Seasonal Influences on Human Placental Transcriptomes Associated with Spontaneous Preterm Birth
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Study Participants and Thermal Exposure Groups Stratification
2.3. Differential Gene Expression (DE) Analysis
2.4. Gene Set Enrichment Analysis (GSEA) and Gene Ontology (GO) Analysis
2.5. ELISA Assay
2.6. Statistical Analysis
3. Results
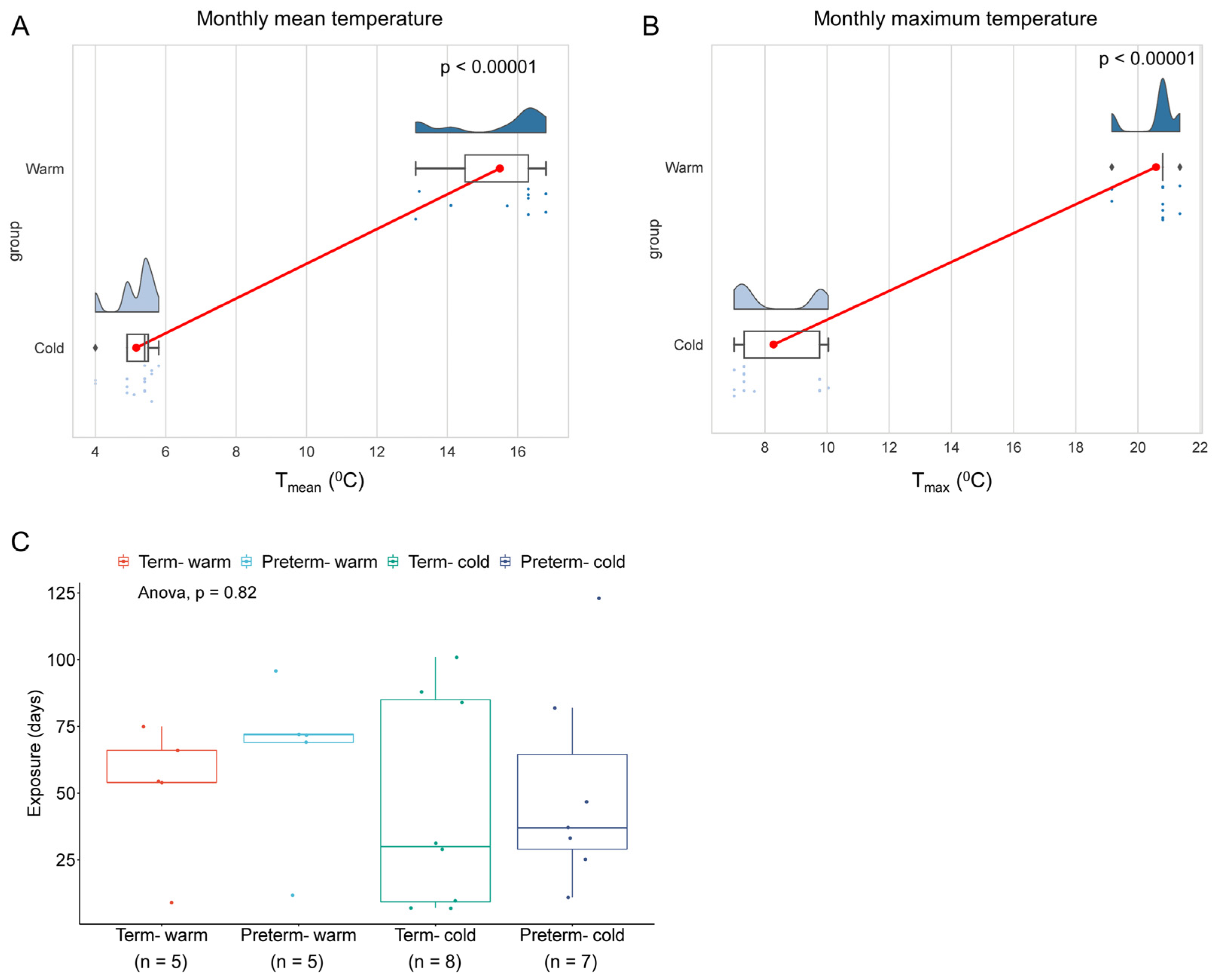
3.1. Study Subjects and Seasonal Temperature Exposures
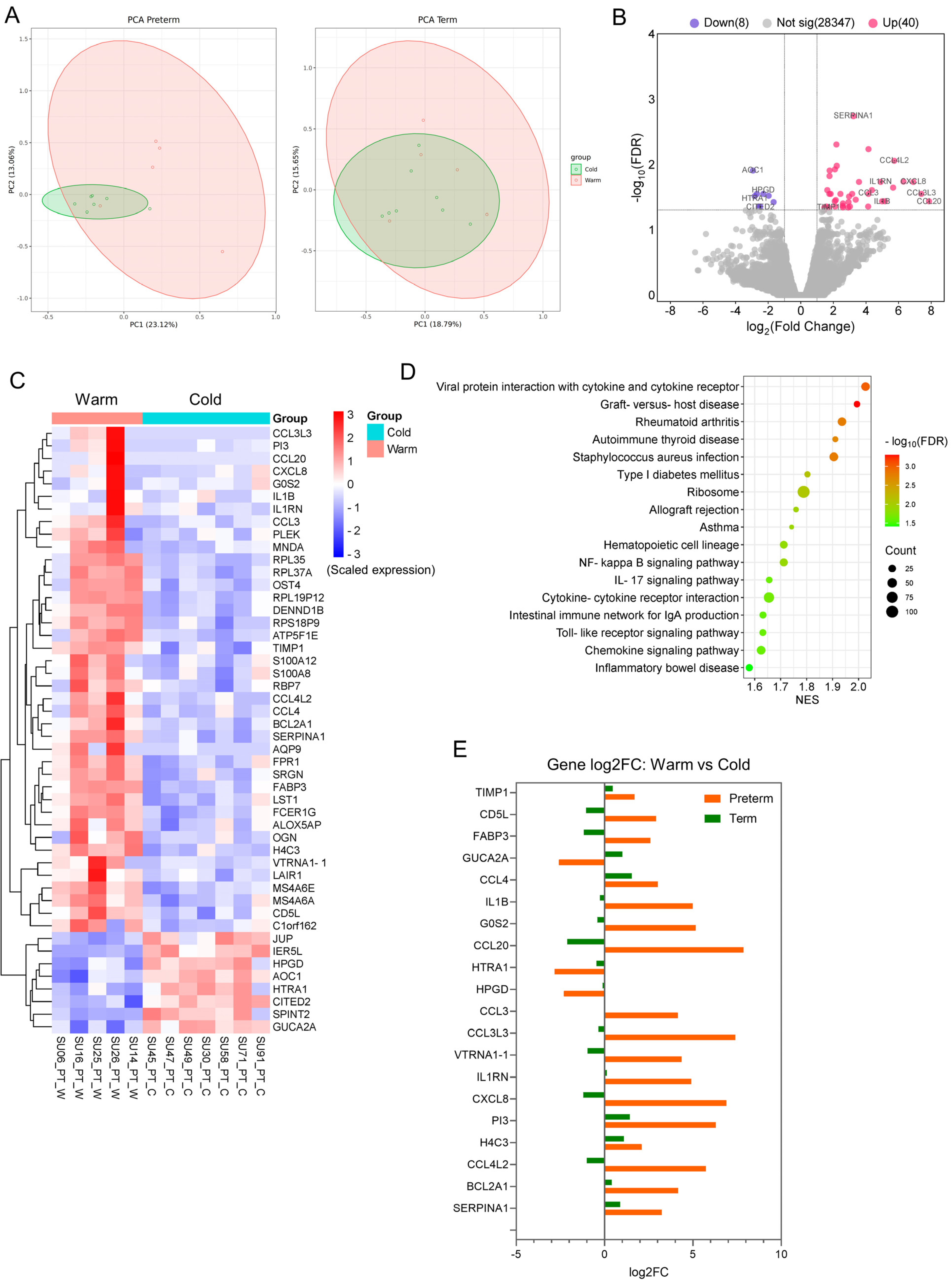
3.2. The Transcriptomic Profiles Are Altered in Preterm Placentas Delivered During Summer Months Compared to Winter Months
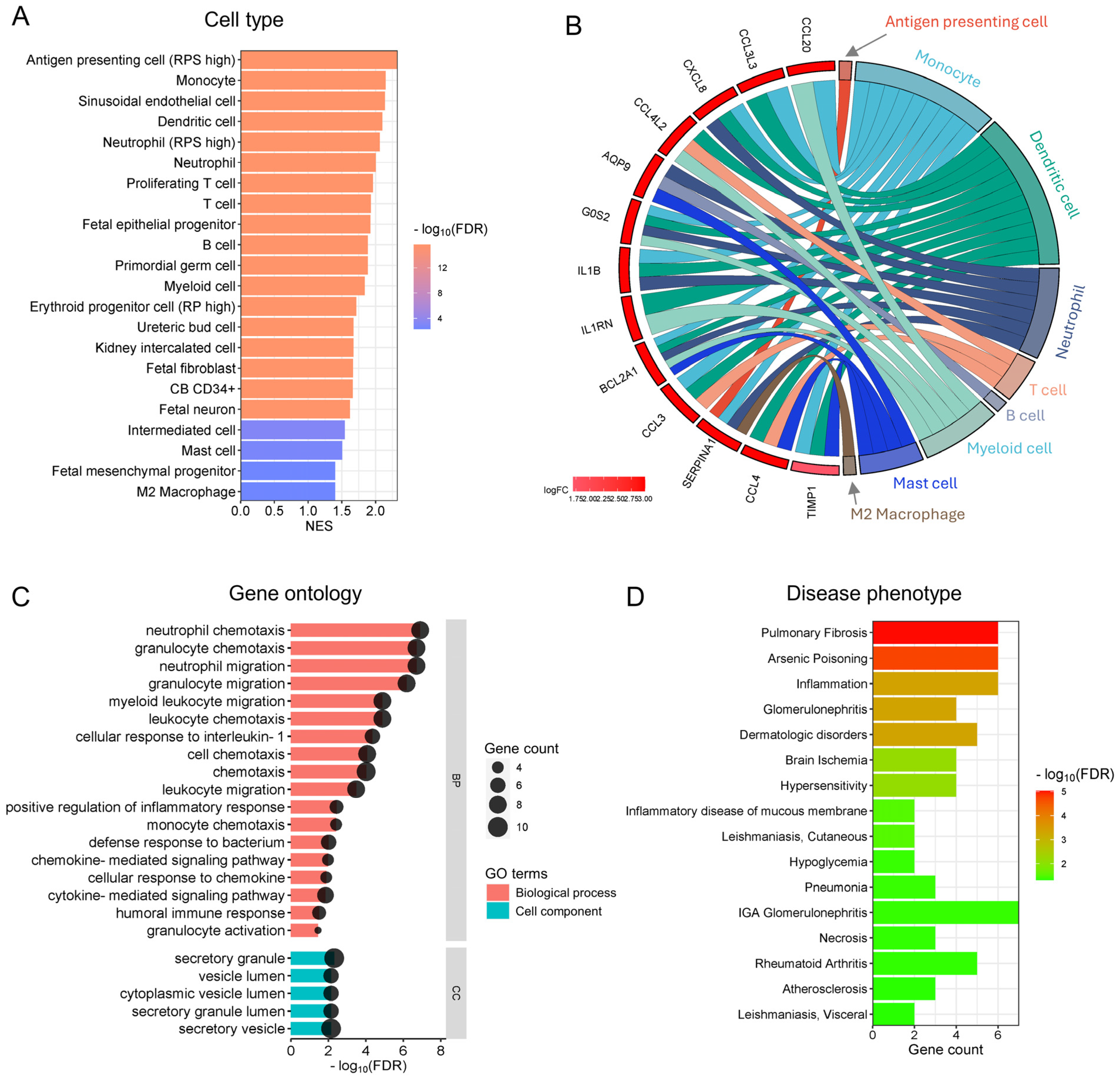
3.3. Functional Analysis Reveals the Enrichment of Inflammatory and Immune Cells and Their Functions in Preterm Placentas Delivered During Summertime
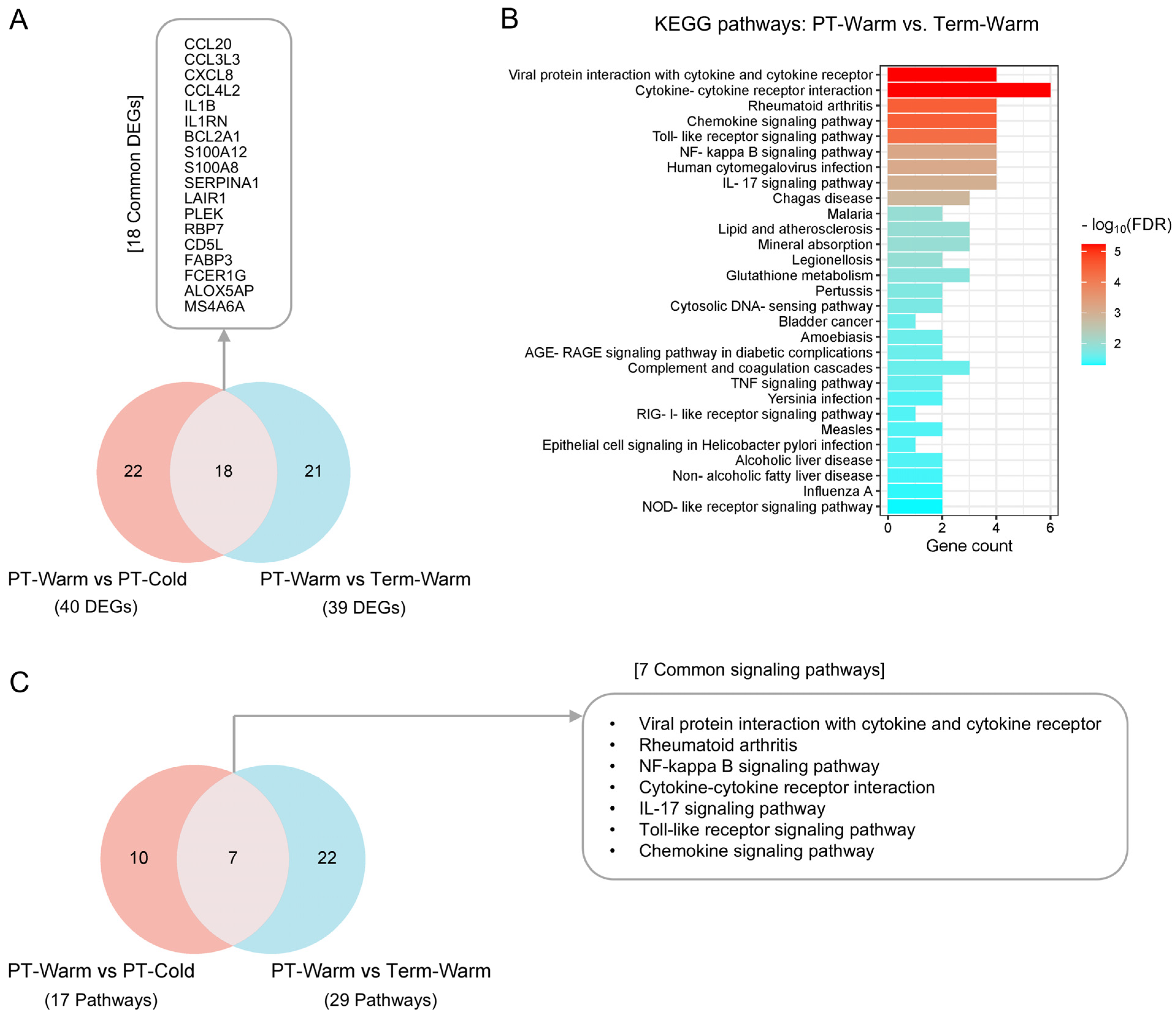
3.4. The Genes and Signalling Pathways Upregulated in Warm-Exposed Preterm Placentas May Mechanistically Link to PTB
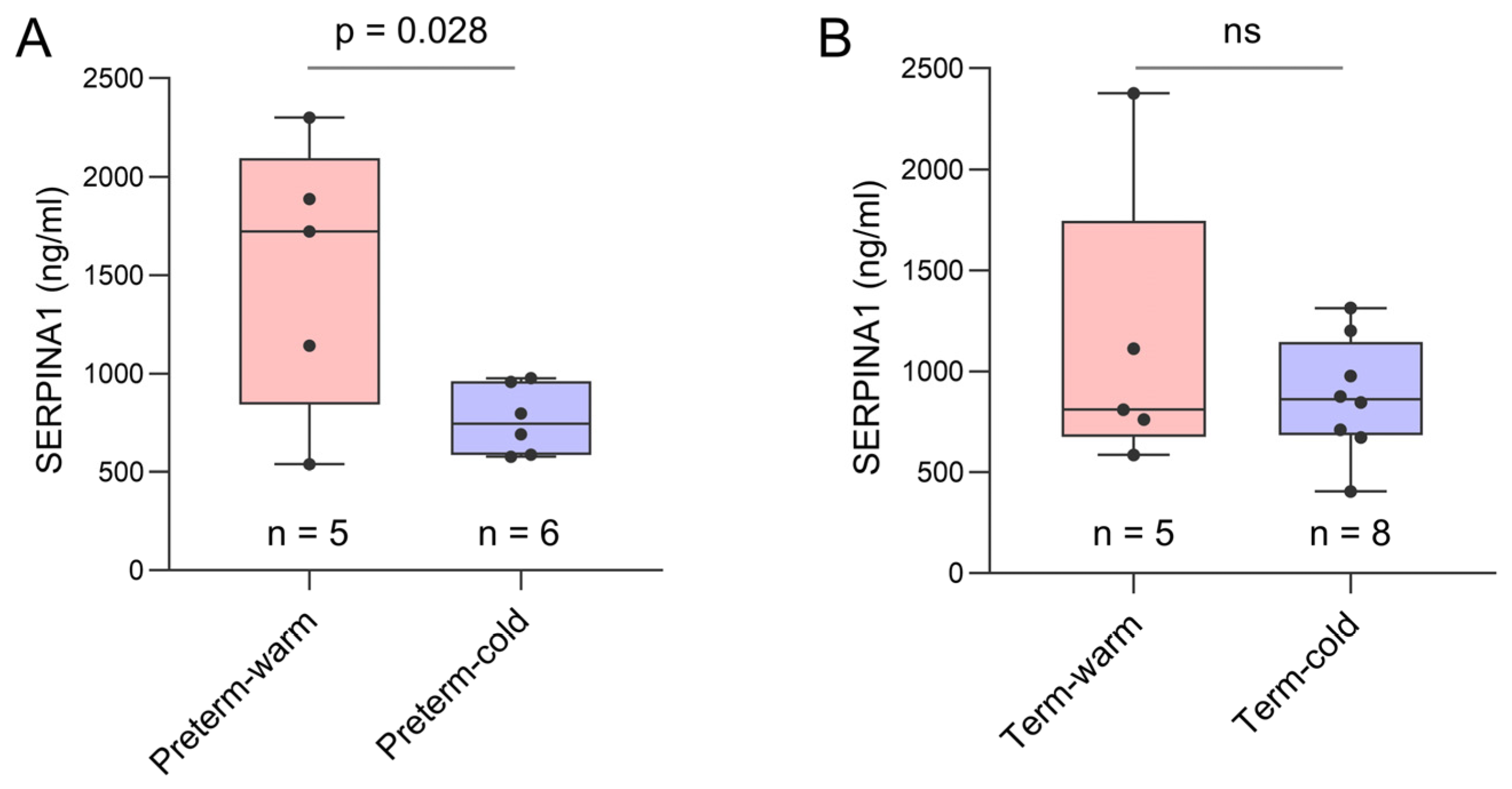
3.5. Key Genes Linked to Placental Inflammatory and Immune Processes Detected in Preterm Births During Summer Months
4. Discussion
Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Watts, N.; Amann, M.; Arnell, N.; Ayeb-Karlsson, S.; Beagley, J.; Belesova, K.; Boykoff, M.; Byass, P.; Cai, W.; Campbell-Lendrum, D.; et al. The 2020 report of The Lancet Countdown on health and climate change: Responding to converging crises. Lancet 2021, 397, 129–170. [Google Scholar] [CrossRef] [PubMed]
- Hoegh-Guldberg, O.; Jacob, D.; Taylor, M.; Guillén Bolaños, T.; Bindi, M.; Brown, S.; Camilloni, I.A.; Diedhiou, A.; Djalante, R.; Ebi, K.; et al. The human imperative of stabilizing global climate change at 1.5 °C. Science 2019, 365, eaaw6974. [Google Scholar] [CrossRef] [PubMed]
- Judge, C.M.; Chasan-Taber, L.; Gensburg, L.; Nasca, P.C.; Marshall, E.G. Physical exposures during pregnancy and congenital cardiovascular malformations. Paediatr. Perinat. Epidemiol. 2004, 18, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Suarez, L.; Felkner, M.; Hendricks, K. The effect of fever, febrile illnesses, and heat exposures on the risk of neural tube defects in a Texas-Mexico border population. Birth Defects Res. Part A Clin. Mol. Teratol. 2004, 70, 815–819. [Google Scholar] [CrossRef]
- Chersich, M.F.; Pham, M.D.; Areal, A.; Haghighi, M.M.; Manyuchi, A.; Swift, C.P.; Wernecke, B.; Robinson, M.; Hetem, R.; Boeckmann, M.; et al. Associations between high temperatures in pregnancy and risk of preterm birth, low birth weight, and stillbirths: Systematic review and meta-analysis. BMJ 2020, 371, m3811. [Google Scholar] [CrossRef]
