Integration of Dynamical Network Biomarkers, Control Theory and Drosophila Model Identifies Vasa/DDX4 as the Potential Therapeutic Targets for Metabolic Syndrome
Abstract
:1. Introduction
2. Materials and Methods
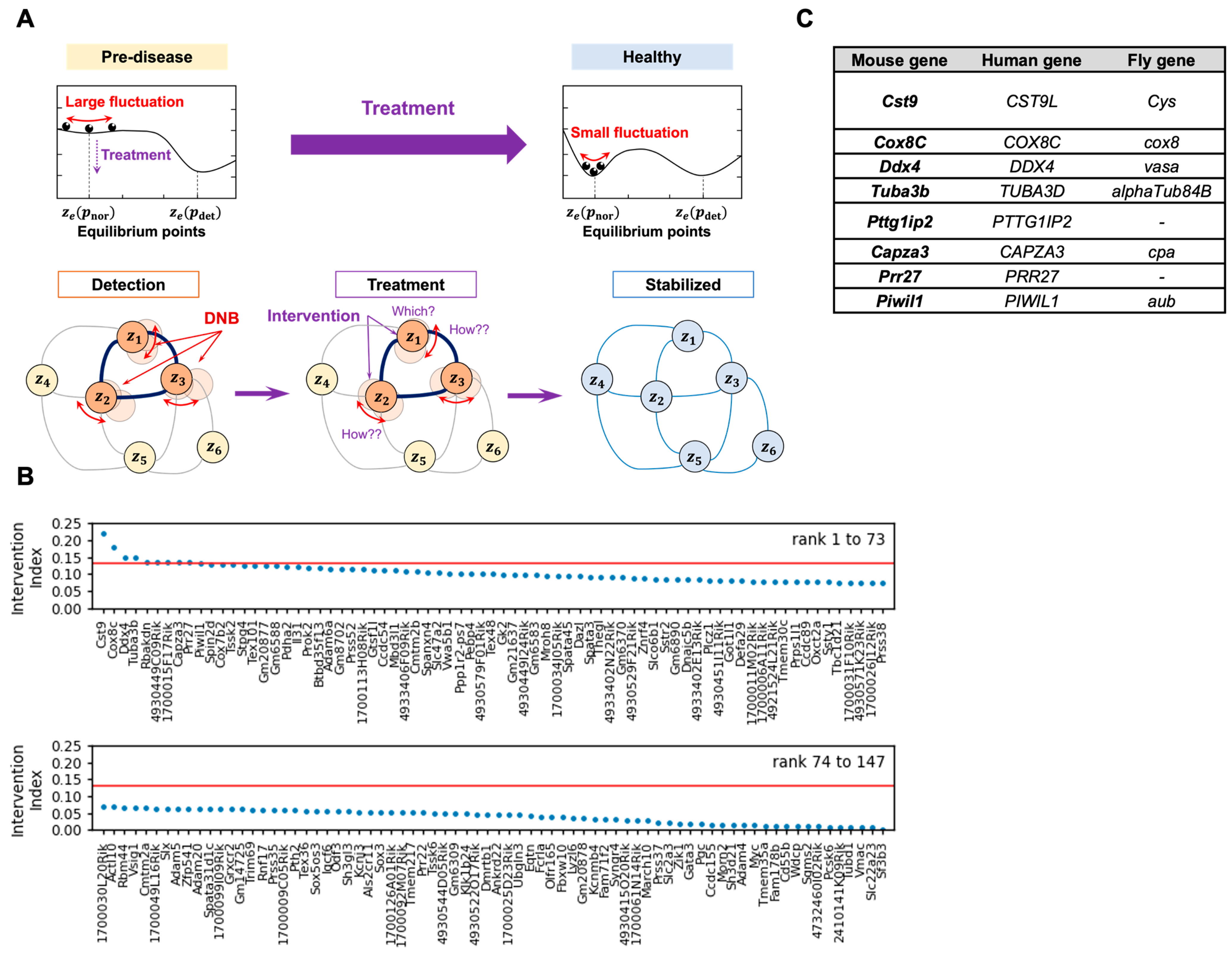
2.1. The DNB Analysis and the DNB Intervention Analysis
2.2. Fly Culture and Stocks
2.3. Starvation Assay
2.4. Bulk RNA Sequencing (RNA-seq) Analysis
2.5. qRT-PCR
2.6. Immunohistochemistry
2.7. Lipid Staining
3. Results
3.1. The DNB Intervention Analysis Selects the Genes to Be Targeted for Therapeutics
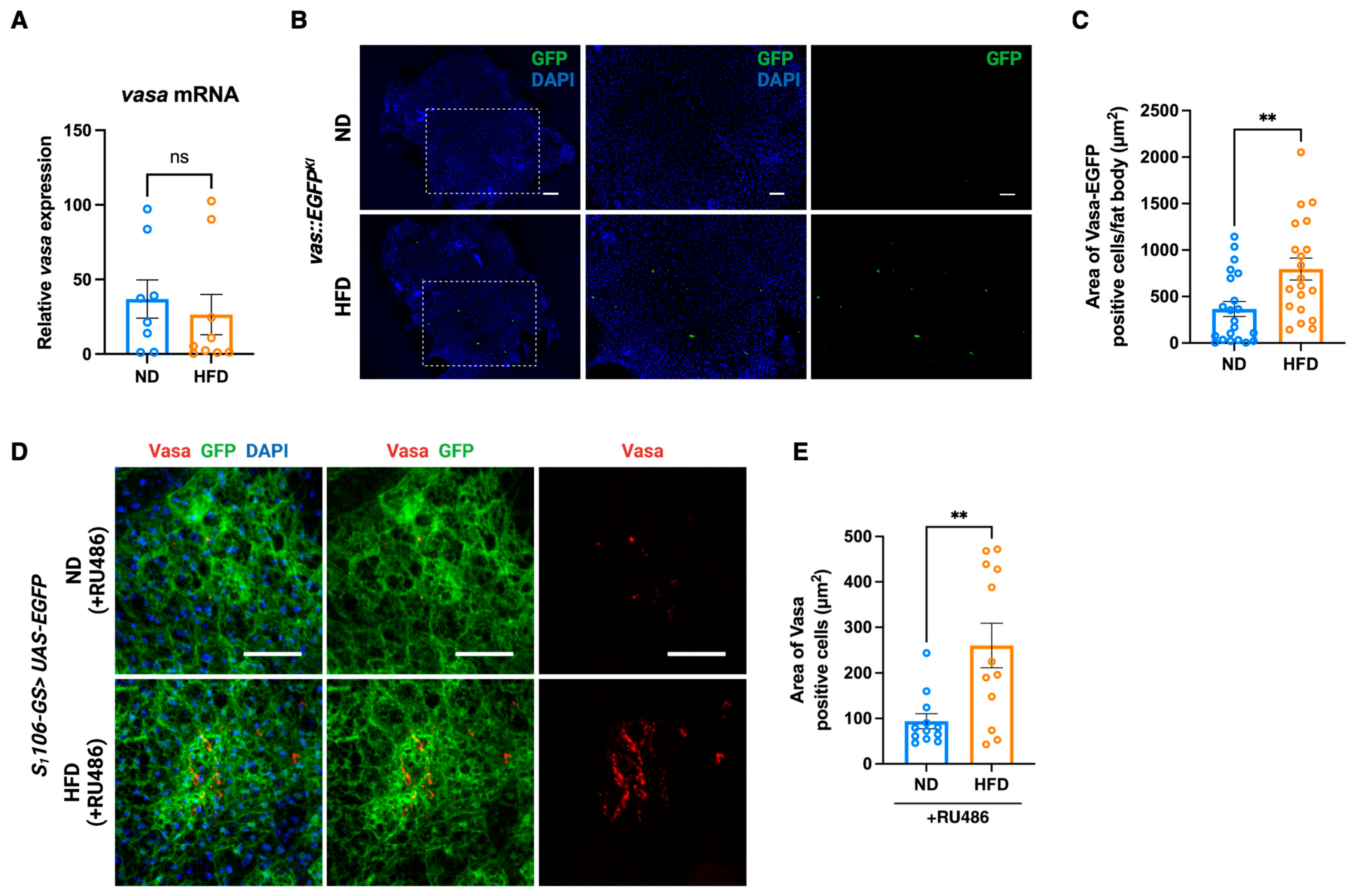
3.2. Fat Body-Specific Knockdown of Vasa Abrogates the Effect of HFD in Fruit Fly
3.3. Vasa Expressing Cells Are Increased in the Fat Body in Response to HFD in Fruit Fly
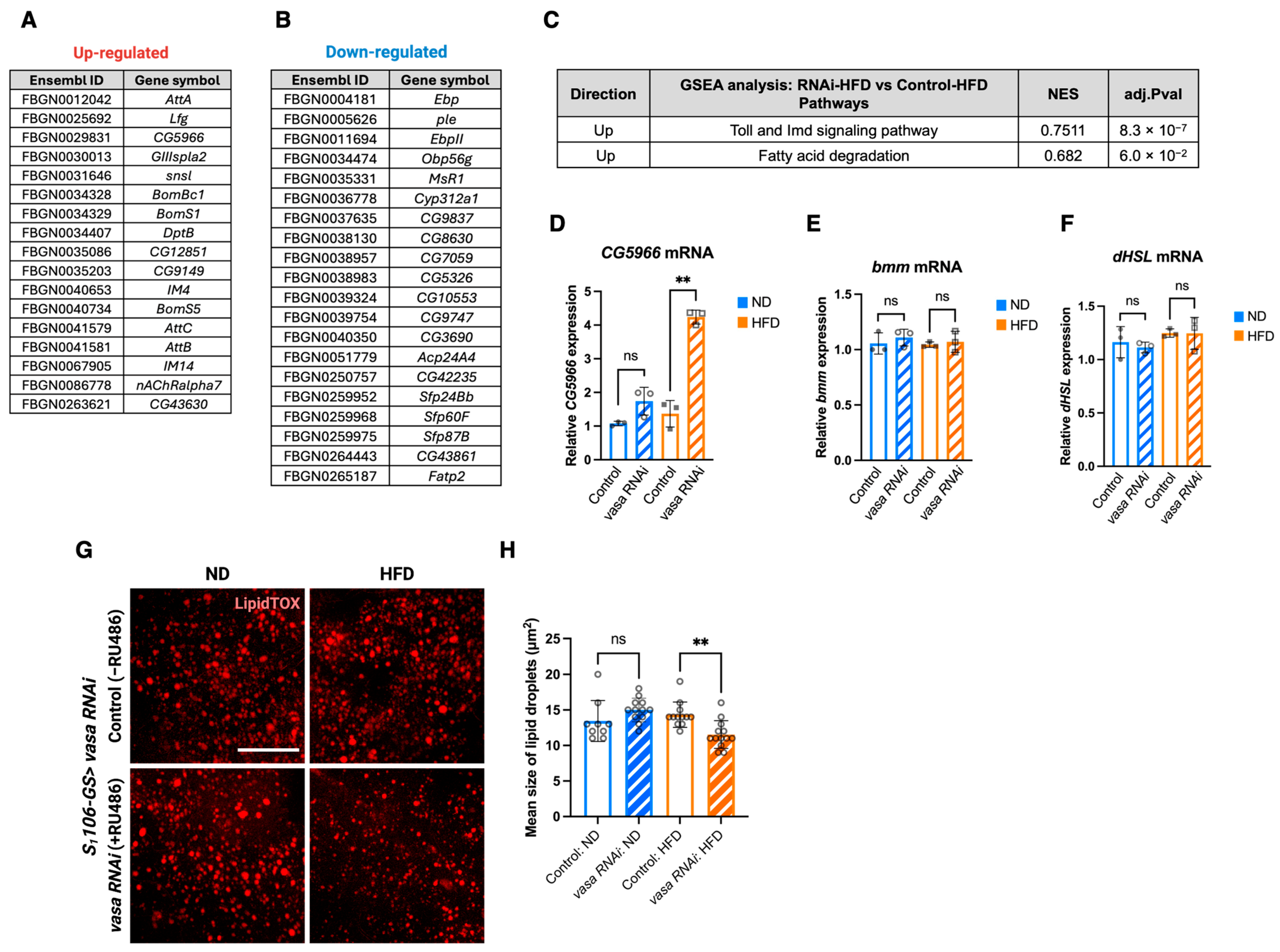
3.4. Fat Body-Specific Knockdown of Vasa Up-Regulates the Lipid Breakdown Pathways Upon HFD
3.5. DDX4/Ddx4 Expressions Are Altered in the Adipose Tissue of Mouse and Human
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Priest, C.; Tontonoz, P. Inter-organ cross-talk in metabolic syndrome. Nat. Metab. 2019, 1, 1177–1188. [Google Scholar] [CrossRef] [PubMed]
- Non-mammalian metabolic models in the spotlight. Nat. Metab. 2022, 4, 1423. [CrossRef]
- Kim, S.K.; Tsao, D.D.; Suh, G.S.B.; Miguel-Aliaga, I. Discovering signaling mechanisms governing metabolism and metabolic diseases with Drosophila. Cell Metab. 2021, 33, 1279–1292. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, N.; Perrimon, N. What fuels the fly: Energy metabolism in Drosophila and its application to the study of obesity and diabetes. Sci. Adv. 2021, 7, 1279–1292. [Google Scholar] [CrossRef]
- Chen, L.; Liu, R.; Liu, Z.P.; Li, M.; Aihara, K. Detecting early-warning signals for sudden deterioration of complex diseases by dynamical network biomarkers. Sci. Rep. 2012, 2, 342. [Google Scholar] [CrossRef] [PubMed]
- Akagi, K.; Koizumi, K.; Kadowaki, M.; Kitajima, I.; Saito, S. New Possibilities for Evaluating the Development of Age-Related Pathologies Using the Dynamical Network Biomarkers Theory. Cells 2023, 12, 2297. [Google Scholar] [CrossRef]
