Figure 1.
ROCK Expression and Localization in Mouse Testes. (
A) Violin plots indicate the expression levels of ROCK1 and ROCK2 in testicular Leydig cells. Transcriptomic data were obtained from the Male Health Atlas (
http://malehealthatlas.cn/, accessed on 6 March 2024.). SPG: spermatogonia; SPC: spermatocytes; SPT: spermatids/sperms; SC: Sertoli cells; LC: Leydig cells; PTM: peritubular myoid cells; EC: Endothelial cells; SMC: vascular smooth muscle cells; Immune: Immune cells. (
B) Western Blot analysis shows the expression of ROCK1 and ROCK2 in the testes from different developmental stages, with a noted decrease in ROCK1 expression in the testes of aged mice, using GAPDH as a loading control. (
C) Quantification of results of Western Blot analysis of ROCK1 from (
B), normalized to PD7 (
n = 3). Statistical significance was determined by unpaired two-tailed
t-test. *
p < 0.05; **
p < 0.01; ***
p < 0.001. (
D) Quantification of results of Western Blot analysis of ROCK2 from (
B), normalized to PD7 (
n = 3). Statistical significance was determined by unpaired two-tailed
t-test. ns: not significant; *
p < 0.05; **
p < 0.01; ***
p < 0.001. (
E) Immunofluorescence results on paraffin sections demonstrate that ROCK1 and ROCK2 (green) are primarily localized in 3β-HSD-positive (purple) Leydig cells (indicated by red triangles) in the testes of PD21 (top row) and PD56 (bottom row) mice, with nuclei stained by DAPI (blue). The dashed boxed areas are magnified. Scale bars: PD21 (top row): 15 µm; PD56 (bottom row): 40 µm.
Figure 1.
ROCK Expression and Localization in Mouse Testes. (
A) Violin plots indicate the expression levels of ROCK1 and ROCK2 in testicular Leydig cells. Transcriptomic data were obtained from the Male Health Atlas (
http://malehealthatlas.cn/, accessed on 6 March 2024.). SPG: spermatogonia; SPC: spermatocytes; SPT: spermatids/sperms; SC: Sertoli cells; LC: Leydig cells; PTM: peritubular myoid cells; EC: Endothelial cells; SMC: vascular smooth muscle cells; Immune: Immune cells. (
B) Western Blot analysis shows the expression of ROCK1 and ROCK2 in the testes from different developmental stages, with a noted decrease in ROCK1 expression in the testes of aged mice, using GAPDH as a loading control. (
C) Quantification of results of Western Blot analysis of ROCK1 from (
B), normalized to PD7 (
n = 3). Statistical significance was determined by unpaired two-tailed
t-test. *
p < 0.05; **
p < 0.01; ***
p < 0.001. (
D) Quantification of results of Western Blot analysis of ROCK2 from (
B), normalized to PD7 (
n = 3). Statistical significance was determined by unpaired two-tailed
t-test. ns: not significant; *
p < 0.05; **
p < 0.01; ***
p < 0.001. (
E) Immunofluorescence results on paraffin sections demonstrate that ROCK1 and ROCK2 (green) are primarily localized in 3β-HSD-positive (purple) Leydig cells (indicated by red triangles) in the testes of PD21 (top row) and PD56 (bottom row) mice, with nuclei stained by DAPI (blue). The dashed boxed areas are magnified. Scale bars: PD21 (top row): 15 µm; PD56 (bottom row): 40 µm.
![Cells 14 01868 g001 Cells 14 01868 g001]()
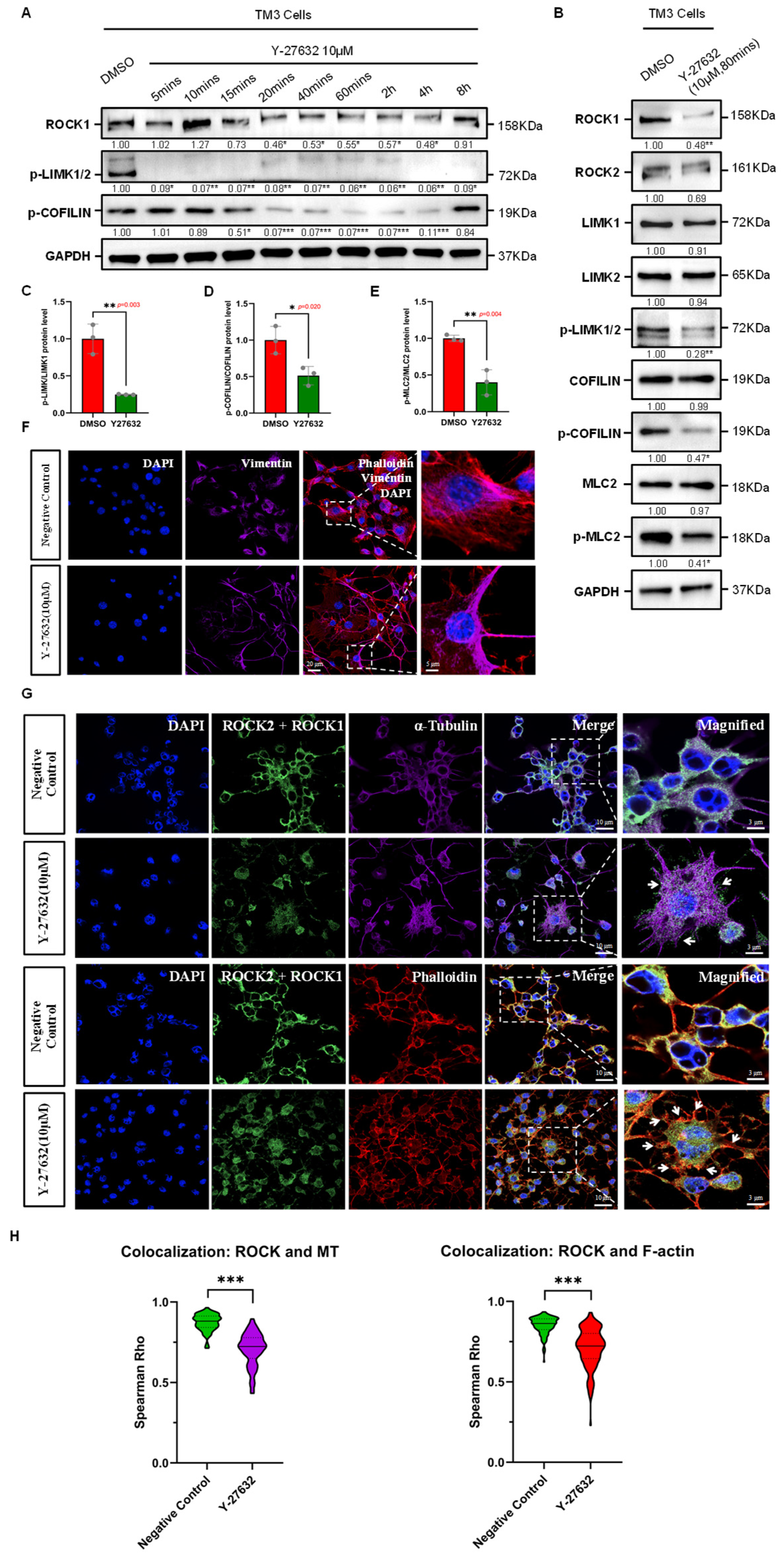
Figure 2.
Effects of ROCK-i on Cytoskeletal Remodeling in Leydig Cells. (A) Effective duration of Y-27632 treatment. Western Blot results show that Y-27632 significantly inhibits ROCK activity in TM3 cells within the effective duration, with a notable decrease in phosphorylation levels of downstream substrates LIMK and COFILIN. The effective duration ranges from 15 min to 4 h, with GAPDH used as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. Statistical significance was determined by unpaired two-tailed t-test. * p < 0.05; ** p < 0.01; *** p < 0.001. (B) The phosphorylation levels of cytoskeletal remodeling regulators LIMK, COFILIN, and MLC2 in TM3 cells are significantly reduced by ROCK-i treatment, with GAPDH used as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. Statistical significance was determined by unpaired two-tailed t-test. * p < 0.05; ** p < 0.01. (C) Quantification of phospho-LIMK/LIMK1 protein ratio from (B), normalized to DMSO group (n = 3). The ratio was significantly reduced in Y-27632 group (p = 0.003). Statistical significance was determined by unpaired two-tailed t-test. ** p < 0.01. (D) Quantification of phospho-COFILIN/COFILIN protein ratio from (B), normalized to DMSO group (n = 3). The ratio was significantly reduced in Y-27632 group (p = 0.020). Statistical significance was determined by unpaired two-tailed t-test. * p < 0.05. (E) Quantification of phospho-MLC2/MLC2 protein ratio from (B), normalized to DMSO group (n = 3). The ratio was significantly reduced in Y-27632 group (p = 0.004). Statistical significance was determined by unpaired two-tailed t-test. ** p < 0.01. (F) Effects of ROCK-i on the intermediate filament cytoskeletal structure in TM3 cells. Red fluorescence indicates Phalloidin-labeled microfilaments; purple fluorescence indicates Vimentin-labeled intermediate filaments; nuclei are stained with DAPI (blue). The dashed box areas are magnified. Scale bars: original areas: 20 µm; magnified areas: 5 µm. (G) Effects of ROCK-i on the microfilament and microtubule cytoskeletal structures in TM3 cells. Green fluorescence indicates ROCK1/2 localization; red fluorescence indicates Phalloidin-labeled microfilaments; purple fluorescence indicates α-Tubulin-labeled microtubules; nuclei are stained with DAPI (blue). The dashed box areas are magnified. Arrows in the top row indicate abnormally aggregated microtubules, while arrows in the bottom row indicate abnormally aggregated microfilaments at cell junctions. Scale bars: original areas: 10 µm; magnified areas: 3 µm. (H) Violin plots showing Spearman correlation coefficients (Spearman Rho) quantifying the colocalization of ROCK1/2 with microtubules and F-actin in TM3 cells. Each group included 50 cells per replicate (n = 3). Y-27632 treatment significantly reduced the colocalization of ROCK1/2 with both microtubules (Negative Control: 0.8724 ± 0.0521; Y-27632: 0.7055 ± 0.1029; p < 0.001, Mann–Whitney U test) and F-actin (Negative Control: 0.8509 ± 0.0536; Y-27632: 0.7110 ± 0.1226; p < 0.001, Mann–Whitney U test). Data are presented as the mean ± SD. *** p < 0.001.
Figure 2.