- Bekkar, B.; Pacheco, S.; Basu, R.; DeNicola, N. Association of Air Pollution and Heat Exposure with Preterm Birth, Low Birth Weight, and Stillbirth in the US: A Systematic Review. JAMA Netw. Open 2020, 3, e208243. [Google Scholar] [CrossRef] [PubMed]
- Gong, Y.; Chai, J.; Yang, M.; Sun, P.; Sun, R.; Dong, W.; Li, Q.; Zhou, D.; Yu, F.; Wang, Y.; et al. Effects of ambient temperature on the risk of preterm birth in offspring of adolescent mothers in rural henan, China. Environ. Res. 2021, 201, 111545. [Google Scholar] [CrossRef] [PubMed]
- Syed, S.; O’Sullivan, T.L.; Phillips, K.P. Extreme Heat and Pregnancy Outcomes: A Scoping Review of the Epidemiological Evidence. Int. J. Environ. Res. Public Health 2022, 19, 2412. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Wang, Q.; Zhao, W.; Ren, Z.; Zhang, H.; Jalaludin, B.; Benmarhnia, T.; Di, J.; Hu, H.; Wang, Y.; et al. Effects of extreme temperature on the risk of preterm birth in China: A population-based multi-center cohort study. Lancet Reg. Health West. Pac. 2022, 24, 100496. [Google Scholar] [CrossRef]
- Wang, Q.; Yin, L.; Wu, H.; Ren, Z.; He, S.; Huang, A.; Huang, C. Effects of gestational ambient extreme temperature exposures on the risk of preterm birth in China: A sibling-matched study based on a multi-center prospective cohort. Sci. Total Environ. 2023, 887, 164135. [Google Scholar] [CrossRef] [PubMed]
- Chawanpaiboon, S.; Vogel, J.P.; Moller, A.B.; Lumbiganon, P.; Petzold, M.; Hogan, D.; Landoulsi, S.; Jampathong, N.; Kongwattanakul, K.; Laopaiboon, M.; et al. Global, regional, and national estimates of levels of preterm birth in 2014: A systematic review and modelling analysis. Lancet Glob. Health 2019, 7, e37–e46. [Google Scholar] [CrossRef]
- Goldenberg, R.L.; Culhane, J.F.; Iams, J.D.; Romero, R. Epidemiology and causes of preterm birth. Lancet 2008, 371, 75–84. [Google Scholar] [CrossRef]
- Bonell, A.; Sonko, B.; Badjie, J.; Samateh, T.; Saidy, T.; Sosseh, F.; Sallah, Y.; Bajo, K.; Murray, K.A.; Hirst, J.; et al. Environmental heat stress on maternal physiology and fetal blood flow in pregnant subsistence farmers in The Gambia, west Africa: An observational cohort study. Lancet Planet. Health 2022, 6, e968–e976. [Google Scholar] [CrossRef] [PubMed]
- Turco, M.Y.; Moffett, A. Development of the human placenta. Development 2019, 146, dev163428. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Dong, M.; Sun, X.; Xiao, J.; Zeng, W.; Hu, J.; Li, X.; Guo, L.; Rong, Z.; et al. Associations of maternal ambient temperature exposures during pregnancy with the placental weight, volume and PFR: A birth cohort study in Guangzhou, China. Environ. Int. 2020, 139, 105682. [Google Scholar] [CrossRef] [PubMed]
