Cellular and Molecular Roles of Human Odorant-Binding Proteins and Related Lipocalins in Olfaction and Neuroinflammation
Highlights
- Human olfactory mucus proteins, including hOBPs and lipocalins, contribute to odorant transport and mucosal defense, highlighting their broader physiological roles beyond classical chemosensory function.
- The soluble carrier protein repertoire of human olfactory mucus is not dominated by the classical OBPs (OBP2A/2B), which are inconsistently detected, but rather by OBP-like lipocalins (LCN1, LCN2, LCN15, ApoD) and BPI-fold proteins. Together, these proteins coordinate odorant solubilization, antimicrobial defense, redox balance, and ECM remodeling.
- Alterations in these proteins and ECM components are linked to age-related and idiopathic smell loss, potentially involving olfactory signal transduction deficits, and have also been implicated in chronic rhinosinusitis and neurodegenerative disorders.
- Emerging evidence highlights the pivotal roles of human olfactory mucus proteins in maintaining sensory signaling, neuronal integrity, and mucosal defense, underscoring the importance of mucus proteomics for understanding olfactory dysfunction and its links to inflammation and neurodegeneration.
- Odorant-binding and carrier proteins functionally interact with ECM components, forming an OBP-like protein–ECM network that sustains olfactory signal transduction and mucosal homeostasis in the human olfactory system.
Abstract
1. Introduction
2. Structural and Functional Diversity of Odorant-Binding Proteins
2.1. Discovery and Evolutionary Divergence of OBPs
2.2. Structural Features of OBPs Across Species
2.3. Mechanistic Comparison of Odorant Binding
3. Proteomic Landscape of the Human Olfactory Cleft
3.1. Odorant-Binding Proteins
3.2. OBP-like Lipocalins
3.3. BPI-Fold/PLUNC Family
3.4. Proteins Supporting Stress Resilience and Barrier Integrity
4. Extracellular Matrix (ECM)–Protein Networks in the Human Olfactory Cleft
4.1. Roles of Soluble and Structural ECM Components in Olfactory Sensation
4.2. ECM Remodeling Circuits
4.3. Direct Interactions Between Carrier Proteins and ECM
5. Pathological Alterations in Olfactory Cleft Proteins
5.1. Smell Loss Disorders (Idiopathic and Age-Related)
5.2. Nasal Inflammatory Diseases
5.3. Infection and Environmental Stress Defense
6. Controversies and Knowledge Gaps
6.1. Controversy over the Essentiality of OBPs in Humans
6.2. Crosstalk Between the Olfactory Mucus Proteome and Neurodegeneration
6.3. Conclusion and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| OBP | Odorant-binding protein |
| OBP2A/2B | Human odorant-binding proteins 2A/2B |
| LCN | Lipocalin |
| LCN1 | Lipocalin-1 |
| LCN2 | Lipocalin-2 (NGAL) |
| LCN15 | Lipocalin-15 |
| ApoD | Apolipoprotein D |
| ApoA1/A2/A4 | Apolipoproteins A-I/A-II/A-IV |
| ApoE | Apolipoprotein E |
| CLU (ApoJ) | Clusterin (Apolipoprotein J) |
| BPI | Bactericidal/permeability-increasing protein |
| LBP | Lipopolysaccharide-binding protein |
| PLUNC | Palate, lung, and nasal epithelium clone protein family |
| BPIFA1 (SPLUNC1) | BPI fold-containing family A member 1 |
| BPIFB2 (LPLUNC2) | BPI fold-containing family B member 2 |
| BPIFB3/BPIFB4 | BPI fold-containing family B members 3/4 |
| ECM | Extracellular matrix |
| OSN | Olfactory sensory neuron |
| OR | Olfactory receptor |
| GPCR | G-protein–coupled receptor |
| EOG | Electro-olfactogram |
| MMP-9 | Matrix metalloproteinase-9 |
| MUPs | Major urinary proteins |
| LOXL2 | Lysyl oxidase-like 2 |
| FAK | Focal adhesion kinase |
| AKT | Protein kinase B |
| GSK3β | Glycogen synthase kinase-3 beta |
| EMT | Epithelial–mesenchymal transition |
| UGT2A1/2 | UDP-glucuronosyltransferases 2A1/2 |
| ALDH | Aldehyde dehydrogenase |
| AKR | Aldo-keto reductase |
| GSTP1 | Glutathione S-transferase P1 |
| CES | Carboxylesterase |
| CYP | Cytochrome P450 |
| HSP | Heat shock protein |
| CRS | Chronic rhinosinusitis |
| CRSwNP | Chronic rhinosinusitis with nasal polyps |
| AR | Allergic rhinitis |
| ENaC | Epithelial sodium channel |
| cVA | cis-Vaccenyl acetate |
| PBP1 | Pheromone-binding protein 1 |
| SNP | Single-nucleotide polymorphism |
| OC | Olfactory cleft |
References
- Heydel, J.M.; Coelho, A.; Thiebaud, N.; Legendre, A.; Le Bon, A.M.; Faure, P.; Neiers, F.; Artur, Y.; Golebiowski, J.; Briand, L. Odorant-binding proteins and xenobiotic metabolizing enzymes: Implications in olfactory perireceptor events. Anat. Rec. 2013, 296, 1333–1345. [Google Scholar] [CrossRef]
- Pelosi, P.; Knoll, W. Odorant-binding proteins of mammals. Biol. Rev. Camb. Philos. Soc. 2022, 97, 20–44. [Google Scholar] [CrossRef] [PubMed]