Effects of ROCK-i on Cytoskeletal Remodeling in Leydig Cells. (A) Effective duration of Y-27632 treatment. Western Blot results show that Y-27632 significantly inhibits ROCK activity in TM3 cells within the effective duration, with a notable decrease in phosphorylation levels of downstream substrates LIMK and COFILIN. The effective duration ranges from 15 min to 4 h, with GAPDH used as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. Statistical significance was determined by unpaired two-tailed t-test. * p < 0.05; ** p < 0.01; *** p < 0.001. (B) The phosphorylation levels of cytoskeletal remodeling regulators LIMK, COFILIN, and MLC2 in TM3 cells are significantly reduced by ROCK-i treatment, with GAPDH used as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. Statistical significance was determined by unpaired two-tailed t-test. * p < 0.05; ** p < 0.01. (C) Quantification of phospho-LIMK/LIMK1 protein ratio from (B), normalized to DMSO group (n = 3). The ratio was significantly reduced in Y-27632 group (p = 0.003). Statistical significance was determined by unpaired two-tailed t-test. ** p < 0.01. (D) Quantification of phospho-COFILIN/COFILIN protein ratio from (B), normalized to DMSO group (n = 3). The ratio was significantly reduced in Y-27632 group (p = 0.020). Statistical significance was determined by unpaired two-tailed t-test. * p < 0.05. (E) Quantification of phospho-MLC2/MLC2 protein ratio from (B), normalized to DMSO group (n = 3). The ratio was significantly reduced in Y-27632 group (p = 0.004). Statistical significance was determined by unpaired two-tailed t-test. ** p < 0.01. (F) Effects of ROCK-i on the intermediate filament cytoskeletal structure in TM3 cells. Red fluorescence indicates Phalloidin-labeled microfilaments; purple fluorescence indicates Vimentin-labeled intermediate filaments; nuclei are stained with DAPI (blue). The dashed box areas are magnified. Scale bars: original areas: 20 µm; magnified areas: 5 µm. (G) Effects of ROCK-i on the microfilament and microtubule cytoskeletal structures in TM3 cells. Green fluorescence indicates ROCK1/2 localization; red fluorescence indicates Phalloidin-labeled microfilaments; purple fluorescence indicates α-Tubulin-labeled microtubules; nuclei are stained with DAPI (blue). The dashed box areas are magnified. Arrows in the top row indicate abnormally aggregated microtubules, while arrows in the bottom row indicate abnormally aggregated microfilaments at cell junctions. Scale bars: original areas: 10 µm; magnified areas: 3 µm. (H) Violin plots showing Spearman correlation coefficients (Spearman Rho) quantifying the colocalization of ROCK1/2 with microtubules and F-actin in TM3 cells. Each group included 50 cells per replicate (n = 3). Y-27632 treatment significantly reduced the colocalization of ROCK1/2 with both microtubules (Negative Control: 0.8724 ± 0.0521; Y-27632: 0.7055 ± 0.1029; p < 0.001, Mann–Whitney U test) and F-actin (Negative Control: 0.8509 ± 0.0536; Y-27632: 0.7110 ± 0.1226; p < 0.001, Mann–Whitney U test). Data are presented as the mean ± SD. *** p < 0.001.
![Cells 14 01868 g002 Cells 14 01868 g002]()
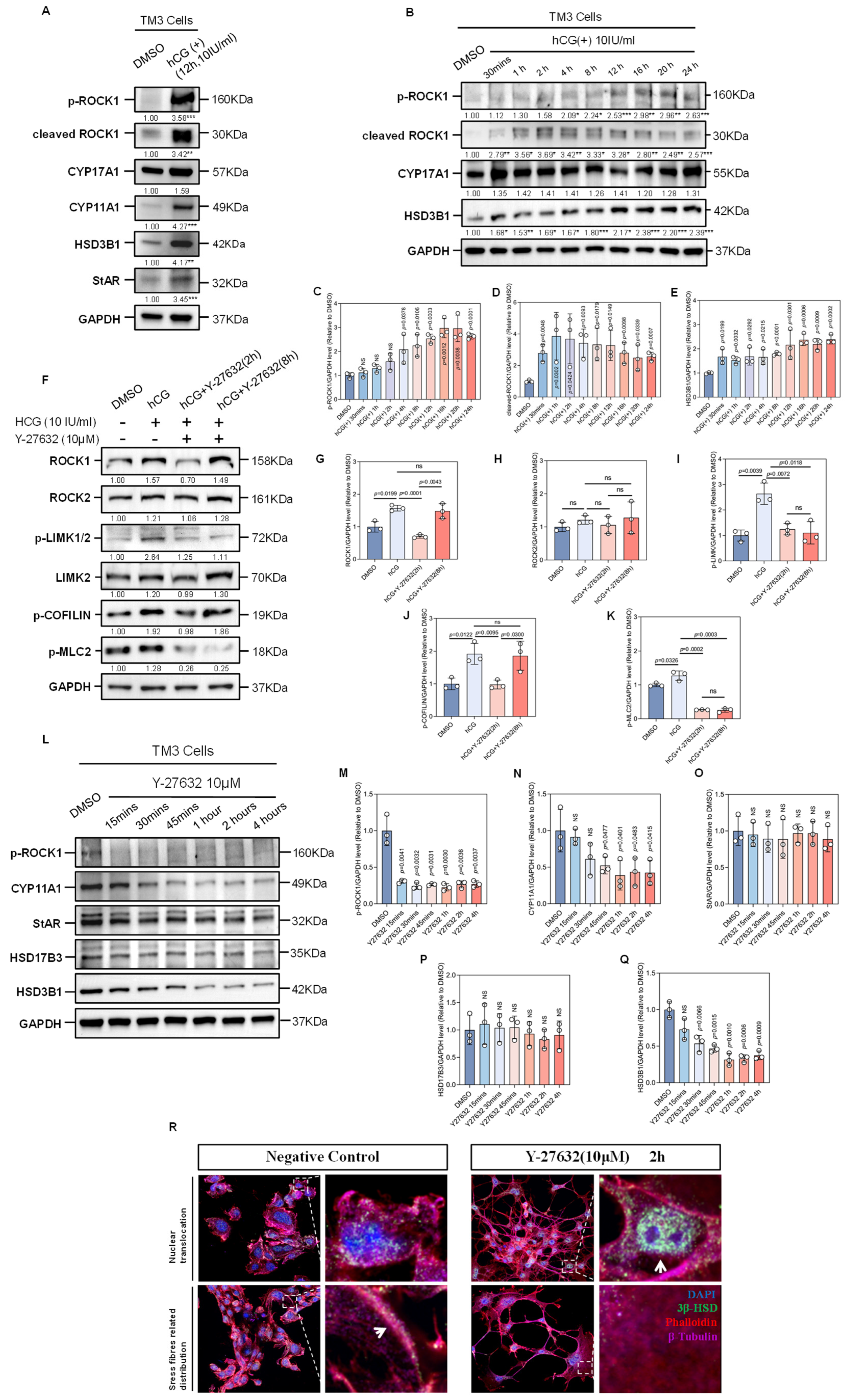
Figure 3.
Effects of hCG and ROCK-i on the Expression of Factors of Microfilament Remodeling and Testosterone Synthesis-Related Enzymes. (A) Western Blot analysis shows the expression of p-ROCK1, cleaved ROCK1, CYP11A1, CYP17A1, HSD3B1, and StAR in TM3 cells before and after hCG treatment, with GAPDH as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. Statistical significance was determined by unpaired two-tailed t-test. ** p < 0.01; *** p < 0.001. (B) Western Blot analysis shows the changes in the expression of p-ROCK1, cleaved ROCK1, CYP17A1, and HSD3B1 over different durations of hCG treatment, with GAPDH as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. Statistical significance was determined by unpaired two-tailed t-test. * p < 0.05; ** p < 0.01; *** p < 0.001. (C) Quantification of results of Western Blot analysis of p-ROCK1 from (B), normalized to DMSO (n = 3). Statistical significance was determined by unpaired two-tailed t-test between hCG group with certain duration and DMSO group. (D) Quantification of results of Western Blot analysis of cleaved ROCK1 from (B), normalized to DMSO (n = 3). Statistical significance was determined by unpaired two-tailed t-test between hCG group with certain duration and DMSO group. (E) Quantification of results of Western Blot analysis of HSD3B1 from (B), normalized to DMSO (n = 3). Statistical significance was determined by unpaired two-tailed t-test between hCG group with certain duration and DMSO group. (F) Western Blot analysis shows the changes in the expression of ROCK1, ROCK2, p-LIMK1/2, LIMK2, p-COFILIN, and p-MLC2 in TM3 cells following hCG stimulation and Y-27632 treatment over time, with Y-27632 treatment applied after 12 h of hCG stimulation, using GAPDH as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. (G) Quantification of results of Western Blot analysis of ROCK1 from (F), normalized to DMSO (n = 3). ns: not significant. (H) Quantification of results of Western Blot analysis of ROCK2 from (F), normalized to DMSO (n = 3). ns: not significant. (I) Quantification of results of Western Blot analysis of p-LIMK from (F), normalized to DMSO (n = 3). ns: not significant. (J) Quantification of results of Western Blot analysis of p-COFILIN from (F), normalized to DMSO (n = 3). ns: not significant. (K) Quantification of results of Western Blot analysis of p-MLC2 from (F), normalized to DMSO (n = 3). ns: not significant. (L) Western Blot analysis shows the changes in the expression of p-ROCK1, CYP11A1, StAR, HSD17B3, and HSD3B1 in TM3 cells over different durations of Y-27632 treatment, with GAPDH as a loading control. (M) Quantification of results of Western Blot analysis of p-ROCK1 from (L), normalized to DMSO (n = 3). (N) Quantification of results of Western Blot analysis of CYP11A1 from (L), normalized to DMSO (n = 3). ns: not significant. (O) Quantification of results of Western Blot analysis of StAR from (L), normalized to DMSO (n = 3). ns: not significant. (P) Quantification of results of Western Blot analysis of HSD17B3 from (L), normalized to DMSO (n = 3). ns: not significant. (Q) Quantification of results of Western Blot analysis of HSD3B1 from (L), normalized to DMSO (n = 3). ns: not significant. (R) Immunofluorescence of cell coverslips shows the changes in the localization pattern of 3β-HSD in TM3 cells. Red fluorescence indicates Phalloidin-labeled microfilaments; purple fluorescence indicates β-Tubulin-labeled microtubules; green fluorescence indicates 3β-HSD; nuclei are stained with DAPI (blue). The dashed box areas are magnified. Arrows in the left panel indicate 3β-HSD distributed along stress fibers, while arrows in the right panel indicate nuclear translocation of 3β-HSD. Scale bars: original areas: 15 µm; magnified areas: 2 µm.