- Galan, H.L.; Hussey, M.J.; Barbera, A.; Ferrazzi, E.; Chung, M.; Hobbins, J.C.; Battaglia, F.C. Relationship of fetal growth to duration of heat stress in an ovine model of placental insufficiency. Am. J. Obstet. Gynecol. 1999, 180, 1278–1282. [Google Scholar] [CrossRef]
- Limesand, S.W.; Regnault, T.R.; Hay, W.W., Jr. Characterization of glucose transporter 8 (GLUT8) in the ovine placenta of normal and growth restricted fetuses. Placenta 2004, 25, 70–77. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Akram, K.M.; Kulkarni, N.S.; Brook, A.; Wyles, M.D.; Anumba, D.O.C. Transcriptomic analysis of the human placenta reveals trophoblast dysfunction and augmented Wnt signalling associated with spontaneous preterm birth. Front. Cell Dev. Biol. 2022, 10, 987740. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Brockway, H.M.; Kallapur, S.G.; Buhimschi, I.A.; Buhimschi, C.S.; Ackerman, W.E.; Muglia, L.J.; Jones, H.N. Unique transcriptomic landscapes identified in idiopathic spontaneous and infection related preterm births compared to normal term births. PLoS ONE 2019, 14, e0225062. [Google Scholar] [CrossRef] [PubMed]
- Ching, T.; Huang, S.; Garmire, L.X. Power analysis and sample size estimation for RNA-Seq differential expression. RNA 2014, 20, 1684–1696. [Google Scholar] [CrossRef]
- Elizarraras, J.M.; Liao, Y.; Shi, Z.; Zhu, Q.; Pico, A.R.; Zhang, B. WebGestalt 2024: Faster gene set analysis and new support for metabolomics and multi-omics. Nucleic Acids Res. 2024, 52, W415–W421. [Google Scholar] [CrossRef]
- Liao, Y.; Wang, J.; Jaehnig, E.J.; Shi, Z.; Zhang, B. WebGestalt 2019: Gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019, 47, W199–W205. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Kirov, S.; Snoddy, J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005, 33, W741–W748. [Google Scholar] [CrossRef] [PubMed]
- Akram, K.M.; Frost, L.I.; Anumba, D.O. Impaired autophagy with augmented apoptosis in a Th1/Th2-imbalanced placental micromilieu is associated with spontaneous preterm birth. Front. Mol. Biosci. 2022, 9, 897228. [Google Scholar] [CrossRef]
- Montojo, J.; Zuberi, K.; Rodriguez, H.; Kazi, F.; Wright, G.; Donaldson, S.L.; Morris, Q.; Bader, G.D. GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 2010, 26, 2927–2928. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visualization and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Greene, C.M.; Marciniak, S.J.; Teckman, J.; Ferrarotti, I.; Brantly, M.L.; Lomas, D.A.; Stoller, J.K.; McElvaney, N.G. α1-Antitrypsin deficiency. Nat. Rev. Dis. Primers 2016, 2, 16051. [Google Scholar] [CrossRef] [PubMed]