- Scheffer, M.; Bascompte, J.; Brock, W.A.; Brovkin, V.; Carpenter, S.R.; Dakos, V.; Held, H.; van Nes, E.H.; Rietkerk, M.; Sugihara, G. Early-warning signals for critical transitions. Nature 2009, 461, 53–59. [Google Scholar] [CrossRef]
- Scheffer, M.; Carpenter, S.R.; Lenton, T.M.; Bascompte, J.; Brock, W.; Dakos, V.; van de Koppel, J.; van de Leemput, I.A.; Levin, S.A.; van Nes, E.H.; et al. Anticipating critical transitions. Science 2012, 338, 344–348. [Google Scholar] [CrossRef]
- Aihara, K.; Liu, R.; Koizumi, K.; Liu, X.; Chen, L. Dynamical network biomarkers: Theory and applications. Gene 2022, 808, 145997. [Google Scholar] [CrossRef]
- Liu, R.; Wang, J.; Ukai, M.; Sewon, K.; Chen, P.; Suzuki, Y.; Wang, H.; Aihara, K.; Okada-Hatakeyama, M.; Chen, L. Hunt for the tipping point during endocrine resistance process in breast cancer by dynamic network biomarkers. J. Mol. Cell Biol. 2019, 11, 649–664. [Google Scholar] [CrossRef]
- Liu, S.; Hu, Y.; Liu, F.; Jiang, Y.; Wang, H.; Wu, X.; Hu, D. Identifying Key Genes as Progression Indicators of Prostate Cancer with Castration Resistance Based on Dynamic Network Biomarker Algorithm and Weighted Gene Correlation Network Analysis. Biomedicines 2024, 12, 649–664. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Li, M.; Tang, W.; Liu, W.; Zhang, S.; Chen, L.; Xia, J. Dynamic network biomarker indicates pulmonary metastasis at the tipping point of hepatocellular carcinoma. Nat. Commun. 2018, 9, 678. [Google Scholar] [CrossRef]
- Zhong, Z.; Li, J.; Zhong, J.; Huang, Y.; Hu, J.; Zhang, P.; Zhang, B.; Jin, Y.; Luo, W.; Liu, R.; et al. MAPKAPK2, a potential dynamic network biomarker of α-synuclein prior to its aggregation in PD patients. Npj Parkinsons Dis. 2023, 9, 678. [Google Scholar] [CrossRef]