Figure 3.
Effects of hCG and ROCK-i on the Expression of Factors of Microfilament Remodeling and Testosterone Synthesis-Related Enzymes. (A) Western Blot analysis shows the expression of p-ROCK1, cleaved ROCK1, CYP11A1, CYP17A1, HSD3B1, and StAR in TM3 cells before and after hCG treatment, with GAPDH as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. Statistical significance was determined by unpaired two-tailed t-test. ** p < 0.01; *** p < 0.001. (B) Western Blot analysis shows the changes in the expression of p-ROCK1, cleaved ROCK1, CYP17A1, and HSD3B1 over different durations of hCG treatment, with GAPDH as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. Statistical significance was determined by unpaired two-tailed t-test. * p < 0.05; ** p < 0.01; *** p < 0.001. (C) Quantification of results of Western Blot analysis of p-ROCK1 from (B), normalized to DMSO (n = 3). Statistical significance was determined by unpaired two-tailed t-test between hCG group with certain duration and DMSO group. (D) Quantification of results of Western Blot analysis of cleaved ROCK1 from (B), normalized to DMSO (n = 3). Statistical significance was determined by unpaired two-tailed t-test between hCG group with certain duration and DMSO group. (E) Quantification of results of Western Blot analysis of HSD3B1 from (B), normalized to DMSO (n = 3). Statistical significance was determined by unpaired two-tailed t-test between hCG group with certain duration and DMSO group. (F) Western Blot analysis shows the changes in the expression of ROCK1, ROCK2, p-LIMK1/2, LIMK2, p-COFILIN, and p-MLC2 in TM3 cells following hCG stimulation and Y-27632 treatment over time, with Y-27632 treatment applied after 12 h of hCG stimulation, using GAPDH as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. (G) Quantification of results of Western Blot analysis of ROCK1 from (F), normalized to DMSO (n = 3). ns: not significant. (H) Quantification of results of Western Blot analysis of ROCK2 from (F), normalized to DMSO (n = 3). ns: not significant. (I) Quantification of results of Western Blot analysis of p-LIMK from (F), normalized to DMSO (n = 3). ns: not significant. (J) Quantification of results of Western Blot analysis of p-COFILIN from (F), normalized to DMSO (n = 3). ns: not significant. (K) Quantification of results of Western Blot analysis of p-MLC2 from (F), normalized to DMSO (n = 3). ns: not significant. (L) Western Blot analysis shows the changes in the expression of p-ROCK1, CYP11A1, StAR, HSD17B3, and HSD3B1 in TM3 cells over different durations of Y-27632 treatment, with GAPDH as a loading control. (M) Quantification of results of Western Blot analysis of p-ROCK1 from (L), normalized to DMSO (n = 3). (N) Quantification of results of Western Blot analysis of CYP11A1 from (L), normalized to DMSO (n = 3). ns: not significant. (O) Quantification of results of Western Blot analysis of StAR from (L), normalized to DMSO (n = 3). ns: not significant. (P) Quantification of results of Western Blot analysis of HSD17B3 from (L), normalized to DMSO (n = 3). ns: not significant. (Q) Quantification of results of Western Blot analysis of HSD3B1 from (L), normalized to DMSO (n = 3). ns: not significant. (R) Immunofluorescence of cell coverslips shows the changes in the localization pattern of 3β-HSD in TM3 cells. Red fluorescence indicates Phalloidin-labeled microfilaments; purple fluorescence indicates β-Tubulin-labeled microtubules; green fluorescence indicates 3β-HSD; nuclei are stained with DAPI (blue). The dashed box areas are magnified. Arrows in the left panel indicate 3β-HSD distributed along stress fibers, while arrows in the right panel indicate nuclear translocation of 3β-HSD. Scale bars: original areas: 15 µm; magnified areas: 2 µm.
![Cells 14 01868 g003 Cells 14 01868 g003]()
Figure 4.
ROCK-i Reverses hCG-Induced F-actin Remodeling and Testosterone Synthesis in Leydig Cells. (A) Immunofluorescence of cell coverslips shows changes in F-actin stress fiber structures in TM3 cells. Y-27632 treatment was applied following 12 h of hCG stimulation. Red fluorescence indicates Phalloidin-labeled microfilaments; green fluorescence indicates β-ACTIN monomeric actin; nuclei are stained with DAPI (blue). The dashed box areas are magnified. Scale bars: original areas: 15 µm; magnified areas: 5 µm. (B) Western Blot analysis shows changes in F-ACTIN expression in cytoskeletal protein fractions in response to hCG stimulation and Y-27632 treatment over time. Y-27632 treatment was applied following 12 h of hCG stimulation, with β-ACTIN used as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. (C) Quantification of F-ACTIN/β-ACTIN protein ratio from (B), normalized to DMSO (n = 3). ns: not significant. * p < 0.05; ** p < 0.01. (D) Immunofluorescence of cell coverslips shows the distribution changes in StAR (green), Phalloidin (red), and Vimentin (gray) in TM3 cells. Nuclei are stained with DAPI (blue). The dashed box areas are magnified. White arrows indicate StAR localization in microfilament stress fiber regions; red arrows indicate StAR localization with Vimentin-labeled intermediate filaments; white dashed circles indicate StAR dispersed distribution influenced by ROCK-i, unrelated to F-actin; red dashed circles indicate StAR distribution unrelated to Vimentin; white lines indicate the regions selected for colocalization analysis. Scale bars: original areas: 15 µm; magnified areas: 2 µm. (E) Fluorescence intensity profiles of StAR (green), F-actin (red), and Vimentin (gray) in TM3 cells. In the Negative Control group, StAR showed colocalization with F-actin (Rho = 0.686) and Vimentin (Rho = 0.488). After Y-27632 treatment, this colocalization was significantly reduced (Rho = 0.140 and 0.289, respectively), indicating cytoskeletal disruption. (F) ROCK-i inhibits hCG-induced testosterone synthesis and secretion. Testosterone levels were 30.30 ± 1.950 pg/mL in the negative control group, 67.37 ± 5.521 pg/mL in the hCG 12 h group, 48.60 ± 3.820 pg/mL in the hCG 12 h + Y27632 2 h group, and 69.07 ± 6.871 pg/mL in the hCG 12 h + Y27632 4 h group (n = 3, data presented as mean ± SEM). ns: not significant; ** p < 0.01; *** p < 0.001. (G) Distribution of p-ROCK1 (green), StAR (purple), and Phalloidin (red) in TM3 cells. Nuclei are stained with DAPI (blue). Y-27632 treatment was applied following 12 h of hCG stimulation. The dashed box areas are magnified. White arrows indicate StAR localization in microfilament stress fiber regions; white triangles indicate p-ROCK1 localization in microfilament stress fiber regions; white dashed circles indicate abnormal StAR distribution at cell junctions; red dashed circles indicate abnormal p-ROCK1 distribution at cell junctions; white lines indicate the regions selected for colocalization analysis. Scale bars: original areas: 15 µm; magnified areas: 2 µm. (H) Fluorescence intensity profiles of StAR (purple), F-actin (red), and p-ROCK1 (green) in TM3 cells. In the hCG group, F-actin showed colocalization with StAR (Rho = 0.483) and p-ROCK1 (Rho = 0.430). In contrast, colocalization was reduced in the hCG + ROCKi group (Rho = 0.126 and 0.196, respectively), indicating that ROCK inhibition disrupts their spatial association.
Figure 4.
ROCK-i Reverses hCG-Induced F-actin Remodeling and Testosterone Synthesis in Leydig Cells. (A) Immunofluorescence of cell coverslips shows changes in F-actin stress fiber structures in TM3 cells. Y-27632 treatment was applied following 12 h of hCG stimulation. Red fluorescence indicates Phalloidin-labeled microfilaments; green fluorescence indicates β-ACTIN monomeric actin; nuclei are stained with DAPI (blue). The dashed box areas are magnified. Scale bars: original areas: 15 µm; magnified areas: 5 µm. (B) Western Blot analysis shows changes in F-ACTIN expression in cytoskeletal protein fractions in response to hCG stimulation and Y-27632 treatment over time. Y-27632 treatment was applied following 12 h of hCG stimulation, with β-ACTIN used as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group. (C) Quantification of F-ACTIN/β-ACTIN protein ratio from (B), normalized to DMSO (n = 3). ns: not significant. * p < 0.05; ** p < 0.01. (D) Immunofluorescence of cell coverslips shows the distribution changes in StAR (green), Phalloidin (red), and Vimentin (gray) in TM3 cells. Nuclei are stained with DAPI (blue). The dashed box areas are magnified. White arrows indicate StAR localization in microfilament stress fiber regions; red arrows indicate StAR localization with Vimentin-labeled intermediate filaments; white dashed circles indicate StAR dispersed distribution influenced by ROCK-i, unrelated to F-actin; red dashed circles indicate StAR distribution unrelated to Vimentin; white lines indicate the regions selected for colocalization analysis. Scale bars: original areas: 15 µm; magnified areas: 2 µm. (E) Fluorescence intensity profiles of StAR (green), F-actin (red), and Vimentin (gray) in TM3 cells. In the Negative Control group, StAR showed colocalization with F-actin (Rho = 0.686) and Vimentin (Rho = 0.488). After Y-27632 treatment, this colocalization was significantly reduced (Rho = 0.140 and 0.289, respectively), indicating cytoskeletal disruption. (F) ROCK-i inhibits hCG-induced testosterone synthesis and secretion. Testosterone levels were 30.30 ± 1.950 pg/mL in the negative control group, 67.37 ± 5.521 pg/mL in the hCG 12 h group, 48.60 ± 3.820 pg/mL in the hCG 12 h + Y27632 2 h group, and 69.07 ± 6.871 pg/mL in the hCG 12 h + Y27632 4 h group (n = 3, data presented as mean ± SEM). ns: not significant; ** p < 0.01; *** p < 0.001. (G) Distribution of p-ROCK1 (green), StAR (purple), and Phalloidin (red) in TM3 cells. Nuclei are stained with DAPI (blue). Y-27632 treatment was applied following 12 h of hCG stimulation. The dashed box areas are magnified. White arrows indicate StAR localization in microfilament stress fiber regions; white triangles indicate p-ROCK1 localization in microfilament stress fiber regions; white dashed circles indicate abnormal StAR distribution at cell junctions; red dashed circles indicate abnormal p-ROCK1 distribution at cell junctions; white lines indicate the regions selected for colocalization analysis. Scale bars: original areas: 15 µm; magnified areas: 2 µm. (H) Fluorescence intensity profiles of StAR (purple), F-actin (red), and p-ROCK1 (green) in TM3 cells. In the hCG group, F-actin showed colocalization with StAR (Rho = 0.483) and p-ROCK1 (Rho = 0.430). In contrast, colocalization was reduced in the hCG + ROCKi group (Rho = 0.126 and 0.196, respectively), indicating that ROCK inhibition disrupts their spatial association.