- Sindhu, S.; Kochumon, S.; Shenouda, S.; Wilson, A.; Al-Mulla, F.; Ahmad, R. The Cooperative Induction of CCL4 in Human Monocytic Cells by TNF-α and Palmitate Requires MyD88 and Involves MAPK/NF-κB Signaling Pathways. Int. J. Mol. Sci. 2019, 20, 4658. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Bystry, R.S.; Aluvihare, V.; Welch, K.A.; Kallikourdis, M.; Betz, A.G. B cells and professional APCs recruit regulatory T cells via CCL4. Nat. Immunol. 2001, 2, 1126–1132. [Google Scholar] [CrossRef]
- Sanchez-Moral, L.; Paul, T.; Martori, C.; Font-Díaz, J.; Sanjurjo, L.; Aran, G.; Téllez, É.; Blanco, J.; Carrillo, J.; Ito, M.; et al. Macrophage CD5L is a target for cancer immunotherapy. eBioMedicine 2023, 91, 104555. [Google Scholar] [CrossRef] [PubMed]
- Sanjurjo, L.; Aran, G.; Téllez, É.; Amézaga, N.; Armengol, C.; López, D.; Prats, C.; Sarrias, M.R. CD5L Promotes M2 Macrophage Polarization through Autophagy-Mediated Upregulation of ID3. Front. Immunol. 2018, 9, 480. [Google Scholar] [CrossRef]
- Schoeps, B.; Frädrich, J.; Krüger, A. Cut loose TIMP-1: An emerging cytokine in inflammation. Trends Cell Biol. 2023, 33, 413–426. [Google Scholar] [CrossRef]
- Díaz, P.; Harris, J.; Rosario, F.J.; Powell, T.L.; Jansson, T. Increased placental fatty acid transporter 6 and binding protein 3 expression and fetal liver lipid accumulation in a mouse model of obesity in pregnancy. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 309, R1569–R1577. [Google Scholar] [CrossRef]
- Cathcart, M.C.; O’Byrne, K.J.; Reynolds, J.V.; O’Sullivan, J.; Pidgeon, G.P. COX-derived prostanoid pathways in gastrointestinal cancer development and progression: Novel targets for prevention and intervention. Biochim. Biophys. Acta 2012, 1825, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Olson, D.M. The role of prostaglandins in the initiation of parturition. Best Pract. Res. Clin. Obstet. Gynaecol. 2003, 17, 717–730. [Google Scholar] [CrossRef]
- Wells, J.C. Thermal environment and human birth weight. J. Theor. Biol. 2002, 214, 413–425. [Google Scholar] [CrossRef]
- Basu, R.; Malig, B.; Ostro, B. High ambient temperature and the risk of preterm delivery. Am. J. Epidemiol. 2010, 172, 1108–1117. [Google Scholar] [CrossRef]
- Yackerson, N.; Piura, B.; Sheiner, E. The influence of meteorological factors on the emergence of preterm delivery and preterm premature rupture of membrane. J. Perinatol. 2008, 28, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Jiao, A.; Sun, Y.; Avila, C.; Chiu, V.; Slezak, J.; Sacks, D.A.; Abatzoglou, J.T.; Molitor, J.; Chen, J.C.; Benmarhnia, T.; et al. Analysis of Heat Exposure During Pregnancy and Severe Maternal Morbidity. JAMA Netw. Open 2023, 6, e2332780. [Google Scholar] [CrossRef]
