Mathematical Modeling of Cell Death and Survival: Toward an Integrated Computational Framework for Multi-Decision Regulatory Dynamics
Highlights
- Existing ordinary differential equation models incorporate no more than two regulated cell death types simultaneously, limiting their capacity to capture the full complexity of pathway crosstalk and coordination.
- Ferroptosis is currently modeled only as an independent process, with no mechanistic links established to other regulated cell death pathways.
- Comprehensive models that integrate molecular mechanisms of regulated cell death, damage-associated molecular pattern release, and subsequent intercellular immune activation are still absent.
Abstract
1. Introduction
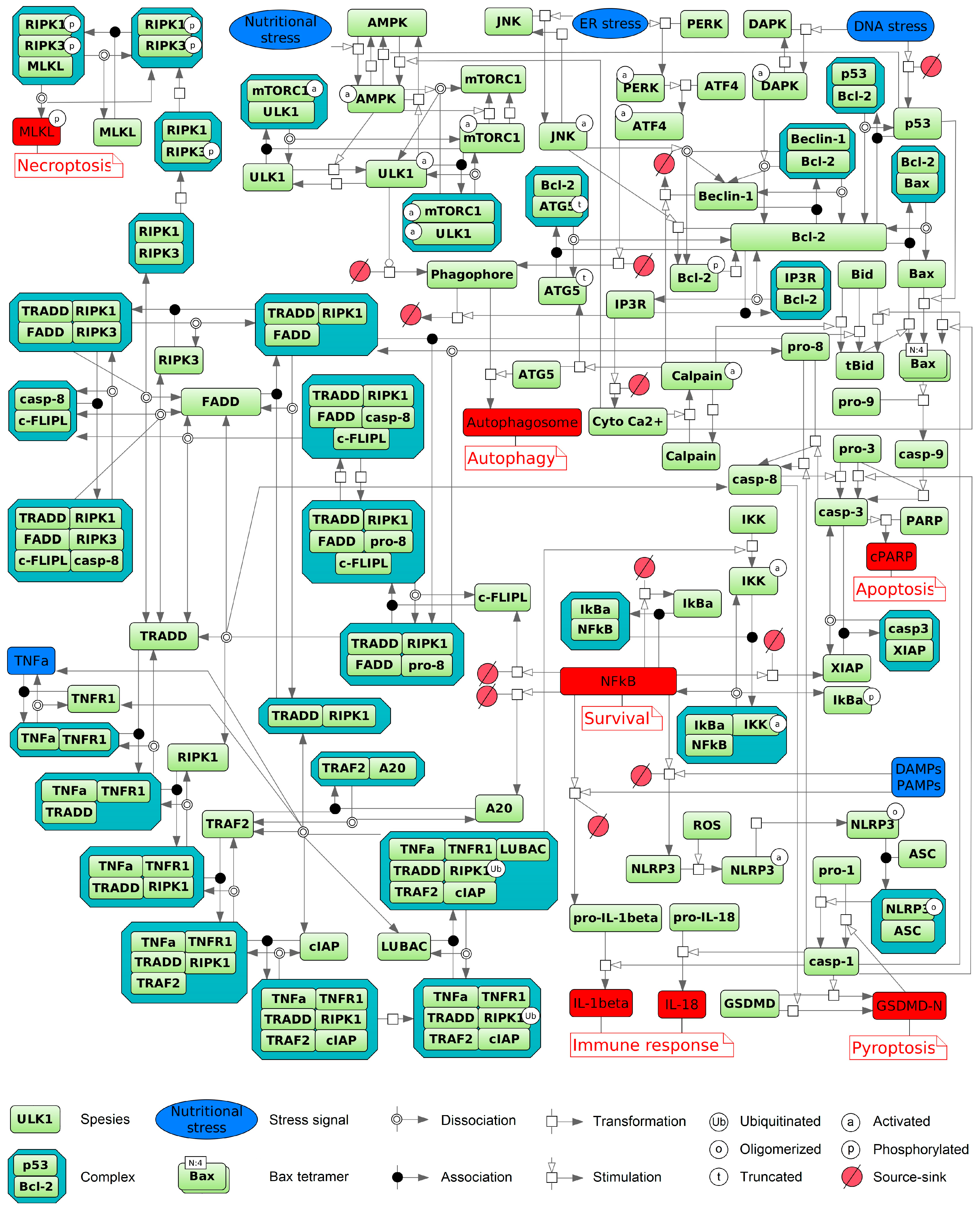
2. Apoptosis
3. Autophagy
4. Ferroptosis
5. Immunogenic Cell Death
6. Necroptosis
7. Pyroptosis
8. Comparative Statistics
9. Integrated Model of the Pathways
10. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AMBRA1 | Activating molecule in beclin-1-regulated autophagy 1 |
| AMP | Adenosine monophosphate |
| AMPK | AMP-activated protein kinase |
| ATF4 | Activating transcription factor 4 |
| ASC | Apoptosis-associated speck-like protein containing a CARD |
| BH3 | BCL-2 homology domain 3 |
| BioUML | Biological universal modeling language |
| c-FLIPs | Cellular FADD-like interleukin (IL)-1β-converting enzyme-inhibitory proteins |
| CARD | Caspase recruitment domain |
| CHOP | C/EBP homologous protein |
| cIAPs | Cellular inhibitor of apoptosis proteins |
| CYLD | Cylindromatosis lysine 63 deubiquitinase |
| DAMPs | Damage-associated molecular patterns |
| DED | Death effector domain |
| DISC | Death-inducing signaling complex |
| DRAM | Damage-regulated autophagy modulator |
| DR | Death receptor |
| ER | Endoplasmic reticulum |
| GADD34 | Growth arrest and DNA damage-inducible 34 |
| GPX4 | Glutathione peroxidase 4 |
| GSDMD | Gasdermin D |
| GSDME | Gasdermin E |
| ICD | Immunogenic cell death |
| IκB | Inhibitor of kappa B |
| IKKs | IκB kinases |
| IL-1β, -18 | Interleukin-1β, -18 |
| iNOS | Inducible nitric oxide synthase |
| IRE1 | Inositol requiring 1 |
| IRF1 | Interferon regulatory factor-1 |
| FADD | Fas-associated protein with death domain |
| MLKL | Mixed lineage kinase domain-like |
| MOMP | Mitochondrial outer membrane permeabilization |
| MPT | Mitochondrial permeability transition |
| mTOR | Mammalian target of rapamycin |
| mTORC1 | mTOR complex 1 |
| NFκB | Nuclear factor κB |
| NLR | NOD-like receptor |
| NLRP3 | NLR family pyrin domain containing 3 |
| NO | Nitric oxide |
| NOD | Nucleotide-binding oligomerization domain |