![Cells 14 01868 g004 Cells 14 01868 g004]()
Figure 5.
ROCK-i Affects Cholesterol Transport to Mitochondria in Leydig Cells. (A) Changes in the colocalization of cholesterol and mitochondria in TM3 cells. Free cholesterol is labeled with the CholEsteryl BODIPY fluorescent probe (green), mitochondria are labeled with the MitoTracker fluorescent probe (purple), and microfilaments are labeled with Phalloidin (red). Nuclei are stained with DAPI (blue). The dashed box areas are magnified. Red arrows indicate colocalized fluorescent signal points; white circles indicate mitochondrial clustering; red circles indicate cholesterol accumulation; yellow lines indicate the regions selected for colocalization analysis. Scale bars: original areas: 15 µm; magnified areas: 2 µm. (B) Quantification of cholesterol–mitochondria colocalization in TM3 cells. Colocalization between BODIPY-labeled cholesterol and MitoTracker-labeled mitochondria was quantified across six groups: Negative Control, HCG 12 h, HCG + ROCKi (1 h, 2 h, 4 h, 8 h). (C) Microfilament-mediated transport of cholesterol to mitochondria. Free cholesterol is labeled with the CholEsteryl BODIPY fluorescent probe (green), mitochondria are labeled with the MitoTracker fluorescent probe (purple), and microfilaments are labeled with Phalloidin (red). Nuclei are stained with DAPI (blue). The dashed box areas are magnified. White arrows indicate cholesterol localization in the microfilament regions; white circles indicate colocalization of cholesterol and mitochondria; white triangles indicate abnormally aggregated fluorescent signals at cell junctions; white dashed circles indicate the free state of cholesterol and mitochondria; yellow lines indicate the regions selected for colocalization analysis. Scale bars: original areas: 10 µm; magnified areas: 1 µm. (D) Fluorescence intensity profiles of BODIPY (green), F-actin (red), and MitoTracker (purple) in TM3 cells across Negative Control and Y-27632 2h groups. (E) qPCR results showing the mRNA expression levels of cholesterol transport-related genes, including Osbp, Orp4, Orp8, Orp9, Orp10, Abca1, and ApoE, in TM3 cells under three experimental conditions: NC (Negative Control), HCG (hCG-treated 12 h), and ROCKi (Y-27632-treated 2 h following hCG treatment). Gene expression was normalized to Gapdh and presented as mean ± SEM (n = 3), with expression in the NC group set to 1. For Orp8 and Orp9, no significant differences were observed between NC, HCG and ROCKi groups. Osbp, Orp4 and Orp10 were significantly upregulated in HCG group compared to NC group, which expression were downregulated in ROCKi group compared to HCG group. In the HCG group, expression of Abca1 and ApoE was significantly decreased compared to NC, whereas these genes were either unchanged or significantly upregulated in the ROCKi group. Statistical significance was determined by unpaired two-tailed t-test. ns: not significant; * p < 0.05; ** p < 0.01; *** p < 0.001.
Figure 5.
ROCK-i Affects Cholesterol Transport to Mitochondria in Leydig Cells. (A) Changes in the colocalization of cholesterol and mitochondria in TM3 cells. Free cholesterol is labeled with the CholEsteryl BODIPY fluorescent probe (green), mitochondria are labeled with the MitoTracker fluorescent probe (purple), and microfilaments are labeled with Phalloidin (red). Nuclei are stained with DAPI (blue). The dashed box areas are magnified. Red arrows indicate colocalized fluorescent signal points; white circles indicate mitochondrial clustering; red circles indicate cholesterol accumulation; yellow lines indicate the regions selected for colocalization analysis. Scale bars: original areas: 15 µm; magnified areas: 2 µm. (B) Quantification of cholesterol–mitochondria colocalization in TM3 cells. Colocalization between BODIPY-labeled cholesterol and MitoTracker-labeled mitochondria was quantified across six groups: Negative Control, HCG 12 h, HCG + ROCKi (1 h, 2 h, 4 h, 8 h). (C) Microfilament-mediated transport of cholesterol to mitochondria. Free cholesterol is labeled with the CholEsteryl BODIPY fluorescent probe (green), mitochondria are labeled with the MitoTracker fluorescent probe (purple), and microfilaments are labeled with Phalloidin (red). Nuclei are stained with DAPI (blue). The dashed box areas are magnified. White arrows indicate cholesterol localization in the microfilament regions; white circles indicate colocalization of cholesterol and mitochondria; white triangles indicate abnormally aggregated fluorescent signals at cell junctions; white dashed circles indicate the free state of cholesterol and mitochondria; yellow lines indicate the regions selected for colocalization analysis. Scale bars: original areas: 10 µm; magnified areas: 1 µm. (D) Fluorescence intensity profiles of BODIPY (green), F-actin (red), and MitoTracker (purple) in TM3 cells across Negative Control and Y-27632 2h groups. (E) qPCR results showing the mRNA expression levels of cholesterol transport-related genes, including Osbp, Orp4, Orp8, Orp9, Orp10, Abca1, and ApoE, in TM3 cells under three experimental conditions: NC (Negative Control), HCG (hCG-treated 12 h), and ROCKi (Y-27632-treated 2 h following hCG treatment). Gene expression was normalized to Gapdh and presented as mean ± SEM (n = 3), with expression in the NC group set to 1. For Orp8 and Orp9, no significant differences were observed between NC, HCG and ROCKi groups. Osbp, Orp4 and Orp10 were significantly upregulated in HCG group compared to NC group, which expression were downregulated in ROCKi group compared to HCG group. In the HCG group, expression of Abca1 and ApoE was significantly decreased compared to NC, whereas these genes were either unchanged or significantly upregulated in the ROCKi group. Statistical significance was determined by unpaired two-tailed t-test. ns: not significant; * p < 0.05; ** p < 0.01; *** p < 0.001.
![Cells 14 01868 g005 Cells 14 01868 g005]()
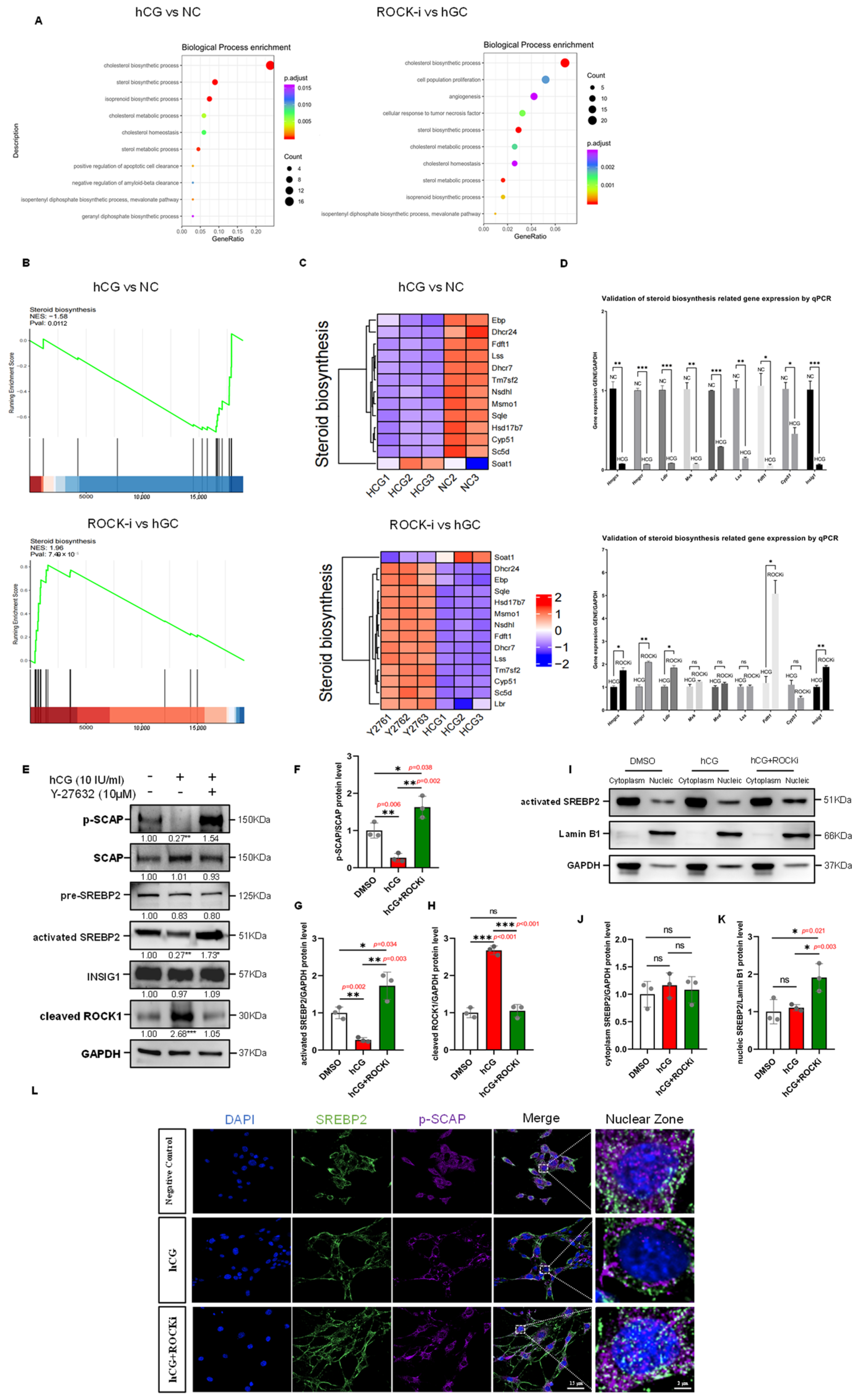
Figure 6.