- Getchell, T.V.; Margolis, F.L.; Getchell, M.L. Perireceptor and receptor events in vertebrate olfaction. Prog. Neurobiol. 1984, 23, 317–345. [Google Scholar] [CrossRef]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef]
- Bignetti, E.; Tirindelli, R.; Rossi, G.L.; Bolognesi, M.; Coda, A.; Gatti, G. Crystallization of an odorant-binding protein from cow nasal mucosa. J. Mol. Biol. 1985, 186, 211–212. [Google Scholar] [CrossRef]
- Pevsner, J.; Sklar, P.B.; Snyder, S.H. Odorant-binding protein: Localization to nasal glands and secretions. Proc. Natl. Acad. Sci. USA 1986, 83, 4942–4946. [Google Scholar] [CrossRef] [PubMed]
- Charkoftaki, G.; Wang, Y.; McAndrews, M.; Bruford, E.A.; Thompson, D.C.; Vasiliou, V.; Nebert, D.W. Update on the human and mouse lipocalin (LCN) gene family, including evidence the mouse Mup cluster is result of an “evolutionary bloom”. Hum. Genom. 2019, 13, 11. [Google Scholar] [CrossRef]
- Graham, L.A.; Davies, P.L. The odorant-binding proteins of Drosophila melanogaster: Annotation and characterization of a divergent gene family. Gene 2002, 292, 43–55. [Google Scholar] [CrossRef]
- Leal, W.S.; Nikonova, L.; Peng, G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999, 464, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Briand, L.; Eloit, C.; Nespoulous, C.; Bezirard, V.; Huet, J.C.; Henry, C.; Blon, F.; Trotier, D.; Pernollet, J.C. Evidence of an odorant-binding protein in the human olfactory mucus: Location, structural characterization, and odorant-binding properties. Biochemistry 2002, 41, 7241–7252. [Google Scholar] [CrossRef]
- Pes, D.; Pelosi, P. Odorant-binding proteins of the mouse. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1995, 112, 471–479. [Google Scholar] [CrossRef]
- Shukla, S.; Nakano-Baker, O.; Godin, D.; MacKenzie, D.; Sarikaya, M. iOBPdb A Database for Experimentally Determined Functional Characterization of Insect Odorant Binding Proteins. Sci. Data 2023, 10, 295. [Google Scholar] [CrossRef]
- Tegoni, M.; Pelosi, P.; Vincent, F.; Spinelli, S.; Campanacci, V.; Grolli, S.; Ramoni, R.; Cambillau, C. Mammalian odorant binding proteins. Biochim. Biophys. Acta 2000, 1482, 229–240. [Google Scholar] [CrossRef]
- Yang, R.; Zhou, J.; Hao, J.; Zhang, T.; Jiang, Y.; Liu, W.; Wang, Y. Olfactory binding proteins: A review across the Insecta. Front. Zool. 2025, 22, 29. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.S.; Smith, D.P. Recent Insights into Insect Olfactory Receptors and Odorant-Binding Proteins. Insects 2022, 13, 926. [Google Scholar] [CrossRef]
- Mombaerts, P.; Wang, F.; Dulac, C.; Chao, S.K.; Nemes, A.; Mendelsohn, M.; Edmondson, J.; Axel, R. Visualizing an olfactory sensory map. Cell 1996, 87, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Acton, S.; Rigotti, A.; Landschulz, K.T.; Xu, S.; Hobbs, H.H.; Krieger, M. Identification of scavenger receptor SR-BI as a high density lipoprotein receptor. Science 1996, 271, 518–520. [Google Scholar] [CrossRef] [PubMed]
- Ha, T.S.; Smith, D.P. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J. Neurosci. 2006, 26, 8727–8733. [Google Scholar] [CrossRef]
- Kaur, A.W.; Ackels, T.; Kuo, T.H.; Cichy, A.; Dey, S.; Hays, C.; Kateri, M.; Logan, D.W.; Marton, T.F.; Spehr, M.; et al. Murine pheromone proteins constitute a context-dependent combinatorial code governing multiple social behaviors. Cell 2014, 157, 676–688. [Google Scholar] [CrossRef]
- Sandler, B.H.; Nikonova, L.; Leal, W.S.; Clardy, J. Sexual attraction in the silkworm moth: Structure of the pheromone-binding-protein-bombykol complex. Chem. Biol. 2000, 7, 143–151. [Google Scholar] [CrossRef]
- Althumiri, N.A.; Basyouni, M.H.; AlMousa, N.; AlJuwaysim, M.F.; Almubark, R.A.; BinDhim, N.F.; Alkhamaali, Z.; Alqahtani, S.A. Obesity in Saudi Arabia in 2020: Prevalence, Distribution, and Its Current Association with Various Health Conditions. Healthcare 2021, 9, 311. [Google Scholar] [CrossRef]
- Hekmat-Scafe, D.S.; Scafe, C.R.; McKinney, A.J.; Tanouye, M.A. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002, 12, 1357–1369. [Google Scholar] [CrossRef]
- Xu, P.X.; Zwiebel, L.J.; Smith, D.P. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol. 2003, 12, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.J.; Huang, W.; Zhang, G.A.; Pickett, J.A.; Field, L.M. “Plus-C” odorant-binding protein genes in two Drosophila species and the malaria mosquito Anopheles gambiae. Gene 2004, 327, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Lautenschlager, C.; Leal, W.S.; Clardy, J. Coil-to-helix transition and ligand release of Bombyx mori pheromone-binding protein. Biochem. Biophys. Res. Commun. 2005, 335, 1044–1050. [Google Scholar] [CrossRef]