- Yonezawa, S.; Haruki, T.; Koizumi, K.; Taketani, A.; Oshima, Y.; Oku, M.; Wada, A.; Sato, T.; Masuda, N.; Tahara, J.; et al. Establishing Monoclonal Gammopathy of Undetermined Significance as an Independent Pre-Disease State of Multiple Myeloma Using Raman Spectroscopy, Dynamical Network Biomarker Theory, and Energy Landscape Analysis. Int. J. Mol. Sci. 2024, 25, 1570. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, K.; Oku, M.; Hayashi, S.; Inujima, A.; Shibahara, N.; Chen, L.; Igarashi, Y.; Tobe, K.; Saito, S.; Kadowaki, M.; et al. Identifying pre-disease signals before metabolic syndrome in mice by dynamical network biomarkers. Sci. Rep. 2019, 9, 8767. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, K.; Oku, M.; Hayashi, S.; Inujima, A.; Shibahara, N.; Chen, L.; Igarashi, Y.; Tobe, K.; Saito, S.; Kadowaki, M.; et al. Suppression of Dynamical Network Biomarker Signals at the Predisease State (Mibyou) before Metabolic Syndrome in Mice by a Traditional Japanese Medicine (Kampo Formula) Bofutsushosan. Evid. Based Complement. Altern. Med. 2020, 2020, 9129134. [Google Scholar] [CrossRef]
- Shen, X.; Sasahara, H.; Morishita, M.; Imura, J.-I.; Oku, M.; Aihara, K. Model-Free Dominant Pole Placement for Restabilizing High-Dimensional Network Systems via Small-Sample-Size Data. IEEE Access 2023, 11, 45572–45585. [Google Scholar] [CrossRef]
- Shen, X.; Sasahara, H.; Imura, J.i.; Oku, M.; Aihara, K. Re-Stabilizing Large-Scale Network Systems Using High-Dimension Low-Sample-Size Data Analysis. IEEE Trans. Emerg. Top. Comput. Intell. 2024, 1–12. [Google Scholar] [CrossRef]
- Sengoku, T.; Nureki, O.; Nakamura, A.; Kobayashi, S.; Yokoyama, S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 2006, 125, 287–300. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]
- Suzuki, W.; Iizuka, S.; Tabuchi, M.; Funo, S.; Yanagisawa, T.; Kimura, M.; Sato, T.; Endo, T.; Kawamura, H. A new mouse model of spontaneous diabetes derived from ddY strain. Exp. Anim. 1999, 48, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Durdevic, Z.; Ephrussi, A. Germ Cell Lineage Homeostasis in Drosophila Requires the Vasa RNA Helicase. Genetics 2019, 213, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Voog, J.; Sandall, S.L.; Hime, G.R.; Resende, L.P.; Loza-Coll, M.; Aslanian, A.; Yates, J.R., 3rd; Hunter, T.; Fuller, M.T.; Jones, D.L. Escargot restricts niche cell to stem cell conversion in the Drosophila testis. Cell Rep. 2014, 7, 722–734. [Google Scholar] [CrossRef]