Steroid Biosynthesis regulated by hCG stimulation and ROCK-i in Leydig Cells through SREBP2-Mediated Transcriptional Control. (A) GO enrichment bubble chart. Setting p-adjust < 0.05 as the threshold and selecting the top 10 pathways, bubble charts were used to visualize these results. The y-axis represents GO terms, the x-axis represents the ratio of genes enriched in each term to the total number of genes, the color indicates the p-adjust value, with redder colors indicating greater significance, and the bubble size represents the number of genes enriched in each term, with larger bubbles indicating more genes. (B) GSEA enrichment analysis results. The y-axis represents the enrichment score (ES), and the Enrichment Score line graph shows the ES value at each position during calculation. The x-axis represents the rank value distribution of all genes after sorting. In the heatmap, red areas correspond to genes highly expressed in the experimental group, and blue areas correspond to genes with low expression in the experimental group. Lines mark the positions of gene set members in the gene rank list. (C) Heatmap analysis of steroid biosynthesis gene set expression levels in the negative control, hCG, and ROCK-i groups. (D) qPCR validation of steroid biosynthesis-related genes (Hmgcs, Hmgcr, Ldlr, Mvk, Mvd, Lss, Fdft1, Cyp51, and Insig1) in TM3 cells under the following conditions: NC (Negative Control), HCG (hCG-treated 12 h), and ROCKi (Y-27632-treated 2 h following hCG treatment). Gene expression was normalized to Gapdh and presented as mean ± SEM (n = 3), with expression in the NC group set to 1. For all these genes, expression levels were significantly downregulated in the hCG group compared to the NC group. In contrast, Hmgcs, Hmgcr, Ldlr, Fdft1, and Insig1 were significantly upregulated in the ROCKi group relative to the hCG group. The expression of Mvk, Mvd, Lss, and Cyp51 showed no significant difference between the ROCKi and hCG groups. Statistical significance was determined by unpaired two-tailed t-test. ns: not significant; * p < 0.05; ** p < 0.01; *** p < 0.001. (E) Western Blot analysis of SREBP2 complex components in the hCG and ROCK-i groups. Y-27632 treatment was applied following 12 h of hCG stimulation, with GAPDH used as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group; * p < 0.05; ** p < 0.01; *** p < 0.001. (F) Quantification of phospho-SCAP/SCAP protein ratio from (E), normalized to DMSO (n = 3). * p < 0.05; ** p < 0.01. (G) Quantification of results of Western Blot analysis of activated-SREBP2 from (E), normalized to DMSO (n = 3). * p < 0.05; ** p < 0.01. (H) Quantification of results of Western Blot analysis of cleaved-ROCK1 from (E), normalized to DMSO (n = 3). ns: not significant; *** p < 0.001. (I) Western Blot analysis of activated SREBP2 in subcellular fractions in the hCG and ROCK-i groups. Y-27632 treatment was applied following 12 h of hCG stimulation. Lamin B1 was used as a nuclear protein loading control, and GAPDH was used as a cytoplasmic protein loading control. (J) Quantification of results of Western Blot analysis of cytoplasm SREBP2 from (I), normalized to DMSO (n = 3). ns: not significant. (K) Quantification of results of Western Blot analysis of nucleic SREBP2 from (I), normalized to DMSO (n = 3). ns: not significant; * p < 0.05. (L) Immunofluorescence of cell coverslips shows changes in nuclear expression of SREBP2 in TM3 cells. Y-27632 treatment was applied following 12 h of hCG stimulation. Green fluorescence indicates activated SREBP2; purple fluorescence indicates p-SCAP; nuclei are stained with DAPI (blue). The dashed box areas are magnified. Scale bars: original areas: 15 µm; magnified areas: 2 µm.
Figure 6.
Steroid Biosynthesis regulated by hCG stimulation and ROCK-i in Leydig Cells through SREBP2-Mediated Transcriptional Control. (A) GO enrichment bubble chart. Setting p-adjust < 0.05 as the threshold and selecting the top 10 pathways, bubble charts were used to visualize these results. The y-axis represents GO terms, the x-axis represents the ratio of genes enriched in each term to the total number of genes, the color indicates the p-adjust value, with redder colors indicating greater significance, and the bubble size represents the number of genes enriched in each term, with larger bubbles indicating more genes. (B) GSEA enrichment analysis results. The y-axis represents the enrichment score (ES), and the Enrichment Score line graph shows the ES value at each position during calculation. The x-axis represents the rank value distribution of all genes after sorting. In the heatmap, red areas correspond to genes highly expressed in the experimental group, and blue areas correspond to genes with low expression in the experimental group. Lines mark the positions of gene set members in the gene rank list. (C) Heatmap analysis of steroid biosynthesis gene set expression levels in the negative control, hCG, and ROCK-i groups. (D) qPCR validation of steroid biosynthesis-related genes (Hmgcs, Hmgcr, Ldlr, Mvk, Mvd, Lss, Fdft1, Cyp51, and Insig1) in TM3 cells under the following conditions: NC (Negative Control), HCG (hCG-treated 12 h), and ROCKi (Y-27632-treated 2 h following hCG treatment). Gene expression was normalized to Gapdh and presented as mean ± SEM (n = 3), with expression in the NC group set to 1. For all these genes, expression levels were significantly downregulated in the hCG group compared to the NC group. In contrast, Hmgcs, Hmgcr, Ldlr, Fdft1, and Insig1 were significantly upregulated in the ROCKi group relative to the hCG group. The expression of Mvk, Mvd, Lss, and Cyp51 showed no significant difference between the ROCKi and hCG groups. Statistical significance was determined by unpaired two-tailed t-test. ns: not significant; * p < 0.05; ** p < 0.01; *** p < 0.001. (E) Western Blot analysis of SREBP2 complex components in the hCG and ROCK-i groups. Y-27632 treatment was applied following 12 h of hCG stimulation, with GAPDH used as a loading control. Data are mean of n = 3. Values were normalized to the DMSO group; * p < 0.05; ** p < 0.01; *** p < 0.001. (F) Quantification of phospho-SCAP/SCAP protein ratio from (E), normalized to DMSO (n = 3). * p < 0.05; ** p < 0.01. (G) Quantification of results of Western Blot analysis of activated-SREBP2 from (E), normalized to DMSO (n = 3). * p < 0.05; ** p < 0.01. (H) Quantification of results of Western Blot analysis of cleaved-ROCK1 from (E), normalized to DMSO (n = 3). ns: not significant; *** p < 0.001. (I) Western Blot analysis of activated SREBP2 in subcellular fractions in the hCG and ROCK-i groups. Y-27632 treatment was applied following 12 h of hCG stimulation. Lamin B1 was used as a nuclear protein loading control, and GAPDH was used as a cytoplasmic protein loading control. (J) Quantification of results of Western Blot analysis of cytoplasm SREBP2 from (I), normalized to DMSO (n = 3). ns: not significant. (K) Quantification of results of Western Blot analysis of nucleic SREBP2 from (I), normalized to DMSO (n = 3). ns: not significant; * p < 0.05. (L) Immunofluorescence of cell coverslips shows changes in nuclear expression of SREBP2 in TM3 cells. Y-27632 treatment was applied following 12 h of hCG stimulation. Green fluorescence indicates activated SREBP2; purple fluorescence indicates p-SCAP; nuclei are stained with DAPI (blue). The dashed box areas are magnified. Scale bars: original areas: 15 µm; magnified areas: 2 µm.
![Cells 14 01868 g006 Cells 14 01868 g006]()
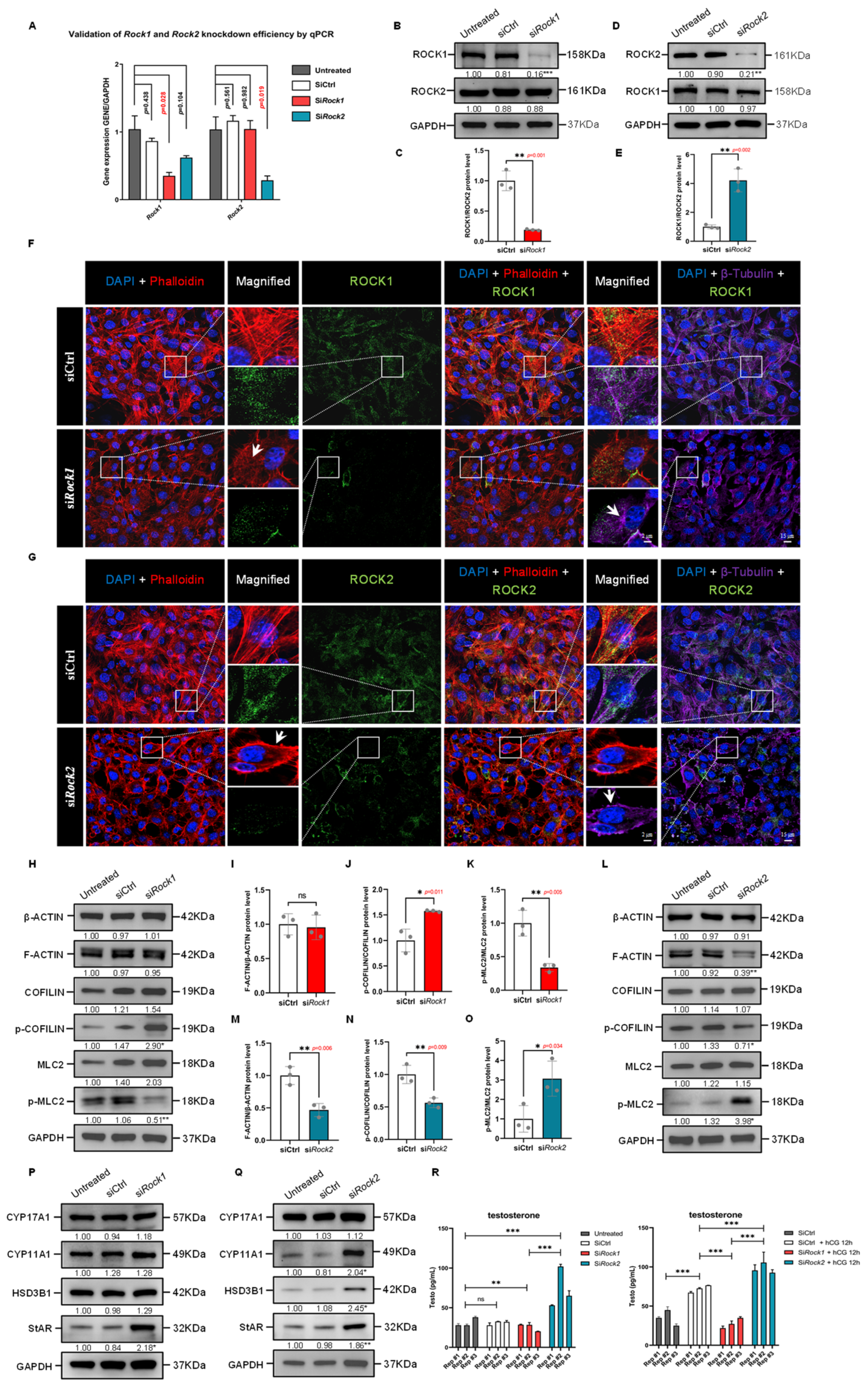
Figure 7.