- Horst, R.; Damberger, F.; Luginbuhl, P.; Guntert, P.; Peng, G.; Nikonova, L.; Leal, W.S.; Wuthrich, K. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc. Natl. Acad. Sci. USA 2001, 98, 14374–14379. [Google Scholar] [CrossRef] [PubMed]
- Smartt, C.T.; Erickson, J.S. Expression of a novel member of the odorant-binding protein gene family in Culex nigripalpus (Diptera: Culicidae). J. Med. Entomol. 2009, 46, 1376–1381. [Google Scholar] [CrossRef]
- Pilka, E.S.; Werner, J.M.; Schwarz-Linek, U.; Pickford, A.R.; Meenan, N.A.; Campbell, I.D.; Potts, J.R. Structural insight into binding of Staphylococcus aureus to human fibronectin. FEBS Lett. 2006, 580, 273–277. [Google Scholar] [CrossRef]
- Rich, D.S. Integrating Oryx into a home care organization’s performance-improvement program. Am. J. Health Syst. Pharm. 2000, 57, 73–75. [Google Scholar] [CrossRef]
- Zubkov, S.; Gronenborn, A.M.; Byeon, I.J.; Mohanty, S. Structural consequences of the pH-induced conformational switch in A. polyphemus pheromone-binding protein: Mechanisms of ligand release. J. Mol. Biol. 2005, 354, 1081–1090. [Google Scholar] [CrossRef]
- Rihani, K.; Ferveur, J.F.; Briand, L. The 40-Year Mystery of Insect Odorant-Binding Proteins. Biomolecules 2021, 11, 509. [Google Scholar] [CrossRef] [PubMed]
- Goncalves, F.; Ribeiro, A.; Silva, C.; Cavaco-Paulo, A. Biotechnological applications of mammalian odorant-binding proteins. Crit. Rev. Biotechnol. 2021, 41, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Tomita, T.; Ishikawa, D.; Noguchi, T.; Katayama, E.; Hashimoto, Y. Assembly of flammutoxin, a cytolytic protein from the edible mushroom Flammulina velutipes, into a pore-forming ring-shaped oligomer on the target cell. Biochem. J. 1998, 333, 129–137. [Google Scholar] [CrossRef]
- Spinelli, S.; Ramoni, R.; Grolli, S.; Bonicel, J.; Cambillau, C.; Tegoni, M. The structure of the monomeric porcine odorant binding protein sheds light on the domain swapping mechanism. Biochemistry 1998, 37, 7913–7918. [Google Scholar] [CrossRef]
- Schiefner, A.; Freier, R.; Eichinger, A.; Skerra, A. Crystal structure of the human odorant binding protein, OBPIIa. Proteins 2015, 83, 1180–1184. [Google Scholar] [CrossRef]
- Wogulis, M.; Morgan, T.; Ishida, Y.; Leal, W.S.; Wilson, D.K. The crystal structure of an odorant binding protein from Anopheles gambiae: Evidence for a common ligand release mechanism. Biochem. Biophys. Res. Commun. 2006, 339, 157–164. [Google Scholar] [CrossRef]
- Leite, N.R.; Krogh, R.; Xu, W.; Ishida, Y.; Iulek, J.; Leal, W.S.; Oliva, G. Structure of an odorant-binding protein from the mosquito Aedes aegypti suggests a binding pocket covered by a pH-sensitive “Lid”. PLoS ONE 2009, 4, e8006. [Google Scholar] [CrossRef]
- Laughlin, J.D.; Ha, T.S.; Jones, D.N.M.; Smith, D.P. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 2008, 133, 1255–1265. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Atkinson, R.; Jones, D.N.; Smith, D.P. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 2005, 45, 193–200. [Google Scholar] [CrossRef]
- Jin, X.; Ha, T.S.; Smith, D.P. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. USA 2008, 105, 10996–11001. [Google Scholar] [CrossRef]
- Tegoni, M.; Ramoni, R.; Bignetti, E.; Spinelli, S.; Cambillau, C. Domain swapping creates a third putative combining site in bovine odorant binding protein dimer. Nat. Struct. Biol. 1996, 3, 863–867. [Google Scholar] [CrossRef]
- Bianchet, M.A.; Bains, G.; Pelosi, P.; Pevsner, J.; Snyder, S.H.; Monaco, H.L.; Amzel, L.M. The three-dimensional structure of bovine odorant binding protein and its mechanism of odor recognition. Nat. Struct. Biol. 1996, 3, 934–939. [Google Scholar] [CrossRef]
- Nagnan-Le Meillour, P.; Joly, A.; Le Danvic, C.; Marie, A.; Zirah, S.; Cornard, J.P. Binding Specificity of Native Odorant-Binding Protein Isoforms Is Driven by Phosphorylation and O-N-Acetylglucosaminylation in the Pig Sus scrofa. Front. Endocrinol. 2018, 9, 816. [Google Scholar] [CrossRef]
- Nagnan-Le Meillour, P.; Vercoutter-Edouart, A.S.; Hilliou, F.; Le Danvic, C.; Levy, F. Proteomic Analysis of Pig (Sus scrofa) Olfactory Soluble Proteome Reveals O-Linked-N-Acetylglucosaminylation of Secreted Odorant-Binding Proteins. Front. Endocrinol. 2014, 5, 202. [Google Scholar] [CrossRef]
- Grolli, S.; Merli, E.; Conti, V.; Scaltriti, E.; Ramoni, R. Odorant binding protein has the biochemical properties of a scavenger for 4-hydroxy-2-nonenal in mammalian nasal mucosa. FEBS J. 2006, 273, 5131–5142. [Google Scholar] [CrossRef]
- Sun, J.S.; Xiao, S.; Carlson, J.R. The diverse small proteins called odorant-binding proteins. R. Soc. Open Biol. 2018, 8, 180208. [Google Scholar] [CrossRef]