| NRF2 | Nuclear factor erythroid-related factor 2 |
| ODEs | Ordinary differential equations |
| PAMPs | Pathogen-associated molecular patterns |
| PARP-1 | Poly (ADP-ribose) polymerase-1 |
| PD-L1, -L2 | Programmed death-ligand 1, 2 |
| PERK | Protein kinase RNA-like ER kinase |
| RCD | Regulated cell death |
| RIPK1, RIPK3 | Receptor-interacting protein kinase 1, 3 |
| RSL3 | RAS-selective lethal 3 |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SBGN | Systems biology graphical notation |
| SBML | Systems biology markup language |
| TLRs | Toll-like receptors |
| TNF | Tumor necrosis factor |
| TNFR1/2 | TNF receptor 1/2 |
| TRADD | TNFR1-associated death domain protein |
| TRAIL | TNF-related apoptosis-inducing ligand |
| TRAIL-R1/R2/R3/R4 | TRAIL receptor 1/2/3/4 |
| TRAF2 | TNF receptor-associated factor 2 |
| ULK1 | Unc-51-like kinase 1 |
| UPR | Unfolded protein response |
References
- Billman, G.E. Homeostasis: The Underappreciated and Far Too Often Ignored Central Organizing Principle of Physiology. Front. Physiol. 2020, 11, 200. [Google Scholar] [CrossRef]
- Wang, S.; Guo, S.; Guo, J.; Du, Q.; Wu, C.; Wu, Y.; Zhang, Y. Cell Death Pathways: Molecular Mechanisms and Therapeutic Targets for Cancer. MedComm 2024, 5, e693. [Google Scholar] [CrossRef]
- Lee, E.; Song, C.-H.; Bae, S.-J.; Ha, K.-T.; Karki, R. Regulated Cell Death Pathways and Their Roles in Homeostasis, Infection, Inflammation, and Tumorigenesis. Exp. Mol. Med. 2023, 55, 1632–1643. [Google Scholar] [CrossRef]
- Kroemer, G.; El-Deiry, W.S.; Golstein, P.; Peter, M.E.; Vaux, D.; Vandenabeele, P.; Zhivotovsky, B.; Blagosklonny, M.V.; Malorni, W.; Knight, R.A.; et al. Classification of Cell Death: Recommendations of the Nomenclature Committee on Cell Death. Cell Death Differ. 2005, 12, 1463–1467. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Galluzzi, L.; Vandenabeele, P.; Abrams, J.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; El-Deiry, W.S.; Golstein, P.; Green, D.R.; et al. Classification of Cell Death: Recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009, 16, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Abrams, J.M.; Alnemri, E.S.; Baehrecke, E.H.; Blagosklonny, M.V.; Dawson, T.M.; Dawson, V.L.; El-Deiry, W.S.; Fulda, S.; et al. Molecular Definitions of Cell Death Subroutines: Recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012, 19, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular Mechanisms of Cell Death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Vitale, I.; Pietrocola, F.; Guilbaud, E.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostini, M.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; et al. Apoptotic Cell Death in Disease—Current Understanding of the NCCD 2023. Cell Death Differ. 2023, 30, 1097–1154. [Google Scholar] [CrossRef]