Differential effects of ROCK1 and ROCK2 knockdown on cytoskeleton organization and steroidogenesis in TM3 cells. (A) qPCR analysis of Rock1 and Rock2 mRNA in TM3 cells treated with siCtrl, siRock1, or siRock2. Expression was normalized to Gapdh and the Untreated group (n = 3). Rock1 expression was significantly reduced by siRock1 (p = 0.028), while Rock2 was significantly reduced by siRock2 (p = 0.019). p values by unpaired t-test. (B) Western Blot analysis of ROCK1 and ROCK2 in TM3 cells transfected with siRock1. GAPDH was used as loading control. ROCK1 levels were significantly reduced in siRock1 cells compared to siCtrl (p < 0.001); ROCK2 levels were unchanged. Data are mean of n = 3. Values were normalized to the Untreated group. *** p < 0.001. (C) Quantification of ROCK1/ROCK2 protein ratio from (B), normalized to siCtrl (n = 3). The ratio was significantly reduced in siRock1 cells (p = 0.001), indicating specific knockdown of ROCK1 without affecting ROCK2. ** p < 0.01. (D) Western Blot analysis of ROCK1 and ROCK2 in TM3 cells transfected with siRock2. GAPDH served as loading control. ROCK2 levels were significantly reduced in siRock2 cells compared to siCtrl (p < 0.01); ROCK1 levels remained unchanged (n = 3). Values were normalized to the Untreated group. ** p < 0.01. (E) Quantification of ROCK1/ROCK2 protein ratio from (D), normalized to siCtrl (n = 3). The ratio was significantly increased in siRock2 cells (p = 0.002), suggesting effective ROCK2 knockdown with no apparent change in ROCK1 levels. ** p < 0.01. (F) Immunofluorescence staining of TM3 cells showing DAPI (blue), F-actin (Phalloidin, red), ROCK1 (green), and β-Tubulin (purple) in siCtrl and siRock1 groups. In siCtrl cells, ROCK1 signal was broadly distributed along both actin filaments and microtubules, with organized cytoskeletal structures. siRock1 cells showed reduced ROCK1 signal, particularly in cytoskeletal regions, accompanied by disrupted actin stress fibers and aberrant microtubule aggregation as arrows indicated. The dashed box areas are magnified. Scale bars: original areas: 20 μm; magnified areas: 5 μm. (G) Immunofluorescence staining of TM3 cells showing DAPI (blue), F-actin (Phalloidin, red), ROCK2 (green), and β-Tubulin (purple) in siCtrl and siRock2 groups. In siCtrl cells, ROCK2 colocalized with organized actin and microtubule networks. siRock2 cells exhibited reduced ROCK2 signal and distinct cytoskeletal alterations, including cortical F-actin formation and clustered microtubules as arrows indicated. The dashed box areas are magnified. Scale bars: original areas: 20 μm; magnified areas: 5 μm. (H) Western Blot analysis of β-ACTIN, F-ACTIN, COFILIN, phospho-COFILIN, MLC2, and phospho-MLC2 in TM3 cells treated with siRock1 (n = 3). Expression was normalized to GAPDH. No significant changes were observed in total β-ACTIN, F-ACTIN, COFILIN, or MLC2 levels. Phospho-COFILIN levels were increased (p < 0.05), while phospho-MLC2 levels were decreased (p < 0.01) in the siRock1 group compared to siCtrl. Values were normalized to the Untreated group. * p < 0.05; ** p < 0.01. (I) Quantification of the F-ACTIN/β-ACTIN protein ratio from (H), normalized to siCtrl (n = 3). No significant difference was observed between siRock1 and siCtrl groups. ns: not significant; *** p < 0.001. (J) Quantification of phospho-COFILIN/COFILIN protein ratio from (H), normalized to siCtrl (n = 3). The ratio was significantly increased in siRock1 cells compared to siCtrl (p = 0.011). * p < 0.05. (K) Quantification of phospho-MLC2/MLC2 protein ratio from (H), normalized to siCtrl (n = 3). The ratio was significantly decreased in siRock1 cells compared to siCtrl (p = 0.005). ** p < 0.01. (L) Western Blot analysis of β-ACTIN, F-ACTIN, COFILIN, phospho-COFILIN, MLC2, and phospho-MLC2 in TM3 cells following ROCK2 knockdown (n = 3). Expression was normalized to GAPDH. No significant changes were observed in β-ACTIN, COFILIN, or MLC2 levels. F-ACTIN expression was reduced in siRock2 cells compared to siCtrl (p < 0.01), accompanied by decreased phospho-COFILIN (p < 0.05) and increased phospho-MLC2 levels (p < 0.05). Values were normalized to the Untreated group. * p < 0.05; ** p < 0.01. (M) Quantification of the F-ACTIN/β-ACTIN protein ratio from (L), normalized to siCtrl (n = 3). The ratio was significantly decreased in siRock2 cells compared to siCtrl (p = 0.006). ** p < 0.01. (N) Quantification of phospho-COFILIN/COFILIN protein ratio from (L), normalized to siCtrl (n = 3). The ratio was significantly decreased in siRock2 cells compared to siCtrl (p = 0.009). ** p < 0.01. (O) Quantification of phospho-MLC2/MLC2 protein ratio from (L), normalized to siCtrl (n = 3). The ratio was significantly increased in siRock2 cells compared to siCtrl (p = 0.034). * p < 0.05. (P) Western Blot analysis of steroidogenic enzymes in TM3 cells following ROCK1 knockdown (n = 3). Expression was normalized to GAPDH. No significant changes were observed in CYP17A1, CYP11A1, or HSD3B1 expression. StAR protein levels were significantly increased in siRock1 cells compared to siCtrl (p < 0.05). Values were normalized to the Untreated group. * p < 0.05. (Q) Western Blot analysis of steroidogenic enzymes in TM3 cells following ROCK2 knockdown. Mean protein levels of CYP11A1, HSD3B1, and StAR were significantly increased in the siRock2 group compared to siCtrl (p < 0.05, p < 0.01 for StAR), while CYP17A1 showed no significant change (n = 3). GAPDH was used as loading control, and values were normalized to the Untreated group. * p < 0.05; ** p < 0.01. (R) ELISA quantification of testosterone levels in TM3 cells following ROCK1 or ROCK2 knockdown, with or without hCG stimulation. Data are presented as mean ± SEM (pg/mL; n = 3). Without hCG treatment, no significant difference was observed between Untreated (31.30 ± 3.35) and siCtrl (30.83 ± 1.52) groups. Testosterone levels were significantly reduced in siRock1 (25.73 ± 2.77, p < 0.01) and increased in siRock2 (73.37 ± 14.74, p < 0.001) compared to Untreated. siRock2 levels were also significantly higher than siRock1 (p < 0.001). With hCG treatment (12 h, 10 IU/mL), testosterone levels increased from 34.93 ± 5.75 (siCtrl) to 72.03 ± 2.72 (siCtrl + hCG, p < 0.001). This hCG-induced increase was significantly suppressed in siRock1 + hCG cells (28.00 ± 3.77, p < 0.001 vs. siCtrl + hCG), and further enhanced in siRock2 + hCG cells (97.93 ± 3.93, p < 0.001 vs. siCtrl + hCG). siRock2 + hCG levels were significantly higher than siRock1 + hCG (p < 0.001). ns: not significant; ** p < 0.01; *** p < 0.001.
Figure 7.