- Oulhen, N.; Morita, S.; Wessel, G.M. Post-transcriptional regulation of factors important for the germ line. Curr. Top. Dev. Biol. 2022, 146, 49–78. [Google Scholar] [CrossRef]
- Li, H.; Janssens, J.; De Waegeneer, M.; Kolluru, S.S.; Davie, K.; Gardeux, V.; Saelens, W.; David, F.P.A.; Brbic, M.; Spanier, K.; et al. Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science 2022, 375, eabk2432. [Google Scholar] [CrossRef]
- Kina, H.; Yoshitani, T.; Hanyu-Nakamura, K.; Nakamura, A. Rapid and efficient generation of GFP-knocked-in Drosophila by the CRISPR-Cas9-mediated genome editing. Dev. Growth Differ. 2019, 61, 265–275. [Google Scholar] [CrossRef]
- Nagao, A.; Sato, K.; Nishida, K.M.; Siomi, H.; Siomi, M.C. Gender-Specific Hierarchy in Nuage Localization of PIWI-Interacting RNA Factors in Drosophila. Front. Genet. 2011, 2, 55. [Google Scholar] [CrossRef] [PubMed]
- Heier, C.; Kuhnlein, R.P. Triacylglycerol Metabolism in Drosophila melanogaster. Genetics 2018, 210, 1163–1184. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L. STREME: Accurate and versatile sequence motif discovery. Bioinformatics 2021, 37, 2834–2840. [Google Scholar] [CrossRef]
- Liu, N.; Han, H.; Lasko, P. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3’ UTR. Genes. Dev. 2009, 23, 2742–2752. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Emont, M.P.; Jacobs, C.; Essene, A.L.; Pant, D.; Tenen, D.; Colleluori, G.; Di Vincenzo, A.; Jorgensen, A.M.; Dashti, H.; Stefek, A.; et al. A single-cell atlas of human and mouse white adipose tissue. Nature 2022, 603, 926–933. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Gao, D.; Zhou, B.; Chen, S.; Wang, L. New discoveries in the field of metabolism by applying single-cell and spatial omics. J. Pharm. Anal. 2023, 13, 711–725. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Mohanty, V.; Dede, M.; Tsai, K.; Daher, M.; Li, L.; Rezvani, K.; Chen, K. Characterizing cancer metabolism from bulk and single-cell RNA-seq data using METAFlux. Nat. Commun. 2023, 14, 4883. [Google Scholar] [CrossRef] [PubMed]