- Jin, X.; Jin, W.; Tong, L.; Zhao, J.; Zhang, L.; Lin, N. Therapeutic Strategies of Targeting Non-Apoptotic Regulated Cell Death (RCD) with Small-Molecule Compounds in Cancer. Acta Pharm. Sin. B 2024, 14, 2815–2853. [Google Scholar] [CrossRef]
- He, R.; Liu, Y.; Fu, W.; He, X.; Liu, S.; Xiao, D.; Tao, Y. Mechanisms and Cross-Talk of Regulated Cell Death and Their Epigenetic Modifications in Tumor Progression. Mol. Cancer 2024, 23, 267. [Google Scholar] [CrossRef]
- Kardynska, M.; Kogut, D.; Pacholczyk, M.; Smieja, J. Mathematical Modeling of Regulatory Networks of Intracellular Processes–Aims and Selected Methods. Comput. Struct. Biotechnol. J. 2023, 21, 1523–1532. [Google Scholar] [CrossRef]
- Daun, S.; Rubin, J.; Vodovotz, Y.; Clermont, G. Equation-Based Models of Dynamic Biological Systems. J. Crit. Care 2008, 23, 585–594. [Google Scholar] [CrossRef]
- Städter, P.; Schälte, Y.; Schmiester, L.; Hasenauer, J.; Stapor, P.L. Benchmarking of Numerical Integration Methods for ODE Models of Biological Systems. Sci. Rep. 2021, 11, 2696. [Google Scholar] [CrossRef] [PubMed]
- Machado, D.; Costa, R.S.; Rocha, M.; Ferreira, E.C.; Tidor, B.; Rocha, I. Modeling Formalisms in Systems Biology. AMB Express 2011, 1, 45. [Google Scholar] [CrossRef]
- Rehm, M.; Huber, H.J.; Hellwig, C.T.; Anguissola, S.; Dussmann, H.; Prehn, J.H.M. Dynamics of Outer Mitochondrial Membrane Permeabilization during Apoptosis. Cell Death Differ. 2009, 16, 613–623. [Google Scholar] [CrossRef]
- Rehm, M.; Prehn, J.H.M. Systems Modelling Methodology for the Analysis of Apoptosis Signal Transduction and Cell Death Decisions. Methods 2013, 61, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Friedman, A.; Lai, X. Combination Therapy for Cancer with Oncolytic Virus and Checkpoint Inhibitor: A Mathematical Model. PLoS ONE 2018, 13, e0192449. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Yoo, J.Y.; Lee, T.J.; Liu, J.; Yu, J.; Caligiuri, M.A.; Kaur, B.; Friedman, A. Complex Role of NK Cells in Regulation of Oncolytic Virus–Bortezomib Therapy. Proc. Natl. Acad. Sci. USA 2018, 115, 4927–4932. [Google Scholar] [CrossRef]
- Wang, W.; Ren, X.; Wang, X. Spatial–Temporal Dynamics of a Novel PDE Model: Applications to Pharmacologic Inhibition of Pyroptosis by Necrosulfonamide. Commun. Nonlinear Sci. Numer. Simul. 2021, 103, 106025. [Google Scholar] [CrossRef]