Differential effects of ROCK1 and ROCK2 knockdown on cytoskeleton organization and steroidogenesis in TM3 cells. (A) qPCR analysis of Rock1 and Rock2 mRNA in TM3 cells treated with siCtrl, siRock1, or siRock2. Expression was normalized to Gapdh and the Untreated group (n = 3). Rock1 expression was significantly reduced by siRock1 (p = 0.028), while Rock2 was significantly reduced by siRock2 (p = 0.019). p values by unpaired t-test. (B) Western Blot analysis of ROCK1 and ROCK2 in TM3 cells transfected with siRock1. GAPDH was used as loading control. ROCK1 levels were significantly reduced in siRock1 cells compared to siCtrl (p < 0.001); ROCK2 levels were unchanged. Data are mean of n = 3. Values were normalized to the Untreated group. *** p < 0.001. (C) Quantification of ROCK1/ROCK2 protein ratio from (B), normalized to siCtrl (n = 3). The ratio was significantly reduced in siRock1 cells (p = 0.001), indicating specific knockdown of ROCK1 without affecting ROCK2. ** p < 0.01. (D) Western Blot analysis of ROCK1 and ROCK2 in TM3 cells transfected with siRock2. GAPDH served as loading control. ROCK2 levels were significantly reduced in siRock2 cells compared to siCtrl (p < 0.01); ROCK1 levels remained unchanged (n = 3). Values were normalized to the Untreated group. ** p < 0.01. (E) Quantification of ROCK1/ROCK2 protein ratio from (D), normalized to siCtrl (n = 3). The ratio was significantly increased in siRock2 cells (p = 0.002), suggesting effective ROCK2 knockdown with no apparent change in ROCK1 levels. ** p < 0.01. (F) Immunofluorescence staining of TM3 cells showing DAPI (blue), F-actin (Phalloidin, red), ROCK1 (green), and β-Tubulin (purple) in siCtrl and siRock1 groups. In siCtrl cells, ROCK1 signal was broadly distributed along both actin filaments and microtubules, with organized cytoskeletal structures. siRock1 cells showed reduced ROCK1 signal, particularly in cytoskeletal regions, accompanied by disrupted actin stress fibers and aberrant microtubule aggregation as arrows indicated. The dashed box areas are magnified. Scale bars: original areas: 20 μm; magnified areas: 5 μm. (G) Immunofluorescence staining of TM3 cells showing DAPI (blue), F-actin (Phalloidin, red), ROCK2 (green), and β-Tubulin (purple) in siCtrl and siRock2 groups. In siCtrl cells, ROCK2 colocalized with organized actin and microtubule networks. siRock2 cells exhibited reduced ROCK2 signal and distinct cytoskeletal alterations, including cortical F-actin formation and clustered microtubules as arrows indicated. The dashed box areas are magnified. Scale bars: original areas: 20 μm; magnified areas: 5 μm. (H) Western Blot analysis of β-ACTIN, F-ACTIN, COFILIN, phospho-COFILIN, MLC2, and phospho-MLC2 in TM3 cells treated with siRock1 (n = 3). Expression was normalized to GAPDH. No significant changes were observed in total β-ACTIN, F-ACTIN, COFILIN, or MLC2 levels. Phospho-COFILIN levels were increased (p < 0.05), while phospho-MLC2 levels were decreased (p < 0.01) in the siRock1 group compared to siCtrl. Values were normalized to the Untreated group. * p < 0.05; ** p < 0.01. (I) Quantification of the F-ACTIN/β-ACTIN protein ratio from (H), normalized to siCtrl (n = 3). No significant difference was observed between siRock1 and siCtrl groups. ns: not significant; *** p < 0.001. (J) Quantification of phospho-COFILIN/COFILIN protein ratio from (H), normalized to siCtrl (n = 3). The ratio was significantly increased in siRock1 cells compared to siCtrl (p = 0.011). * p < 0.05. (K) Quantification of phospho-MLC2/MLC2 protein ratio from (H), normalized to siCtrl (n = 3). The ratio was significantly decreased in siRock1 cells compared to siCtrl (p = 0.005). ** p < 0.01. (L) Western Blot analysis of β-ACTIN, F-ACTIN, COFILIN, phospho-COFILIN, MLC2, and phospho-MLC2 in TM3 cells following ROCK2 knockdown (n = 3). Expression was normalized to GAPDH. No significant changes were observed in β-ACTIN, COFILIN, or MLC2 levels. F-ACTIN expression was reduced in siRock2 cells compared to siCtrl (p < 0.01), accompanied by decreased phospho-COFILIN (p < 0.05) and increased phospho-MLC2 levels (p < 0.05). Values were normalized to the Untreated group. * p < 0.05; ** p < 0.01. (M) Quantification of the F-ACTIN/β-ACTIN protein ratio from (L), normalized to siCtrl (n = 3). The ratio was significantly decreased in siRock2 cells compared to siCtrl (p = 0.006). ** p < 0.01. (N) Quantification of phospho-COFILIN/COFILIN protein ratio from (L), normalized to siCtrl (n = 3). The ratio was significantly decreased in siRock2 cells compared to siCtrl (p = 0.009). ** p < 0.01. (O) Quantification of phospho-MLC2/MLC2 protein ratio from (L), normalized to siCtrl (n = 3). The ratio was significantly increased in siRock2 cells compared to siCtrl (p = 0.034). * p < 0.05. (P) Western Blot analysis of steroidogenic enzymes in TM3 cells following ROCK1 knockdown (n = 3). Expression was normalized to GAPDH. No significant changes were observed in CYP17A1, CYP11A1, or HSD3B1 expression. StAR protein levels were significantly increased in siRock1 cells compared to siCtrl (p < 0.05). Values were normalized to the Untreated group. * p < 0.05. (Q) Western Blot analysis of steroidogenic enzymes in TM3 cells following ROCK2 knockdown. Mean protein levels of CYP11A1, HSD3B1, and StAR were significantly increased in the siRock2 group compared to siCtrl (p < 0.05, p < 0.01 for StAR), while CYP17A1 showed no significant change (n = 3). GAPDH was used as loading control, and values were normalized to the Untreated group. * p < 0.05; ** p < 0.01. (R) ELISA quantification of testosterone levels in TM3 cells following ROCK1 or ROCK2 knockdown, with or without hCG stimulation. Data are presented as mean ± SEM (pg/mL; n = 3). Without hCG treatment, no significant difference was observed between Untreated (31.30 ± 3.35) and siCtrl (30.83 ± 1.52) groups. Testosterone levels were significantly reduced in siRock1 (25.73 ± 2.77, p < 0.01) and increased in siRock2 (73.37 ± 14.74, p < 0.001) compared to Untreated. siRock2 levels were also significantly higher than siRock1 (p < 0.001). With hCG treatment (12 h, 10 IU/mL), testosterone levels increased from 34.93 ± 5.75 (siCtrl) to 72.03 ± 2.72 (siCtrl + hCG, p < 0.001). This hCG-induced increase was significantly suppressed in siRock1 + hCG cells (28.00 ± 3.77, p < 0.001 vs. siCtrl + hCG), and further enhanced in siRock2 + hCG cells (97.93 ± 3.93, p < 0.001 vs. siCtrl + hCG). siRock2 + hCG levels were significantly higher than siRock1 + hCG (p < 0.001). ns: not significant; ** p < 0.01; *** p < 0.001.
![Cells 14 01868 g007 Cells 14 01868 g007]()
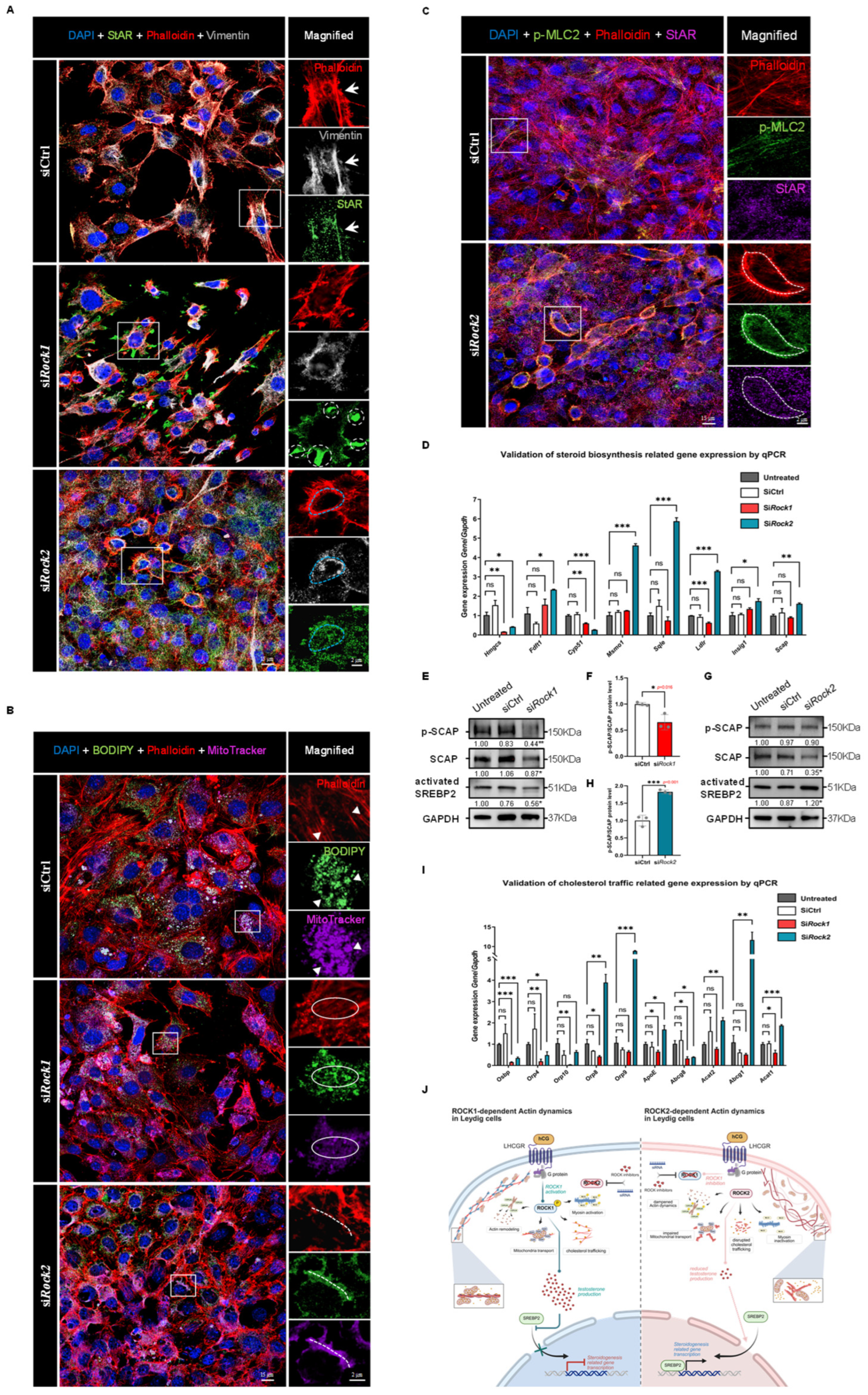
Figure 8.
Knockdown of ROCK1 and ROCK2 reveals opposing effects on cytoskeleton-dependent cholesterol trafficking and SREBP2-driven steroidogenic gene expression. (
A) Immunofluorescence staining of TM3 cells showing DAPI (blue), F-actin (Phalloidin, red), StAR (green), and Vimentin (gray) in siCtrl, si
Rock1, and si
Rock2 groups. In siCtrl cells, StAR signal was distributed along cytoskeletal regions marked by F-actin and Vimentin (arrows). In si
Rock1 cells, StAR appeared mislocalized and clustered near cell–cell junctions, with limited colocalization with cytoskeletal structures (white circles). In contrast, si
Rock2 cells showed enhanced colocalization of StAR with Vimentin and cortical F-actin (blue circles). The box areas are magnified. Scale bars: original areas: 15 μm; magnified areas: 2 μm. (
B) Immunofluorescence staining of TM3 cells showing DAPI (blue), F-actin (Phalloidin, red), cholesterol (BODIPY, green), and mitochondria (MitoTracker, purple) in siCtrl, si
Rock1, and si
Rock2 groups. In siCtrl cells, BODIPY and MitoTracker signals were colocalized along stress fiber regions marked by F-actin (triangles). In si
Rock1 cells, disrupted F-actin structures were associated with loss of BODIPY–MitoTracker colocalization (circles). In si
Rock2 cells, cortical F-actin rings showed enhanced colocalization with both cholesterol and mitochondria (dotted lines). The box areas are magnified. Scale bars: original areas: 15 μm; magnified areas: 2 μm. (
C) Immunofluorescence staining of TM3 cells showing DAPI (blue), F-actin (Phalloidin, red), phospho-MLC2 (green), and StAR (purple) in siCtrl and si
Rock2 groups. In siCtrl cells, p-MLC2 signal was weak and did not appreciably overlap with StAR staining. In si
Rock2 cells, p-MLC2 was enriched in cortical F-actin regions and colocalized with StAR signals (circles), suggesting spatial coupling between activated MLC2 and mitochondrial cholesterol transport. The box areas are magnified. Scale bars: original areas: 15 μm; magnified areas: 2 μm. (
D) qPCR analysis of cholesterol biosynthesis-related genes in TM3 cells following ROCK1 or ROCK2 knockdown. Gene expression was normalized to
Gapdh and the Untreated group (set as 1;
n = 3). No significant differences were observed between Untreated and siCtrl groups.