- Van de Sande, B.; Lee, J.S.; Mutasa-Gottgens, E.; Naughton, B.; Bacon, W.; Manning, J.; Wang, Y.; Pollard, J.; Mendez, M.; Hill, J.; et al. Applications of single-cell RNA sequencing in drug discovery and development. Nat. Rev. Drug Discov. 2023, 22, 496–520. [Google Scholar] [CrossRef]
- Baenas, N.; Wagner, A.E. Drosophila melanogaster as a Model Organism for Obesity and Type-2 Diabetes Mellitus by Applying High-Sugar and High-Fat Diets. Biomolecules 2022, 12, 496–520. [Google Scholar] [CrossRef]
- Woodcock, K.J.; Kierdorf, K.; Pouchelon, C.A.; Vivancos, V.; Dionne, M.S.; Geissmann, F. Macrophage-derived upd3 cytokine causes impaired glucose homeostasis and reduced lifespan in Drosophila fed a lipid-rich diet. Immunity 2015, 42, 133–144. [Google Scholar] [CrossRef]
- Birse, R.T.; Choi, J.; Reardon, K.; Rodriguez, J.; Graham, S.; Diop, S.; Ocorr, K.; Bodmer, R.; Oldham, S. High-fat-diet-induced obesity and heart dysfunction are regulated by the TOR pathway in Drosophila. Cell Metab. 2010, 12, 533–544. [Google Scholar] [CrossRef]
- Zechner, R. FAT FLUX: Enzymes, regulators, and pathophysiology of intracellular lipolysis. EMBO Mol. Med. 2015, 7, 359–362. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Yadav, S.R.; Kannan, N.N. A role for the circadian photoreceptor CRYPTOCHROME in regulating triglyceride metabolism in Drosophila. G3 Genes|Genomes|Genetics 2024, 11, jkae220. [Google Scholar] [CrossRef]
- Blumrich, A.; Vogler, G.; Dresen, S.; Diop, S.B.; Jaeger, C.; Leberer, S.; Grune, J.; Wirth, E.K.; Hoeft, B.; Renko, K.; et al. Fat-body brummer lipase determines survival and cardiac function during starvation in Drosophila melanogaster. iScience 2021, 24, 102288. [Google Scholar] [CrossRef] [PubMed]