- Villar, J.; Cavoretto, P.I.; Barros, F.C.; Romero, R.; Papageorghiou, A.T.; Kennedy, S.H. Etiologically Based Functional Taxonomy of the Preterm Birth Syndrome. Clin. Perinatol. 2024, 51, 475–495. [Google Scholar] [CrossRef] [PubMed]
- Buthmann, J.; Huang, D.; Casaccia, P.; O’Neill, S.; Nomura, Y.; Liu, J. Prenatal Exposure to a Climate-Related Disaster Results in Changes of the Placental Transcriptome and Infant Temperament. Front. Genet. 2022, 13, 887619. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Park, C.H.; Lee, J.; Lee, N.; Zhang, R.; Huesing, C.; Reijnders, D.; Sones, J.; Münzberg, H.; Redman, L.; et al. Maternal cold exposure induces distinct transcriptome changes in the placenta and fetal brown adipose tissue in mice. BMC Genom. 2021, 22, 500. [Google Scholar] [CrossRef]
- Gómez-Chávez, F.; Correa, D.; Navarrete-Meneses, P.; Cancino-Diaz, J.C.; Cancino-Diaz, M.E.; Rodríguez-Martínez, S. NF-κB and Its Regulators During Pregnancy. Front. Immunol. 2021, 12, 679106. [Google Scholar] [CrossRef] [PubMed]
- Elliott, C.L.; Allport, V.C.; Loudon, J.A.; Wu, G.D.; Bennett, P.R. Nuclear factor-kappa B is essential for up-regulation of interleukin-8 expression in human amnion and cervical epithelial cells. Mol. Hum. Reprod. 2001, 7, 787–790. [Google Scholar] [CrossRef] [PubMed]
- Lindström, T.M.; Bennett, P.R. The role of nuclear factor kappa B in human labour. Reproduction 2005, 130, 569–581. [Google Scholar] [CrossRef]
- Noguchi, T.; Sado, T.; Naruse, K.; Shigetomi, H.; Onogi, A.; Haruta, S.; Kawaguchi, R.; Nagai, A.; Tanase, Y.; Yoshida, S.; et al. Evidence for activation of Toll-like receptor and receptor for advanced glycation end products in preterm birth. Mediat. Inflamm. 2010, 2010, 490406. [Google Scholar] [CrossRef]
- Zarember, K.A.; Godowski, P.J. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J. Immunol. 2002, 168, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, T.M.; Fahey, J.V.; Wright, J.A.; Wira, C.R. Innate immunity in the human female reproductive tract: Antiviral response of uterine epithelial cells to the TLR3 agonist poly(I:C). J. Immunol. 2005, 174, 992–1002. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Nakashima, A.; Hidaka, T.; Okabe, M.; Bac, N.D.; Ina, S.; Yoneda, S.; Shiozaki, A.; Sumi, S.; Tsuneyama, K.; et al. A role for IL-17 in induction of an inflammation at the fetomaternal interface in preterm labour. J. Reprod. Immunol. 2010, 84, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Starodubtseva, N.; Nizyaeva, N.; Baev, O.; Bugrova, A.; Gapaeva, M.; Muminova, K.; Kononikhin, A.; Frankevich, V.; Nikolaev, E.; Sukhikh, G. SERPINA1 Peptides in Urine as A Potential Marker of Preeclampsia Severity. Int. J. Mol. Sci. 2020, 21, 914. [Google Scholar] [CrossRef]
