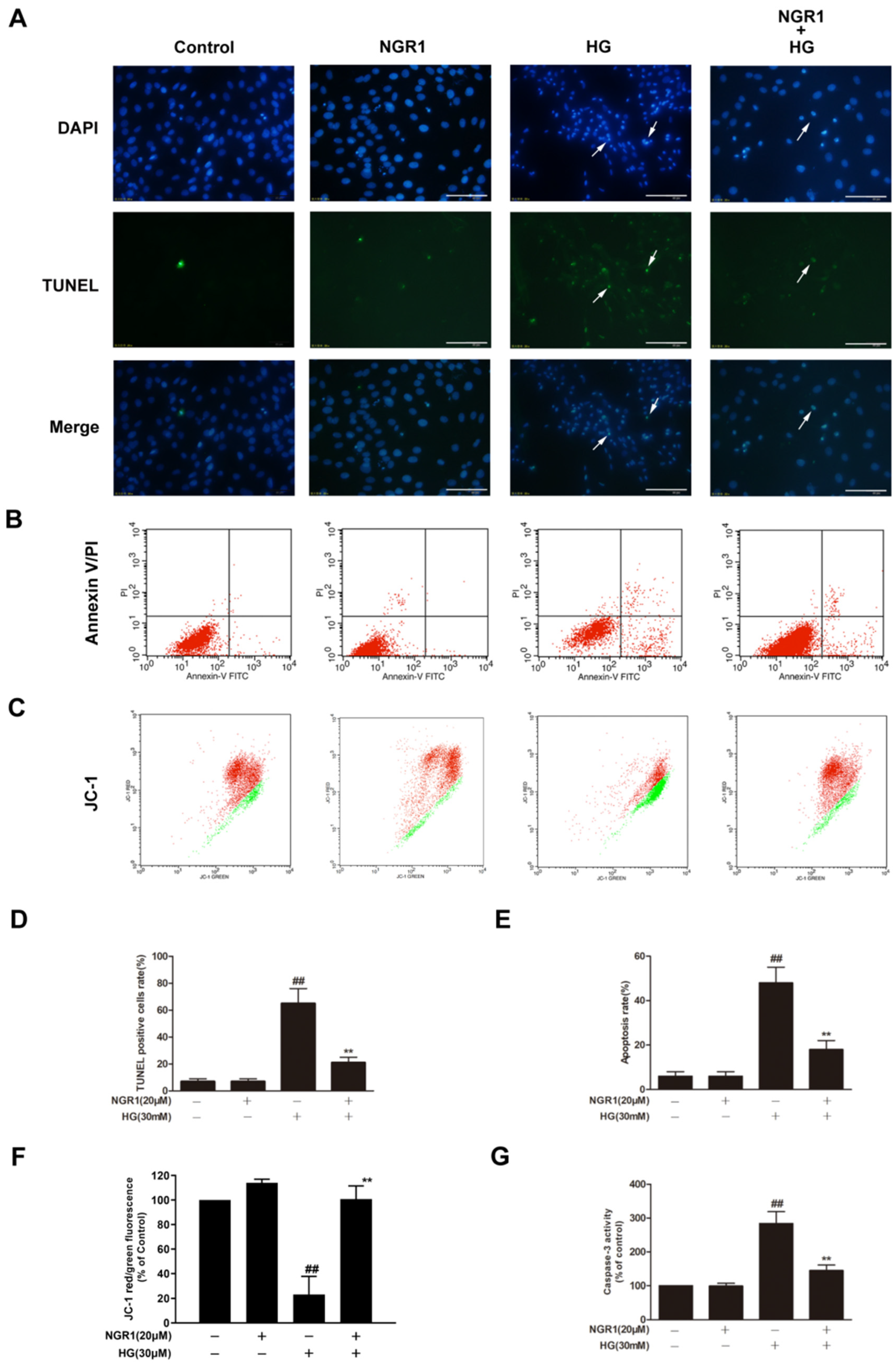
- Zhou, P.; Xie, W.; Meng, X.; Zhai, Y.; Dong, X.; Zhang, X.; Sun, G.; Sun, X. Notoginsenoside R1 Ameliorates Diabetic Retinopathy through PINK1-Dependent Activation of Mitophagy. Cells 2019, 8, 213. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, P.; Xie, W.; Meng, X.; Zhai, Y.; Dong, X.; Zhang, X.; Sun, G.; Sun, X. Correction: Zhou et al. Notoginsenoside R1 Ameliorates Diabetic Retinopathy through PINK1-Dependent Activation of Mitophagy. Cells 2019, 8, 213. Cells 2025, 14, 573. https://doi.org/10.3390/cells14080573
Zhou P, Xie W, Meng X, Zhai Y, Dong X, Zhang X, Sun G, Sun X. Correction: Zhou et al. Notoginsenoside R1 Ameliorates Diabetic Retinopathy through PINK1-Dependent Activation of Mitophagy. Cells 2019, 8, 213. Cells. 2025; 14(8):573. https://doi.org/10.3390/cells14080573
Chicago/Turabian StyleZhou, Ping, Weijie Xie, Xiangbao Meng, Yadong Zhai, Xi Dong, Xuelian Zhang, Guibo Sun, and Xiaobo Sun. 2025. "Correction: Zhou et al. Notoginsenoside R1 Ameliorates Diabetic Retinopathy through PINK1-Dependent Activation of Mitophagy. Cells 2019, 8, 213" Cells 14, no. 8: 573. https://doi.org/10.3390/cells14080573
APA StyleZhou, P., Xie, W., Meng, X., Zhai, Y., Dong, X., Zhang, X., Sun, G., & Sun, X. (2025). Correction: Zhou et al. Notoginsenoside R1 Ameliorates Diabetic Retinopathy through PINK1-Dependent Activation of Mitophagy. Cells 2019, 8, 213. Cells, 14(8), 573. https://doi.org/10.3390/cells14080573