- Lasko, P.F.; Ashburner, M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature 1988, 335, 611–617. [Google Scholar] [CrossRef] [PubMed]
- Hay, B.; Jan, L.Y.; Jan, Y.N. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 1988, 55, 577–587. [Google Scholar] [CrossRef]
- Carrera, P.; Johnstone, O.; Nakamura, A.; Casanova, J.; Jackle, H.; Lasko, P. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol. Cell 2000, 5, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, O.; Lasko, P. Interaction with eIF5B is essential for Vasa function during development. Development 2004, 131, 4167–4178. [Google Scholar] [CrossRef]
- Xiol, J.; Spinelli, P.; Laussmann, M.A.; Homolka, D.; Yang, Z.; Cora, E.; Coute, Y.; Conn, S.; Kadlec, J.; Sachidanandam, R.; et al. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 2014, 157, 1698–1711. [Google Scholar] [CrossRef]
- Poon, J.; Wessel, G.M.; Yajima, M. An unregulated regulator: Vasa expression in the development of somatic cells and in tumorigenesis. Dev. Biol. 2016, 415, 24–32. [Google Scholar] [CrossRef]
- Tapescu, I.; Cherry, S. DDX RNA helicases: Key players in cellular homeostasis and innate antiviral immunity. J. Virol. 2024, 98, e0004024. [Google Scholar] [CrossRef]
- Linder, P.; Jankowsky, E. From unwinding to clamping—The DEAD box RNA helicase family. Nat. Rev. Mol. Cell Biol. 2011, 12, 505–516. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, M.; Cai, Z.; Liu, H.; Zhong, W.; Hao, Q.; Cheng, D.; Hu, X.; Hou, J.; Xu, P.; et al. RNA-binding protein DDX1 is responsible for fatty acid-mediated repression of insulin translation. Nucleic Acids Res. 2018, 46, 12052–12066. [Google Scholar] [CrossRef]
- Hokimoto, S.; Funakoshi-Tago, M.; Tago, K. Identification of DDX5 as an indispensable activator of the glucocorticoid receptor in adipocyte differentiation. FEBS J. 2023, 290, 988–1007. [Google Scholar] [CrossRef] [PubMed]
- Ning, D.; Jin, J.; Fang, Y.; Du, P.; Yuan, C.; Chen, J.; Huang, Q.; Cheng, K.; Mo, J.; Xu, L.; et al. DEAD-Box Helicase 17 exacerbates non-alcoholic steatohepatitis via transcriptional repression of cyp2c29, inducing hepatic lipid metabolism disorder and eliciting the activation of M1 macrophages. Clin. Transl. Med. 2024, 14, e1529. [Google Scholar] [CrossRef] [PubMed]