- Mai, Z.; Liu, H. Boolean Network-Based Analysis of the Apoptosis Network: Irreversible Apoptosis and Stable Surviving. J. Theor. Biol. 2009, 259, 760–769. [Google Scholar] [CrossRef]
- Saez-Rodriguez, J.; Alexopoulos, L.G.; Epperlein, J.; Samaga, R.; Lauffenburger, D.A.; Klamt, S.; Sorger, P.K. Discrete Logic Modelling as a Means to Link Protein Signalling Networks with Functional Analysis of Mammalian Signal Transduction. Mol. Syst. Biol. 2009, 5, 331. [Google Scholar] [CrossRef] [PubMed]
- Schlatter, R.; Schmich, K.; Avalos Vizcarra, I.; Scheurich, P.; Sauter, T.; Borner, C.; Ederer, M.; Merfort, I.; Sawodny, O. ON/OFF and Beyond—A Boolean Model of Apoptosis. PLoS Comput. Biol. 2009, 5, e1000595. [Google Scholar] [CrossRef]
- Calzone, L.; Tournier, L.; Fourquet, S.; Thieffry, D.; Zhivotovsky, B.; Barillot, E.; Zinovyev, A. Mathematical Modelling of Cell-Fate Decision in Response to Death Receptor Engagement. PLoS Comput. Biol. 2010, 6, e1000702. [Google Scholar] [CrossRef]
- Checcoli, A.; Pol, J.G.; Naldi, A.; Noel, V.; Barillot, E.; Kroemer, G.; Thieffry, D.; Calzone, L.; Stoll, G. Dynamical Boolean Modeling of Immunogenic Cell Death. Front. Physiol. 2020, 11, 590479. [Google Scholar] [CrossRef]
- Montagud, A.; Béal, J.; Tobalina, L.; Traynard, P.; Subramanian, V.; Szalai, B.; Alföldi, R.; Puskás, L.; Valencia, A.; Barillot, E.; et al. Patient-Specific Boolean Models of Signalling Networks Guide Personalised Treatments. eLife 2022, 11, e72626. [Google Scholar] [CrossRef]
- Gupta, S.; Silveira, D.A.; Mombach, J.C.M.; Hashimoto, R.F. DNA Damage-Induced Ferroptosis: A Boolean Model Regulating P53 and Non-Coding RNAs in Drug Resistance. Proteomes 2025, 13, 6. [Google Scholar] [CrossRef]
- Chen, C.; Cui, J.; Lu, H.; Wang, R.; Zhang, S.; Shen, P. Modeling of the Role of a Bax-Activation Switch in the Mitochondrial Apoptosis Decision. Biophys. J. 2007, 92, 4304–4315. [Google Scholar] [CrossRef]
- Apte, A.; Bonchev, D.; Fong, S. Cellular Automata Modeling of FASL-Initiated Apoptosis. Chem. Biodivers. 2010, 7, 1163–1172. [Google Scholar] [CrossRef]
- Feuerwerker, S.; Cockrell, R.C.; An, G. Characterizing the Crosstalk Between Programmed Cell Death Pathways in Cytokine Storm with an Agent-Based Model. Surg. Infect. 2023, 24, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Surendran, A.; Jenner, A.L.; Karimi, E.; Fiset, B.; Quail, D.F.; Walsh, L.A.; Craig, M. Agent-Based Modelling Reveals the Role of the Tumor Microenvironment on the Short-Term Success of Combination Temozolomide/Immune Checkpoint Blockade to Treat Glioblastoma. J. Pharmacol. Exp. Ther. 2023, 387, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Spencer, S.L.; Sorger, P.K. Measuring and Modeling Apoptosis in Single Cells. Cell 2011, 144, 926–939. [Google Scholar] [CrossRef]