- Tiensuu, H.; Haapalainen, A.M.; Tissarinen, P.; Pasanen, A.; Määttä, T.A.; Huusko, J.M.; Ohlmeier, S.; Bergmann, U.; Ojaniemi, M.; Muglia, L.J.; et al. Human placental proteomics and exon variant studies link AAT/SERPINA1 with spontaneous preterm birth. BMC Med. 2022, 20, 141. [Google Scholar] [CrossRef]
- Tissarinen, P.; Tiensuu, H.; Haapalainen, A.M.; Ronkainen, E.; Laatio, L.; Vääräsmäki, M.; Öhman, H.; Hallman, M.; Rämet, M. Maternal serum alpha-1 antitrypsin levels in spontaneous preterm and term pregnancies. Sci. Rep. 2024, 14, 10819. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, W.; Li, C. Recent Advances in Genetic and Epigenetic Modulation of Animal Exposure to High Temperature. Front. Genet. 2020, 11, 653. [Google Scholar] [CrossRef] [PubMed]
- Tessadori, F.; Duran, K.; Knapp, K.; Fellner, M.; Smithson, S.; Beleza Meireles, A.; Elting, M.W.; Waisfisz, Q.; O’Donnell-Luria, A.; Nowak, C.; et al. Recurrent de novo missense variants across multiple histone H4 genes underlie a neurodevelopmental syndrome. Am. J. Hum. Genet. 2022, 109, 750–758. [Google Scholar] [CrossRef] [PubMed]
- Wood, E.M.; Hornaday, K.K.; Slater, D.M. Prostaglandins in biofluids in pregnancy and labour: A systematic review. PLoS ONE 2021, 16, e0260115. [Google Scholar] [CrossRef] [PubMed]
- Palliser, H.K.; Kelleher, M.A.; Welsh, T.N.; Zakar, T.; Hirst, J.J. Mechanisms leading to increased risk of preterm birth in growth-restricted guinea pig pregnancies. Reprod. Sci. 2014, 21, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.J.; Fortier, M.A.; López Bernal, A. Prostaglandin pathway gene expression in human placenta, amnion and choriodecidua is differentially affected by preterm and term labour and by uterine inflammation. BMC Pregnancy Childbirth 2014, 14, 241. [Google Scholar] [CrossRef] [PubMed]


| Term (n = 13) | p Values | Preterm (n = 12) | p Values | |||
|---|---|---|---|---|---|---|
| Warm (n = 5) | Cold (n = 8) | Warm (n = 5) | Cold (n = 7) | |||
| Monthly mean weather temp at delivery (°C) | 16.06 ± 1.12 | 5.18 ± 0.53 | 0.0008 | 14.92 ± 1.63 | 5.14 ± 0.61 | 0.0025 |
| Monthly max weather temp at delivery (°C) | 20.69 ± 0.90 | 8.2 ± 1.31 | 0.0016 | 20.47 ± 0.73 | 8.37 ± 1.40 | 0.0013 |