- Boone, C.; Lewis, S.C. Bridging lipid metabolism and mitochondrial genome maintenance. J. Biol. Chem. 2024, 300, 107498. [Google Scholar] [CrossRef]
- Lee, S.H.; Lee, S.K.; Paik, D.; Min, K.J. Overexpression of fatty-acid-beta-oxidation-related genes extends the lifespan of Drosophila melanogaster. Oxid. Med. Cell Longev. 2012, 2012, 854502. [Google Scholar] [CrossRef]
- Styhler, S.; Nakamura, A.; Swan, A.; Suter, B.; Lasko, P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 1998, 125, 1569–1578. [Google Scholar] [CrossRef]
- Spike, C.; Meyer, N.; Racen, E.; Orsborn, A.; Kirchner, J.; Kuznicki, K.; Yee, C.; Bennett, K.; Strome, S. Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics 2008, 178, 1973–1987. [Google Scholar] [CrossRef]
- Hartung, O.; Forbes, M.M.; Marlow, F.L. Zebrafish vasa is required for germ-cell differentiation and maintenance. Mol. Reprod. Dev. 2014, 81, 946–961. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Hong, N.; Xu, H.; Yi, M.; Li, C.; Gui, J.; Hong, Y. Medaka vasa is required for migration but not survival of primordial germ cells. Mech. Dev. 2009, 126, 366–381. [Google Scholar] [CrossRef]
- Tanaka, S.S.; Toyooka, Y.; Akasu, R.; Katoh-Fukui, Y.; Nakahara, Y.; Suzuki, R.; Yokoyama, M.; Noce, T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes. Dev. 2000, 14, 841–853. [Google Scholar] [CrossRef]
- Renault, A.D. vasa is expressed in somatic cells of the embryonic gonad in a sex-specific manner in Drosophila melanogaster. Biol. Open 2012, 1, 1043–1048. [Google Scholar] [CrossRef]
- Landis, G.N.; Doherty, D.V.; Yen, C.A.; Wang, L.; Fan, Y.; Wang, I.; Vroegop, J.; Wang, T.; Wu, J.; Patel, P.; et al. Metabolic Signatures of Life Span Regulated by Mating, Sex Peptide, and Mifepristone/RU486 in Female Drosophila melanogaster. J. Gerontol. A Biol. Sci. Med. Sci. 2021, 76, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Vinayagam, A.; Gibson, T.E.; Lee, H.J.; Yilmazel, B.; Roesel, C.; Hu, Y.; Kwon, Y.; Sharma, A.; Liu, Y.Y.; Perrimon, N.; et al. Controllability analysis of the directed human protein interaction network identifies disease genes and drug targets. Proc. Natl. Acad. Sci. USA 2016, 113, 4976–4981. [Google Scholar] [CrossRef] [PubMed]
- Devkota, P.; Wuchty, S. Controllability analysis of molecular pathways points to proteins that control the entire interaction network. Sci. Rep. 2020, 10, 2943. [Google Scholar] [CrossRef]
- Markowetz, F. How to understand the cell by breaking it: Network analysis of gene perturbation screens. PLoS Comput. Biol. 2010, 6, e1000655. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akagi, K.; Jin, Y.-J.; Koizumi, K.; Oku, M.; Ito, K.; Shen, X.; Imura, J.-i.; Aihara, K.; Saito, S. Integration of Dynamical Network Biomarkers, Control Theory and Drosophila Model Identifies Vasa/DDX4 as the Potential Therapeutic Targets for Metabolic Syndrome. Cells 2025, 14, 415. https://doi.org/10.3390/cells14060415
Akagi K, Jin Y-J, Koizumi K, Oku M, Ito K, Shen X, Imura J-i, Aihara K, Saito S. Integration of Dynamical Network Biomarkers, Control Theory and Drosophila Model Identifies Vasa/DDX4 as the Potential Therapeutic Targets for Metabolic Syndrome. Cells. 2025; 14(6):415. https://doi.org/10.3390/cells14060415
Chicago/Turabian StyleAkagi, Kazutaka, Ying-Jie Jin, Keiichi Koizumi, Makito Oku, Kaisei Ito, Xun Shen, Jun-ichi Imura, Kazuyuki Aihara, and Shigeru Saito. 2025. "Integration of Dynamical Network Biomarkers, Control Theory and Drosophila Model Identifies Vasa/DDX4 as the Potential Therapeutic Targets for Metabolic Syndrome" Cells 14, no. 6: 415. https://doi.org/10.3390/cells14060415
APA StyleAkagi, K., Jin, Y.-J., Koizumi, K., Oku, M., Ito, K., Shen, X., Imura, J.-i., Aihara, K., & Saito, S. (2025). Integration of Dynamical Network Biomarkers, Control Theory and Drosophila Model Identifies Vasa/DDX4 as the Potential Therapeutic Targets for Metabolic Syndrome. Cells, 14(6), 415. https://doi.org/10.3390/cells14060415