- Huber, H.J.; Duessmann, H.; Wenus, J.; Kilbride, S.M.; Prehn, J.H.M. Mathematical Modelling of the Mitochondrial Apoptosis Pathway. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2011, 1813, 608–615. [Google Scholar] [CrossRef]
- Schleich, K.; Lavrik, I.N. Mathematical Modeling of Apoptosis. Cell Commun. Signal. 2013, 11, 44. [Google Scholar] [CrossRef]
- Wu, L.; Jin, W.; Yu, H.; Liu, B. Modulating Autophagy to Treat Diseases: A Revisited Review on in Silico Methods. J. Adv. Res. 2024, 58, 175–191. [Google Scholar] [CrossRef]
- Galhuber, M.; Thedieck, K. ODE-Based Models of Signaling Networks in Autophagy. Curr. Opin. Syst. Biol. 2024, 39, 100519. [Google Scholar] [CrossRef]
- Mahlbacher, G.E.; Reihmer, K.C.; Frieboes, H.B. Mathematical Modeling of Tumor-Immune Cell Interactions. J. Theor. Biol. 2019, 469, 47–60. [Google Scholar] [CrossRef]
- Bekker, R.A.; Kim, S.; Pilon-Thomas, S.; Enderling, H. Mathematical Modeling of Radiotherapy and Its Impact on Tumor Interactions with the Immune System. Neoplasia 2022, 28, 100796. [Google Scholar] [CrossRef] [PubMed]
- Ng, A.M.; MacKinnon, K.M.; Cook, A.A.; D’Alonzo, R.A.; Rowshanfarzad, P.; Nowak, A.K.; Gill, S.; Ebert, M.A. Mechanistic in Silico Explorations of the Immunogenic and Synergistic Effects of Radiotherapy and Immunotherapy: A Critical Review. Phys. Eng. Sci. Med. 2024, 47, 1291–1306. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, M.; Clarke, R.; Kraikivski, P. Mathematical Models of Death Signaling Networks. Entropy 2022, 24, 1402. [Google Scholar] [CrossRef]
- Savageau, M.A. Biochemical Systems Theory: Operational Differences among Variant Representations and Their Significance. J. Theor. Biol. 1991, 151, 509–530. [Google Scholar] [CrossRef] [PubMed]
- Tyson, J.J.; Baumann, W.T.; Chen, C.; Verdugo, A.; Tavassoly, I.; Wang, Y.; Weiner, L.M.; Clarke, R. Dynamic Modelling of Oestrogen Signalling and Cell Fate in Breast Cancer Cells. Nat. Rev. Cancer 2011, 11, 523–532. [Google Scholar] [CrossRef]
- Zinovyev, A.; Fourquet, S.; Tournier, L.; Calzone, L.; Barillot, E. Cell Death and Life in Cancer: Mathematical Modeling of Cell Fate Decisions. In Advances in Systems Biology; Goryanin, I.I., Goryachev, A.B., Eds.; Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2012; Volume 736, pp. 261–274. ISBN 978-1-4419-7209-5. [Google Scholar]