- Lacazette, E.; Gachon, A.-M.; Pitiot, G. A novel human odorant-binding protein gene family resulting from genomic duplicons at 9q34: Differential expression in the oral and genital spheres. Human. Mol. Genet. 2000, 9, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Debat, H.; Eloit, C.; Blon, F.; Sarazin, B.; Henry, C.; Huet, J.C.; Trotier, D.; Pernollet, J.C. Identification of human olfactory cleft mucus proteins using proteomic analysis. J. Proteome Res. 2007, 6, 1985–1996. [Google Scholar] [CrossRef] [PubMed]
- Tcatchoff, L.; Nespoulous, C.; Pernollet, J.-C.; Briand, L. A single lysyl residue defines the binding specificity of a human odorant-binding protein for aldehydes. FEBS Lett. 2006, 580, 2102–2108. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, S.; Hasegawa, T.; Maeno, K.; Motoyama, A.; Denda, M. OBP2A regulates epidermal barrier function and protects against cytotoxic small hydrophobic molecules. iScience 2024, 27, 111093. [Google Scholar] [CrossRef]
- Sano, R.; Takahashi, Y.; Fukuda, H.; Harada, M.; Hayakawa, A.; Okawa, T.; Kubo, R.; Takeshita, H.; Tsukada, J.; Kominato, Y. A cell-specific regulatory region of the human ABO blood group gene regulates the neighborhood gene encoding odorant binding protein 2B. Sci. Rep. 2021, 11, 7325. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, K.; Wang, H.; Jaen, C.; Haneoka, M.; Saito, N.; Nakamura, J.; Adappa, N.D.; Cohen, N.A.; Dalton, P. The human olfactory cleft mucus proteome and its age-related changes. Sci. Rep. 2018, 8, 17170. [Google Scholar] [CrossRef]
- Wolf, A.; Liesinger, L.; Spoerk, S.; Schittmayer, M.; Lang-Loidolt, D.; Birner-Gruenberger, R.; Tomazic, P.V. Olfactory cleft proteome does not reflect olfactory performance in patients with idiopathic and postinfectious olfactory disorder: A pilot study. Sci. Rep. 2018, 8, 17554. [Google Scholar] [CrossRef]
- Ijichi, C.; Kondo, K.; Kobayashi, M.; Shirasawa, A.; Shimbo, K.; Nakata, K.; Maruyama, Y.; Ihara, Y.; Kawato, Y.; Mannen, T.; et al. Lipocalin 15 in the olfactory mucus is a biomarker for Bowman’s gland activity. Sci. Rep. 2022, 12, 9984. [Google Scholar] [CrossRef]
- Goetz, D.H.; Holmes, M.A.; Borregaard, N.; Bluhm, M.E.; Raymond, K.N.; Strong, R.K. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 2002, 10, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Glasgow, B.J.; Abduragimov, A.R. Lipocalin-1 is the acceptor protein for phospholipid transfer protein in tears. Biochem. Biophys. Res. Commun. 2021, 548, 35–38. [Google Scholar] [CrossRef]
- Seshadri, S.; Lin, D.C.; Rosati, M.; Carter, R.G.; Norton, J.E.; Suh, L.; Kato, A.; Chandra, R.K.; Harris, K.E.; Chu, H.W.; et al. Reduced expression of antimicrobial PLUNC proteins in nasal polyp tissues of patients with chronic rhinosinusitis. Allergy 2012, 67, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, M.; Hong, Y.; Bu, X.; Luan, G.; Wang, Y.; Li, Y.; Lou, H.; Wang, C.; Zhang, L. Reduced Expression of Antimicrobial Protein Secretory Leukoprotease Inhibitor and Clusterin in Chronic Rhinosinusitis with Nasal Polyps. J. Immunol. Res. 2021, 2021, 1057186. [Google Scholar] [CrossRef]
- Yoo, F.; Soler, Z.M.; Mulligan, J.K.; Storck, K.A.; Lamira, J.M.; Pasquini, W.N.; Hill, J.B.; Noonan, T.E.; Washington, B.J.; Schlosser, R.J. Olfactory cleft mucus proteins associated with olfactory dysfunction in a cohort without chronic rhinosinusitis. Int. Forum Allergy Rhinol. 2019, 9, 1151–1158. [Google Scholar] [CrossRef]
- Tong, J.; Gu, Q. Expression and Clinical Significance of Mucin Gene in Chronic Rhinosinusitis. Curr. Allergy Asthma Rep. 2020, 20, 63. [Google Scholar] [CrossRef]
- Tai, J.; Shin, J.M.; Park, J.; Han, M.; Kim, T.H. Oxidative Stress and Antioxidants in Chronic Rhinosinusitis with Nasal Polyps. Antioxidants 2023, 12, 195. [Google Scholar] [CrossRef]
- Neiers, F.; Jarriault, D.; Menetrier, F.; Faure, P.; Briand, L.; Heydel, J.M. The odorant metabolizing enzyme UGT2A1: Immunolocalization and impact of the modulation of its activity on the olfactory response. PLoS ONE 2021, 16, e0249029. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shen, Y.; Li, K.; Zhang, P.; Wang, G.; Gao, L.; Bai, C. Role of matrix metalloproteinase-9 in lipopolysaccharide-induced mucin production in human airway epithelial cells. Arch. Biochem. Biophys. 2009, 486, 111–118. [Google Scholar] [CrossRef]
- Carr, V.M.; Menco, B.P.M.; Yankova, M.P.; Morimoto, R.I.; Farbman, A.I. Odorants as cell-type specific activators of a heat shock response in the rat olfactory mucosa. J. Comp. Neurol. 2001, 432, 425–439. [Google Scholar] [CrossRef]
- Hegg, C.C.; Lucero, M.T. Purinergic receptor antagonists inhibit odorant-induced heat shock protein 25 induction in mouse olfactory epithelium. Glia 2006, 53, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Smadja, D.M.; Chocron, A.F.; Abreu, M.M. HSP47 at the Crossroads of Thrombosis and Collagen Dynamics: Unlocking Therapeutic Horizons and Debates. TH Open 2025, 9, a25994925. [Google Scholar] [CrossRef]