Hmgcs and
Cyp51 expression was significantly decreased in both si
Rock1 and si
Rock2 groups.
Fdft1,
Msmo1,
Sqle,
Ldlr,
Insig1, and
Scap were all significantly upregulated in si
Rock2 cells, while their expression remained unchanged or decreased in si
Rock1 cells. These results suggest distinct effects of ROCK1 and ROCK2 knockdown on transcriptional regulation of cholesterol biosynthesis pathways. ns: not significant; *
p < 0.05; **
p < 0.01; ***
p < 0.001. (
E) Western Blot analysis of SCAP, phospho-SCAP (p-SCAP), and activated SREBP2 in TM3 cells following ROCK1 knockdown (
n = 3). GAPDH served as loading control, and signal intensities were normalized to the Untreated group (set as 1). SCAP, p-SCAP, and activated SREBP2 levels were significantly decreased in si
Rock1 cells compared to siCtrl (
p < 0.05,
p < 0.01, and
p < 0.05, respectively). *
p < 0.05; **
p < 0.01. (
F) Quantification of phospho-SCAP/SCAP protein ratio from (
E), normalized to siCtrl (
n = 3). The ratio was significantly decreased in si
Rock1 cells compared to siCtrl (
p = 0.016). *
p < 0.05. (
G) Western Blot analysis of SCAP, phospho-SCAP (p-SCAP), and activated SREBP2 in TM3 cells following ROCK2 knockdown (
n = 3). GAPDH was used as loading control, and signal intensities were normalized to the Untreated group (set as 1). While SCAP levels were slightly decreased and p-SCAP levels remained unchanged, activated SREBP2 was significantly increased in si
Rock2 cells compared to siCtrl (
p < 0.05). *
p < 0.05. (
H) Quantification of phospho-SCAP/SCAP protein ratio from (
G), normalized to siCtrl (
n = 3). The ratio was significantly increased in si
Rock2 cells compared to siCtrl (
p < 0.001). ***
p < 0.001. (
I) qPCR analysis was performed to assess the mRNA levels of genes involved in cholesterol transport, including
Osbp,
Orp4,
Orp10,
Orp8,
Orp9,
ApoE,
Abcg8,
Acat2,
Abcg1, and
Acat1, across four groups: Untreated, SiCtrl, Si
Rock1, and Si
Rock2. Gene expression was normalized to
Gapdh and presented as mean ± SEM (
n = 3), with expression in the Untreated group set to 1. No significant differences were observed between Untreated and SiCtrl groups.
Osbp and
Orp4 were significantly downregulated in both Si
Rock1 and Si
Rock2 groups. In the Si
Rock1 group, expression of
Orp10,
Orp8,
ApoE, and
Acat1 was significantly decreased compared to Untreated, whereas these genes were either unchanged or significantly upregulated in the Si
Rock2 group. Notably,
Orp8,
Orp9,
ApoE,
Acat2,
Abcg1, and
Acat1 were significantly upregulated in the Si
Rock2 group. These changes suggest distinct regulatory roles of ROCK1 and ROCK2 in cholesterol mobilization. Statistical significance was determined by unpaired two-tailed
t-test. ns: not significant; *
p < 0.05; **
p < 0.01; ***
p < 0.001. (
J) Proposed model illustrating the distinct roles of ROCK1 and ROCK2 in regulating testosterone biosynthesis via cytoskeletal remodeling. ROCK1 promotes cytoskeletal support for cholesterol trafficking and enhances SCAP phosphorylation and SREBP2 activation, thereby facilitating steroidogenic gene expression. However, excessive testosterone production triggered by ROCK2 knockdown feeds back to suppress SCAP-SREBP2 signaling. In contrast, ROCK2 normally restrains actomyosin dynamics and SCAP-SREBP2 activation; its knockdown relieves this suppression, boosting both cholesterol transport and steroidogenic gene expression. These findings highlight a dual regulatory mechanism whereby ROCK1 and ROCK2 exert opposing influences on testosterone synthesis and its feedback control of cholesterol biosynthesis. Created in BioRender. Xu, K. (2025)
https://BioRender.com/mwrghma (accessed on 30 July 2025).
Figure 8.
Knockdown of ROCK1 and ROCK2 reveals opposing effects on cytoskeleton-dependent cholesterol trafficking and SREBP2-driven steroidogenic gene expression. (
A) Immunofluorescence staining of TM3 cells showing DAPI (blue), F-actin (Phalloidin, red), StAR (green), and Vimentin (gray) in siCtrl, si
Rock1, and si
Rock2 groups. In siCtrl cells, StAR signal was distributed along cytoskeletal regions marked by F-actin and Vimentin (arrows). In si
Rock1 cells, StAR appeared mislocalized and clustered near cell–cell junctions, with limited colocalization with cytoskeletal structures (white circles). In contrast, si
Rock2 cells showed enhanced colocalization of StAR with Vimentin and cortical F-actin (blue circles). The box areas are magnified. Scale bars: original areas: 15 μm; magnified areas: 2 μm. (
B) Immunofluorescence staining of TM3 cells showing DAPI (blue), F-actin (Phalloidin, red), cholesterol (BODIPY, green), and mitochondria (MitoTracker, purple) in siCtrl, si
Rock1, and si
Rock2 groups. In siCtrl cells, BODIPY and MitoTracker signals were colocalized along stress fiber regions marked by F-actin (triangles). In si
Rock1 cells, disrupted F-actin structures were associated with loss of BODIPY–MitoTracker colocalization (circles). In si
Rock2 cells, cortical F-actin rings showed enhanced colocalization with both cholesterol and mitochondria (dotted lines). The box areas are magnified. Scale bars: original areas: 15 μm; magnified areas: 2 μm. (
C) Immunofluorescence staining of TM3 cells showing DAPI (blue), F-actin (Phalloidin, red), phospho-MLC2 (green), and StAR (purple) in siCtrl and si
Rock2 groups. In siCtrl cells, p-MLC2 signal was weak and did not appreciably overlap with StAR staining. In si
Rock2 cells, p-MLC2 was enriched in cortical F-actin regions and colocalized with StAR signals (circles), suggesting spatial coupling between activated MLC2 and mitochondrial cholesterol transport. The box areas are magnified. Scale bars: original areas: 15 μm; magnified areas: 2 μm. (
D) qPCR analysis of cholesterol biosynthesis-related genes in TM3 cells following ROCK1 or ROCK2 knockdown. Gene expression was normalized to
Gapdh and the Untreated group (set as 1;
n = 3). No significant differences were observed between Untreated and siCtrl groups.
Hmgcs and
Cyp51 expression was significantly decreased in both si
Rock1 and si
Rock2 groups.
Fdft1,
Msmo1,
Sqle,
Ldlr,
Insig1, and
Scap were all significantly upregulated in si
Rock2 cells, while their expression remained unchanged or decreased in si
Rock1 cells. These results suggest distinct effects of ROCK1 and ROCK2 knockdown on transcriptional regulation of cholesterol biosynthesis pathways. ns: not significant; *
p < 0.05; **
p < 0.01; ***
p < 0.001. (
E) Western Blot analysis of SCAP, phospho-SCAP (p-SCAP), and activated SREBP2 in TM3 cells following ROCK1 knockdown (
n = 3). GAPDH served as loading control, and signal intensities were normalized to the Untreated group (set as 1). SCAP, p-SCAP, and activated SREBP2 levels were significantly decreased in si
Rock1 cells compared to siCtrl (
p < 0.05,
p < 0.01, and
p < 0.05, respectively). *
p < 0.05; **
p < 0.01. (
F) Quantification of phospho-SCAP/SCAP protein ratio from (
E), normalized to siCtrl (
n = 3). The ratio was significantly decreased in si
Rock1 cells compared to siCtrl (
p = 0.016). *
p < 0.05. (
G) Western Blot analysis of SCAP, phospho-SCAP (p-SCAP), and activated SREBP2 in TM3 cells following ROCK2 knockdown (
n = 3). GAPDH was used as loading control, and signal intensities were normalized to the Untreated group (set as 1). While SCAP levels were slightly decreased and p-SCAP levels remained unchanged, activated SREBP2 was significantly increased in si
Rock2 cells compared to siCtrl (
p < 0.05). *
p < 0.05. (
H) Quantification of phospho-SCAP/SCAP protein ratio from (
G), normalized to siCtrl (
n = 3). The ratio was significantly increased in si
Rock2 cells compared to siCtrl (
p < 0.001). ***
p < 0.001. (
I) qPCR analysis was performed to assess the mRNA levels of genes involved in cholesterol transport, including
Osbp,
Orp4,
Orp10,
Orp8,
Orp9,
ApoE,
Abcg8,
Acat2,
Abcg1, and
Acat1, across four groups: Untreated, SiCtrl, Si
Rock1, and Si
Rock2. Gene expression was normalized to
Gapdh and presented as mean ± SEM (
n = 3), with expression in the Untreated group set to 1. No significant differences were observed between Untreated and SiCtrl groups.
Osbp and
Orp4 were significantly downregulated in both Si
Rock1 and Si
Rock2 groups. In the Si
Rock1 group, expression of
Orp10,
Orp8,
ApoE, and
Acat1 was significantly decreased compared to Untreated, whereas these genes were either unchanged or significantly upregulated in the Si
Rock2 group. Notably,
Orp8,
Orp9,
ApoE,
Acat2,
Abcg1, and
Acat1 were significantly upregulated in the Si
Rock2 group. These changes suggest distinct regulatory roles of ROCK1 and ROCK2 in cholesterol mobilization. Statistical significance was determined by unpaired two-tailed
t-test. ns: not significant; *
p < 0.05; **
p < 0.01; ***
p < 0.001. (
J) Proposed model illustrating the distinct roles of ROCK1 and ROCK2 in regulating testosterone biosynthesis via cytoskeletal remodeling. ROCK1 promotes cytoskeletal support for cholesterol trafficking and enhances SCAP phosphorylation and SREBP2 activation, thereby facilitating steroidogenic gene expression. However, excessive testosterone production triggered by ROCK2 knockdown feeds back to suppress SCAP-SREBP2 signaling. In contrast, ROCK2 normally restrains actomyosin dynamics and SCAP-SREBP2 activation; its knockdown relieves this suppression, boosting both cholesterol transport and steroidogenic gene expression. These findings highlight a dual regulatory mechanism whereby ROCK1 and ROCK2 exert opposing influences on testosterone synthesis and its feedback control of cholesterol biosynthesis. Created in BioRender. Xu, K. (2025)
https://BioRender.com/mwrghma (accessed on 30 July 2025).
![Cells 14 01868 g008 Cells 14 01868 g008]()