| Mean exposure duration prior delivery (days) | 51.6 ± 25.4 | 44.63 ± 39.77 | 0.8057 | 64.2 ± 31.15 | 51.14 ± 38.64 | 0.5025 |
| Gestational age at delivery (weeks) | 39.9 ± 1.1 | 39.7 ± 1.0 | ns | 30.9 ± 4.1 | 33.3 ± 3.6 | ns |
| Maternal age (years) | 32.4 ± 5.5 | 32.0 ± 4.6 | ns | 29.9 ± 8.3 | 26.2 ± 5.2 | ns |
| BMI at delivery (kg/m2) | 25.6 ± 2.9 | 27.2 ± 4.8 | ns | 32.2 ± 18.2 | 26.7 ± 4.9 | ns |
| Placental weight (g) | 603.9 ± 141.3 | 729 ± 122.5 | ns | 512 ± 167.9 | 447.3 ± 102.5 | ns |
| Baby birth weight (kg) | 3.3 ± 0.4 | 3.6 ± 0.2 | ns | 1.7 ± 0.7 | 2.0 ± 0.7 | ns |
| Foetal sex, n (%) | ||||||
| Male | 3 (60%) | 6 (75%) | ns # | 4 (80%) | 4 (57%) | ns # |
| Female | 2 (40%) | 2 (25%) | 1 (20%) | 3 (43%) | ||
| Maternal ethnicity, n (%) | ||||||
| White Caucasian | 5 (100%) | 6 (75%) | - | 4 (80%) | 7 (100%) | - |
| Black | 0 (0%) | 2 (25%) | 0 (0%) | 0 (0%) | ||
| Asian and others | 0 (0%) | 0 (0%) | 1 (20%) | 0 (0%) | ||
| Gene Symbol | Gene Name | log2FC | FDR |
|---|---|---|---|
| Upregulated DEGs | |||
| CCL20 | C-C motif chemokine ligand 20 | 7.868273398 | 0.036752803 |
| CCL3L3 | C-C motif chemokine ligand 3 like 3 | 7.395239339 | 0.028298209 |
| CXCL8 | C-X-C motif chemokine ligand 8 | 6.90348151 | 0.018563659 |
| PI3 | peptidase inhibitor 3 | 6.299703044 | 0.018354829 |
| CCL4L2 | C-C motif chemokine ligand 4 like 2 | 5.735427056 | 0.00883849 |
| AQP9 | aquaporin 9 | 5.664779974 | 0.02266962 |
| G0S2 | G0/G1 switch 2 | 5.165543412 | 0.036752803 |
| IL1B | interleukin 1 beta | 4.988398402 | 0.036752803 |
| IL1RN | interleukin 1 receptor antagonist | 4.898668294 | 0.018563659 |
| VTRNA1-1 | vault RNA 1-1 | 4.361248506 | 0.024808644 |
| BCL2A1 | BCL2-related protein A1 | 4.153647206 | 0.005843209 |
| CCL3 | C-C motif chemokine ligand 3 | 4.150877295 | 0.028298209 |
| OGN | osteoglycin | 4.133529937 | 0.044324798 |
| S100A12 | S100 calcium-binding protein A12 | 3.570600928 | 0.018563659 |
| S100A8 | S100 calcium-binding protein A8 | 3.371015213 | 0.034733036 |
| SERPINA1 | serpin family A member 1 | 3.229324877 | 0.001820221 |
| LAIR1 | leukocyte-associated immunoglobulin like receptor 1 | 3.150781231 | 0.028298209 |
| CCL4 | C-C motif chemokine ligand 4 | 3.013097884 | 0.044324798 |
| PLEK | pleckstrin | 2.964301423 | 0.031052031 |
| RBP7 | retinol-binding protein 7 | 2.927368252 | 0.0378486 |
| CD5L | CD5 molecule like | 2.906771556 | 0.04583665 |
| FABP3 | fatty acid-binding protein 3 | 2.585251055 | 0.045703663 |
| MS4A6E | membrane spanning 4-domains A6E | 2.584792006 | 0.039727763 |
| C1orf162 | chromosome 1 open reading frame 162 | 2.576776932 | 0.043170704 |
| MNDA | myeloid cell nuclear differentiation antigen | 2.431885444 | 0.028298209 |