- Wen, Z.; Liu, D.; Zhu, H.; Sun, X.; Xiao, Y.; Lin, Z.; Zhang, A.; Ye, C.; Gao, J. Deficiency for Lcn8 causes epididymal sperm maturation defects in mice. Biochem. Biophys. Res. Commun. 2021, 548, 7–13. [Google Scholar] [CrossRef]
- Suzuki, K.; Lareyre, J.J.; Sanchez, D.; Gutierrez, G.; Araki, Y.; Matusik, R.J.; Orgebin-Crist, M.C. Molecular evolution of epididymal lipocalin genes localized on mouse chromosome 2. Gene 2004, 339, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wang, P.; Wang, X.; Du, W.; Yang, H.H.; Liu, Y.; Cui, S.N.; Huang, W.; Peng, T.; Chen, J.; et al. Lipocalin 10 is essential for protection against inflammation-triggered vascular leakage by activating LDL receptor-related protein 2-slingshot homologue 1 signalling pathway. Cardiovasc. Res. 2023, 119, 1981–1996. [Google Scholar] [CrossRef]
- Glasgow, B.J.; Abduragimov, A.R.; Farahbakhsh, Z.T.; Faull, K.F.; Hubbell, W.L. Tear lipocalins bind a broad array of lipid ligands. Curr. Eye Res. 1995, 14, 363–372. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Kao, M.C.; Fang, J.Y.; Zouboulis, C.C.; Zhang, L.; Gallo, R.L.; Huang, C.M. Antimicrobial property of lauric acid against Propionibacterium acnes: Its therapeutic potential for inflammatory acne vulgaris. J. Invest. Dermatol. 2009, 129, 2480–2488. [Google Scholar] [CrossRef] [PubMed]
- Kabara, J.J.; Swieczkowski, D.M.; Conley, A.J.; Truant, J.P. Fatty acids and derivatives as antimicrobial agents. Antimicrob. Agents Chemother. 1972, 2, 23–28. [Google Scholar] [CrossRef]
- Lechner, M.; Wojnar, P.; Redl, B. Human tear lipocalin acts as an oxidative-stress-induced scavenger of potentially harmful lipid peroxidation products in a cell culture system. Biochem. J. 2001, 356, 129–135. [Google Scholar] [CrossRef]
- Fattori, B.; Castagna, M.; Megna, G.; Casani, A.; Pelosi, P. Immunohistochemical localisation of tear lipocalin in human nasal mucosa. Rhinology 1998, 36, 101–103. [Google Scholar]
- Fluckinger, M.; Haas, H.; Merschak, P.; Glasgow, B.J.; Redl, B. Human tear lipocalin exhibits antimicrobial activity by scavenging microbial siderophores. Antimicrob. Agents Chemother. 2004, 48, 3367–3372. [Google Scholar] [CrossRef] [PubMed]
- Gasymov, O.K.; Abduragimov, A.R.; Glasgow, B.J. Ligand binding site of tear lipocalin: Contribution of a trigonal cluster of charged residues probed by 8-anilino-1-naphthalenesulfonic acid. Biochemistry 2008, 47, 1414–1424. [Google Scholar] [CrossRef]
- Bachman, M.A.; Miller, V.L.; Weiser, J.N. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog. 2009, 5, e1000622. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.; Kondo, K.; Ihara, S.; Ijichi, C.; Sato, K.; Touhara, K. Fibronectin in the olfactory mucus increases sensitivity of olfactory receptor response to odorants. Sci. Adv. 2025, 11, eadu7271. [Google Scholar] [CrossRef]
- Soler, Z.M.; Schlosser, R.J.; Mulligan, J.K.; Smith, T.L.; Mace, J.C.; Ramakrishan, V.R.; Norris-Caneda, K.; Bethard, J.R.; Ball, L.E. Olfactory cleft mucus proteome in chronic rhinosinusitis: A case-control pilot study. Int. Forum Allergy Rhinol. 2021, 11, 1162–1176. [Google Scholar] [CrossRef]
- Zeng, C.; Spielman, A.I.; Vowels, B.R.; Leyden, J.J.; Biemann, K.; Preti, G. A human axillary odorant is carried by apolipoprotein D. Proc. Natl. Acad. Sci. USA 1996, 93, 6626–6630. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, H.; Luo, G.; Xu, N.; Pan, Z. Association between the ABCC11 gene polymorphism and the expression of apolipoprotein D by the apocrine glands in axillary osmidrosis. Mol. Med. Rep. 2015, 11, 4463–4467. [Google Scholar] [CrossRef]
- Fyfe-Desmarais, G.; Desmarais, F.; Rassart, E.; Mounier, C. Apolipoprotein D in Oxidative Stress and Inflammation. Antioxidants 2023, 12, 1027. [Google Scholar] [CrossRef]
- Ganfornina, M.D.; Do Carmo, S.; Lora, J.M.; Torres-Schumann, S.; Vogel, M.; Allhorn, M.; Gonzalez, C.; Bastiani, M.J.; Rassart, E.; Sanchez, D. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell 2008, 7, 506–515. [Google Scholar] [CrossRef]
- Dassati, S.; Waldner, A.; Schweigreiter, R. Apolipoprotein D takes center stage in the stress response of the aging and degenerative brain. Neurobiol. Aging 2014, 35, 1632–1642. [Google Scholar] [CrossRef] [PubMed]
- Bingle, C.D.; Craven, C.J. PLUNC: A novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Human. Mol. Genet. 2002, 11, 937–943. [Google Scholar] [CrossRef] [PubMed]
- Andrault, J.-B.; Gaillard, I.; Giorgi, D.; Rouquier, S. Expansion of the BPI family by duplication on human chromosome 20: Characterization of the RY gene cluster in 20q11. 21 encoding olfactory transporters/antimicrobial-like peptides. Genomics 2003, 82, 172–184. [Google Scholar] [CrossRef]
- Delorme-Axford, E.; Morosky, S.; Bomberger, J.; Stolz, D.B.; Jackson, W.T.; Coyne, C.B. BPIFB3 regulates autophagy and coxsackievirus B replication through a noncanonical pathway independent of the core initiation machinery. mBio 2014, 5, e02147. [Google Scholar] [CrossRef]
- Bingle, L.; Bingle, C.D. Distribution of human PLUNC/BPI fold-containing (BPIF) proteins. Biochem. Soc. Trans. 2011, 39, 1023–1027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Nie, X.; Xiao, B.; Xiang, J.; Shen, S.; Gong, J.; Zhou, M.; Zhu, S.; Zhou, J.; Qian, J.; et al. Identification of tissue-specific genes in nasopharyngeal epithelial tissue and differentially expressed genes in nasopharyngeal carcinoma by suppression subtractive hybridization and cDNA microarray. Genes. Chromosomes Cancer 2003, 38, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Yew, P.Y.; Mushiroda, T.; Kiyotani, K.; Govindasamy, G.K.; Yap, L.F.; Teo, S.H.; Lim, P.V.; Govindaraju, S.; Ratnavelu, K.; Sam, C.K.; et al. Identification of a functional variant in SPLUNC1 associated with nasopharyngeal carcinoma susceptibility among Malaysian Chinese. Mol. Carcinog. 2012, 51 (Suppl. 1), E74–E82. [Google Scholar] [CrossRef]
- Zhang, W.; Zeng, Z.; Wei, F.; Chen, P.; Schmitt, D.C.; Fan, S.; Guo, X.; Liang, F.; Shi, L.; Liu, Z.; et al. SPLUNC1 is associated with nasopharyngeal carcinoma prognosis and plays an important role in all-trans-retinoic acid-induced growth inhibition and differentiation in nasopharyngeal cancer cells. FEBS J. 2014, 281, 4815–4829. [Google Scholar] [CrossRef]
- Taha, E.A.; Ono, K.; Eguchi, T. Roles of extracellular HSPs as biomarkers in immune surveillance and immune evasion. Int. J. Mol. Sci. 2019, 20, 4588. [Google Scholar] [CrossRef] [PubMed]
- Herrera, J.; Henke, C.A.; Bitterman, P.B. Extracellular matrix as a driver of progressive fibrosis. J. Clin. Invest. 2018, 128, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Sevilla, C.A.; Dalecki, D.; Hocking, D.C. Regional fibronectin and collagen fibril co-assembly directs cell proliferation and microtissue morphology. PLoS ONE 2013, 8, e77316. [Google Scholar] [CrossRef]
- Kicic, A.; Hallstrand, T.S.; Sutanto, E.N.; Stevens, P.T.; Kobor, M.S.; Taplin, C.; Pare, P.D.; Beyer, R.P.; Stick, S.M.; Knight, D.A. Decreased fibronectin production significantly contributes to dysregulated repair of asthmatic epithelium. Am. J. Respir. Crit. Care Med. 2010, 181, 889–898. [Google Scholar] [CrossRef]
- Herard, A.L.; Pierrot, D.; Hinnrasky, J.; Kaplan, H.; Sheppard, D.; Puchelle, E.; Zahm, J.M. Fibronectin and its alpha 5 beta 1-integrin receptor are involved in the wound-repair process of airway epithelium. Am. J. Physiol. 1996, 271, L726–L733. [Google Scholar] [CrossRef]
- Fernandez, C.A.; Yan, L.; Louis, G.; Yang, J.; Kutok, J.L.; Moses, M.A. The matrix metalloproteinase-9/neutrophil gelatinase-associated lipocalin complex plays a role in breast tumor growth and is present in the urine of breast cancer patients. Clin. Cancer Res. 2005, 11, 5390–5395. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Shukla, M.; Cowland, J.B.; Malemud, C.J.; Haqqi, T.M. Neutrophil gelatinase-associated lipocalin is expressed in osteoarthritis and forms a complex with matrix metalloproteinase 9. Arthritis Rheum. 2007, 56, 3326–3335. [Google Scholar] [CrossRef]
- Cavalcanti, L.; Francati, S.; Ferraguti, G.; Fanfarillo, F.; Peluso, D.; Barbato, C.; Greco, A.; Minni, A.; Petrella, C. Lipocalin-2, Matrix Metalloproteinase-9, and MMP-9/NGAL Complex in Upper Aerodigestive Tract Carcinomas: A Pilot Study. Cells 2025, 14, 506. [Google Scholar] [CrossRef]
- Xia, Q.; Du, Z.; Chen, M.; Zhou, X.; Bai, W.; Zheng, X.; Lin, L.; Zhao, Y.; Ding, J.; Wu, Z.; et al. A protein complex of LCN2, LOXL2 and MMP9 facilitates tumour metastasis in oesophageal cancer. Mol. Oncol. 2023, 17, 2451–2471. [Google Scholar] [CrossRef]
- Cheng, J.; Chen, J.; Zhao, Y.; Yang, J.; Xue, K.; Wang, Z. MicroRNA-761 suppresses remodeling of nasal mucosa and epithelial-mesenchymal transition in mice with chronic rhinosinusitis through LCN2. Stem Cell Res. Ther. 2020, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Paradis, V.; Mathurin, P.; Ratziu, V.; Poynard, T.; Bedossa, P. Binding of apolipoprotein A-I and acetaldehyde-modified apolipoprotein A-I to liver extracellular matrix. Hepatology 1996, 23, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
- Carrijo-Carvalho, L.C.; Maria, D.A.; Ventura, J.S.; Morais, K.L.; Melo, R.L.; Rodrigues, C.J.; Chudzinski-Tavassi, A.M. A lipocalin-derived Peptide modulating fibroblasts and extracellular matrix proteins. J. Toxicol. 2012, 2012, 325250. [Google Scholar] [CrossRef] [PubMed]