| FCER1G | Fc epsilon receptor Ig | 2.206550903 | 0.010524081 |
| RPL35 | ribosomal protein L35 | 2.184858984 | 0.004952226 |
| LST1 | leukocyte specific transcript 1 | 2.145185426 | 0.035237296 |
| ALOX5AP | arachidonate 5-lipoxygenase activating protein | 2.103598893 | 0.044324798 |
| H4C3 | H4 clustered histone 3 | 2.094398124 | 0.011879223 |
| FPR1 | formyl peptide receptor 1 | 2.082935671 | 0.036752803 |
| MS4A6A | membrane spanning 4-domains A6A | 1.832523756 | 0.028298209 |
| RPS18P9 | ribosomal protein S18 pseudogene 9 | 1.768847282 | 0.012395102 |
| RPL19P12 | ribosomal protein L19 pseudogene 12 | 1.760969738 | 0.015050412 |
| RPL37A | ribosomal protein L37a | 1.75489983 | 0.028298209 |
| TIMP1 | TIMP metallopeptidase inhibitor 1 | 1.700953541 | 0.04583665 |
| DENND1B | DENN domain containing 1B | 1.627129554 | 0.024808644 |
| SRGN | serglycin | 1.573143772 | 0.044307338 |
| ATP5F1E | ATP synthase F1 subunit epsilon | 1.443927266 | 0.04583665 |
| OST4 | oligosaccharyltransferase complex subunit 4, non-catalytic | 1.434266824 | 0.049762171 |
| Downregulated DEGs | |||
| AOC1 | amine oxidase copper containing 1 | −2.934503787 | 0.012395102 |
| HTRA1 | HtrA serine peptidase 1 | −2.826833202 | 0.030657503 |
| IER5L | immediate early response 5 like | −2.694111567 | 0.028298209 |
| GUCA2A | guanylate cyclase activator 2A | −2.577359202 | 0.044324798 |
| CITED2 | Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 2 | −2.48474137 | 0.044324798 |
| HPGD | 15-hydroxyprostaglandin dehydrogenase | −2.301054733 | 0.028298209 |
| JUP | junction plakoglobin | −1.970586571 | 0.030191088 |
| SPINT2 | serine peptidase inhibitor, Kunitz type 2 | −1.666023242 | 0.0378486 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akram, K.M.; Dodd, E.; Anumba, D.O.C. Seasonal Influences on Human Placental Transcriptomes Associated with Spontaneous Preterm Birth. Cells 2025, 14, 303. https://doi.org/10.3390/cells14040303
Akram KM, Dodd E, Anumba DOC. Seasonal Influences on Human Placental Transcriptomes Associated with Spontaneous Preterm Birth. Cells. 2025; 14(4):303. https://doi.org/10.3390/cells14040303
Chicago/Turabian StyleAkram, Khondoker M., Eleanor Dodd, and Dilly O. C. Anumba. 2025. "Seasonal Influences on Human Placental Transcriptomes Associated with Spontaneous Preterm Birth" Cells 14, no. 4: 303. https://doi.org/10.3390/cells14040303
APA StyleAkram, K. M., Dodd, E., & Anumba, D. O. C. (2025). Seasonal Influences on Human Placental Transcriptomes Associated with Spontaneous Preterm Birth. Cells, 14(4), 303. https://doi.org/10.3390/cells14040303