- Cattaneo, M.; Aleksova, A.; Malovini, A.; Avolio, E.; Thomas, A.; Alvino, V.V.; Kilcooley, M.; Pieronne-Deperrois, M.; Ouvrard-Pascaud, A.; Maciag, A.; et al. BPIFB4 and its longevity-associated haplotype protect from cardiac ischemia in humans and mice. Cell Death Dis. 2023, 14, 523. [Google Scholar] [CrossRef]
- Alvino, V.V.; Slater, S.; Qiu, Y.; Cattaneo, M.; Mohammed, K.A.K.; Gate, S.; Sekar, V.; Puca, A.A.; Madeddu, P. Healthy longevity-associated protein improves cardiac function in murine models of cardiomyopathy with preserved ejection fraction. Cardiovasc. Diabetol. 2024, 23, 397. [Google Scholar] [CrossRef]
- Cattaneo, M.; Beltrami, A.P.; Thomas, A.C.; Spinetti, G.; Alvino, V.V.; Avolio, E.; Veneziano, C.; Rolle, I.G.; Sponga, S.; Sangalli, E.; et al. The longevity-associated BPIFB4 gene supports cardiac function and vascularization in ageing cardiomyopathy. Cardiovasc. Res. 2023, 119, 1583–1595. [Google Scholar] [CrossRef]
- Stowers, L.; Logan, D.W. LUSH shapes up for a starring role in olfaction. Cell 2008, 133, 1137–1139. [Google Scholar] [CrossRef]
- Lautenschlager, C.; Leal, W.S.; Clardy, J. Bombyx mori pheromone-binding protein binding nonpheromone ligands: Implications for pheromone recognition. Structure 2007, 15, 1148–1154. [Google Scholar] [CrossRef]
- Shelton, J.F.; Shastri, A.J.; Fletez-Brant, K.; Me, C.-T.; Aslibekyan, S.; Auton, A. The UGT2A1/UGT2A2 locus is associated with COVID-19-related loss of smell or taste. Nat. Genet. 2022, 54, 121–124. [Google Scholar] [CrossRef]
- Getchell, T.V.; Krishna, N.R.; Sparks, D.L.; Dhooper, N.; Getchell, M.L. Human olfactory receptor neurons express heat shock protein 70: Age-related trends. Ann. Otol. Rhinol. Laryngol. 1995, 104, 47–56. [Google Scholar] [CrossRef]
- Crum, T.S.; Gleixner, A.M.; Posimo, J.M.; Mason, D.M.; Broeren, M.T.; Heinemann, S.D.; Wipf, P.; Brodsky, J.L.; Leak, R.K. Heat shock protein responses to aging and proteotoxicity in the olfactory bulb. J. Neurochem. 2015, 133, 780–794. [Google Scholar] [CrossRef]
- Sziraki, A.; Lu, Z.; Lee, J.; Banyai, G.; Anderson, S.; Abdulraouf, A.; Metzner, E.; Liao, A.; Banfelder, J.; Epstein, A.; et al. A global view of aging and Alzheimer’s pathogenesis-associated cell population dynamics and molecular signatures in human and mouse brains. Nat. Genet. 2023, 55, 2104–2116. [Google Scholar] [CrossRef]
- Zhu, H.; Qu, S.; Gong, M.; Xiang, Y.; Gan, S.; Teng, Y.; Ye, D. Mechanisms, diagnosis, and treatment of olfactory dysfunction in rhinosinusitis. Eur. J. Med. Res. 2025, 30, 474. [Google Scholar] [CrossRef] [PubMed]
- Fagotti, A.; Lucentini, L.; Simoncelli, F.; La Porta, G.; Brustenga, L.; Bizzarri, I.; Trio, S.; Isidori, C.; Di Rosa, I.; di Cara, G. HSP70 upregulation in nasal mucosa of symptomatic children with allergic rhinitis and potential risk of asthma development. Sci. Rep. 2022, 12, 14104. [Google Scholar] [CrossRef] [PubMed]
- Tomazic, P.V.; Birner-Gruenberger, R.; Leitner, A.; Darnhofer, B.; Spoerk, S.; Lang-Loidolt, D. Apolipoproteins have a potential role in nasal mucus of allergic rhinitis patients: A proteomic study. Laryngoscope 2015, 125, E91–E96. [Google Scholar] [CrossRef] [PubMed]
- Olender, T.; Keydar, I.; Pinto, J.M.; Tatarskyy, P.; Alkelai, A.; Chien, M.S.; Fishilevich, S.; Restrepo, D.; Matsunami, H.; Gilad, Y.; et al. The human olfactory transcriptome. BMC Genom. 2016, 17, 619. [Google Scholar] [CrossRef]

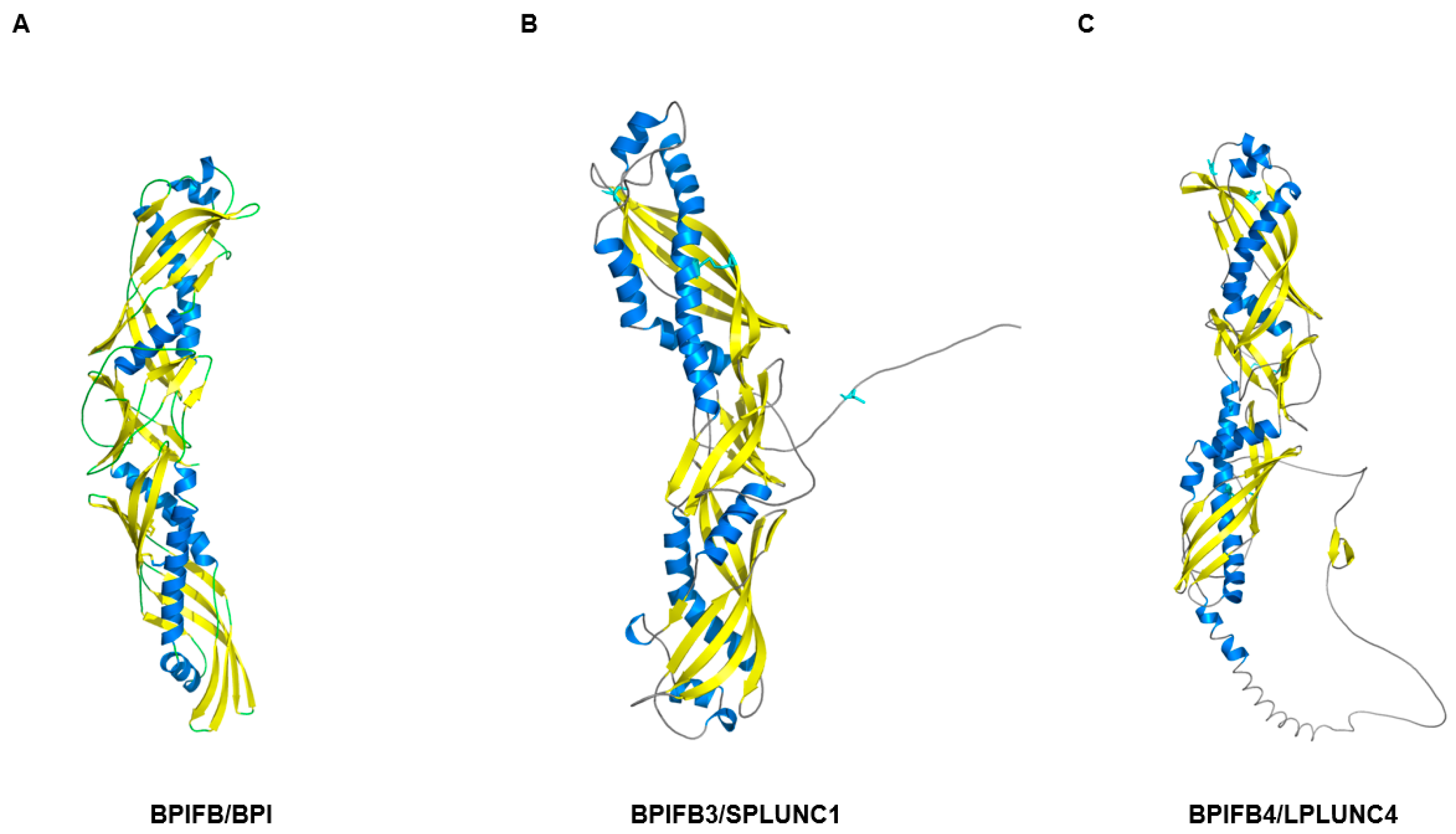
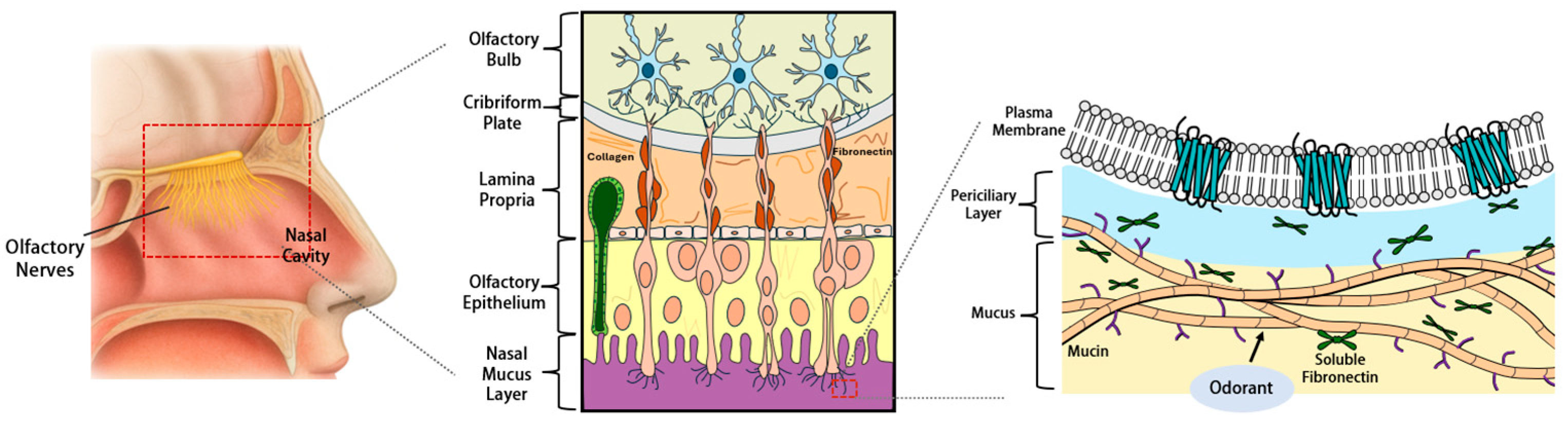
| Protein Category a | Representative Proteins (Gene Symbols) b | Primary Functions in Olfactory Mucus c | Detection and Characteristics d | Primary Detection Method e | Key References f |
|---|---|---|---|---|---|
| Classical OBP | OBP2A, OBP2B | Binding and transport of hydrophobic odorants | Variably reported in OC mucus depending on study; some ELISA-positive reports, often undetected in proteomics | Genomic identification; ELISA | [47,50,51] |
| Lipocalin (OBP-like) | LCN15 | Odorant binding | Highly expressed in OC mucus, secreted from Bowman’s glands, linked to age, inflammation, and olfactory dysfunction | LC-MS/MS proteomics of olfactory mucus | [54] |
| LCN2 | Antibacterial/Defense | Linked to inflammatory OC mucus | LC-MS/MS proteomics | [55] | |
| LCN1 | Chemical defense | Reported in some proteomic studies | LC-MS/MS proteomics | [56] | |
| PLUNC/BPI-fold | BPIFB4, BPIFA1(SPLUNC1), BPIFB2 | Innate defense, Surface tension regulation, Ion balance | Repeatedly detected in OC/non-OC mucus proteomes (BPIFB4 especially abundant) | LC-MS/MS proteomics of nasal/olfactory mucus | [57,58] |
| Apolipoprotein | APOA1/2/4, APOB100, APOE, CLU | Lipid transport, Protein quality control, Anti-inflammatory defense | Repeatedly reported across cohorts; APOA1/2 strongly link to AR | LC-MS/MS proteomics | [59,60,61] |
| Peri-receptor Enzymes (ODE) | UGT2A1/2, ALDHs, GSTP1, CES, CYPs | Odorant metabolism, Detoxification | Repeatedly identified in AR/OC/CRS mucus proteomes; included in multiple proteomic datasets | LC-MS/MS proteomics | [62] |
| Barrier/Defense proteins | SLPI, MUC5B | Mucosal barrier maintenance, Antimicrobial activity | Detected in AR/CRS OC mucus; interpreted as barrier weakness | LC-MS/MS proteomics | [58,63] |
| Chaperones/Stress proteins | HSP70, HSP27/25 | Protein quality control, Stress response | Altered expression under inflammatory and stress conditions | Immunocytochemistry; Immunohistochemistry | [64,65,66] |
| Subfamily | Full Protein Name | Alias | Identity | Similarity |
|---|---|---|---|---|
| BPIFD1 | Bactericidal permeability-increasing protein precursor | BPI | 100 | 100 |
| BPIFD2 | Lipopolysaccharide-binding protein precursor | LBP | 45.2 | 65.2 |
| BPIFE | Phospholipid transfer protein | PLTP, HDLCQ9 | 25.8 | 45.4 |
| BPIFF | Cholesteryl ester transfer protein | CETP, HDLCQ10 | 23 | 40.9 |
| BPIFA1 | BPI fold–containing family A member 1 | SPLUNC1/PLUNC | 12.9 | 21 |
| BPIFA2 | BPI fold–containing family A member 2 | LPLUNC2, BPIL1, C20orf184, RYSR, | 9.6 | 18 |
| BPIFA3 | BPI fold–containing family A member 3 | SPLUNC3, C20orf71 | 9.9 | 16.2 |
| BPIFB1 | BPI fold–containing family B member 1 | LPLUNC1, C20orf114 | 19.2 | 36.3 |
| BPIFB2 | BPI fold–containing family B member 2 | LPLUNC2, BPIL1, C20orf184, RYSR | 21.9 | 43 |
| BPIFB3 | BPI fold–containing family B member 3 | LPLUNC3, C20orf185, RYA3 | 20 | 38.1 |
| BPIFB4 | BPI fold–containing family B member 4 | LPLUNC4, C20orf186, RY2G5 | 14.7 | 28.4 |
| BPIFB6 | BPI fold–containing family B member 6 | BPIL3, LPLUNC6 | 22.1 | 39.2 |
| BPIFC | BPI fold–containing family C protein | BPIL2 | 26.6 | 46.4 |
| Disease/Condition a | Affected Proteins or Genes b | Expression Change (↑/↓) c | Tissue/Sample Source d | Key References e |
|---|---|---|---|---|
| Aging | LCN15 | ↓ (aged; idiopathic loss tendency | Human, nasal mucosa | [54] |
| HSP70 | ↓ in ORNs (not in sustentacular cells or Bowman’s glands) | Human, olfactory mucosa (immunocytochemistry) | [110] | |
| Alzheimer’s disease | HSP70 | ↓ in ORNs (not in sustentacular cells or Bowman’s glands) | Human, Human, olfactory mucosa (immunocytochemistry) | [110] |
| Viral infection | UGT2A1/2 | SNP (rs7688383 T allele) ↑ risk of COVID-19-related smell/taste loss | Human, genome-wide association study | [109] |
| BPIFB3 | ↓ → CVB replication ↑; ↑ → replication ↓ | Human cell lines (HBMEC, U2OS, HeLa, 786-O; Coxsackievirus B infection model | [87] | |
| Bacterial infection | LCN2 | Infection → LCN2 ↑ → bacterial growth ↓ | In vitro (E. coli growth assay); mouse nasal infection model (K. pneumoniae KPPR1) | [55] |
| Allergic rhinitis | APOA1, APOA2, APOE, APOJ | ↑ | Human, nasal mucosa and nasal fluid | [101] |
| HSP70 | ↑ | Human, nasal mucosa (mRNA) | [114] | |
| CRS/CRSwNP | MUC3, MUC6 | ↓ | Human, nasal mucosa | [60] |
| MUC1, MUC2, MUC4, MUC5AC, MUC5B, MUC8 | ↑ | Human, nasal mucosa | [60] | |
| Fibronectin | ↓ (also in Idiopathic olfactory disorder) | Human, olfactory mucosa | [78] | |
| LCN2 | ↑ in CRSwNP (human); ↓ by miR-761 (mouse) | Human, CRSwNP tissue; Mouse, nasal mucosa | [63,101] | |
| BPIFA1/SPLUNC1, BPIFB1, BPIFB2/LPLUNC2, BPIL-1, SLPI, CLU, LTF, LYZ | ↓ | Human, CRSwNP tissue | [57,58] | |
| Environmental stress | HSP25 | ↑ (odorant-induced) | Mouse, olfacoty mucosa | [65] |
| HSP70 | ↑ (odorant-induced) | Rat, olfacoty mucosa | [64] | |
| Axillary osmidrosis | ApoD | ↑ | Human, apocrine gland(mRNA) | [80,81] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha, J.; Kim, H.; Kim, H.; Jang, Y. Cellular and Molecular Roles of Human Odorant-Binding Proteins and Related Lipocalins in Olfaction and Neuroinflammation. Cells 2025, 14, 1859. https://doi.org/10.3390/cells14231859
Ha J, Kim H, Kim H, Jang Y. Cellular and Molecular Roles of Human Odorant-Binding Proteins and Related Lipocalins in Olfaction and Neuroinflammation. Cells. 2025; 14(23):1859. https://doi.org/10.3390/cells14231859
Chicago/Turabian StyleHa, Juchan, Hyojin Kim, Hyungsup Kim, and Yongwoo Jang. 2025. "Cellular and Molecular Roles of Human Odorant-Binding Proteins and Related Lipocalins in Olfaction and Neuroinflammation" Cells 14, no. 23: 1859. https://doi.org/10.3390/cells14231859
APA StyleHa, J., Kim, H., Kim, H., & Jang, Y. (2025). Cellular and Molecular Roles of Human Odorant-Binding Proteins and Related Lipocalins in Olfaction and Neuroinflammation. Cells, 14(23), 1859. https://doi.org/10.3390/cells14231859