- Parmar, J.H.; Cook, K.L.; Shajahan-Haq, A.N.; Clarke, P.A.G.; Tavassoly, I.; Clarke, R.; Tyson, J.J.; Baumann, W.T. Modelling the Effect of GRP78 on Anti-Oestrogen Sensitivity and Resistance in Breast Cancer. Interface Focus 2013, 3, 20130012. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, B.; Oltvai, Z.N.; Bayır, H.; Silverman, G.A.; Pak, S.C.; Perlmutter, D.H.; Bahar, I. Quantitative Assessment of Cell Fate Decision between Autophagy and Apoptosis. Sci. Rep. 2017, 7, 17605. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhong, C.-Q.; Wu, R.; Xu, X.; Yang, Z.-H.; Cai, S.; Wu, X.; Chen, X.; Yin, Z.; He, Q.; et al. RIP1-Dependent Linear and Nonlinear Recruitments of Caspase-8 and RIP3 Respectively to Necrosome Specify Distinct Cell Death Outcomes. Protein Cell 2021, 12, 858–876. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, P.; Xu, F.; Liu, Z.; Zhu, L.; Jin, J.; Qi, H.; Shuai, J.; Li, X. Cell Death Modes Are Specified by the Crosstalk Dynamics within Pyroptotic and Apoptotic Signaling. Chaos Interdiscip. J. Nonlinear Sci. 2021, 31, 093103. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, P.; Yin, Z.; Xu, F.; Yang, Z.-H.; Jin, J.; Qu, J.; Liu, Z.; Qi, H.; Yao, C.; et al. Caspase-1 and Gasdermin D Afford the Optimal Targets with Distinct Switching Strategies in NLRP1b Inflammasome-Induced Cell Death. Research 2022, 2022, 2022/9838341. [Google Scholar] [CrossRef]
- Mustafa, M.; Ahmad, R.; Tantry, I.Q.; Ahmad, W.; Siddiqui, S.; Alam, M.; Abbas, K.; Moinuddin; Hassan, M.d.I.; Habib, S.; et al. Apoptosis: A Comprehensive Overview of Signaling Pathways, Morphological Changes, and Physiological Significance and Therapeutic Implications. Cells 2024, 13, 1838. [Google Scholar] [CrossRef]
- Kroemer, G.; Petit, P.; Zamzami, N.; Vayssière, J.; Mignotte, B. The Biochemistry of Programmed Cell Death. FASEB J. 1995, 9, 1277–1287. [Google Scholar] [CrossRef]
- Pallardy, M.; Perrin-Wolff, M.; Biola, A. Cellular Stress and Apoptosis. Toxicol. Vitr. 1997, 11, 573–578. [Google Scholar] [CrossRef]
- Fulda, S.; Debatin, K.-M. Extrinsic versus Intrinsic Apoptosis Pathways in Anticancer Chemotherapy. Oncogene 2006, 25, 4798–4811. [Google Scholar] [CrossRef]
- Kashyap, D.; Garg, V.K.; Goel, N. Intrinsic and Extrinsic Pathways of Apoptosis: Role in Cancer Development and Prognosis. In Advances in Protein Chemistry and Structural Biology; Elsevier: Amsterdam, The Netherlands, 2021; Volume 125, pp. 73–120. ISBN 978-0-323-85315-6. [Google Scholar]
- Lavrik, I.N. Caspases: Pharmacological Manipulation of Cell Death. J. Clin. Investig. 2005, 115, 2665–2672. [Google Scholar] [CrossRef] [PubMed]
- Julien, O.; Wells, J.A. Caspases and Their Substrates. Cell Death Differ. 2017, 24, 1380–1389. [Google Scholar] [CrossRef]
- Harvey, K.J.; Blomquist, J.F.; Ucker, D.S. Commitment and Effector Phases of the Physiological Cell Death Pathway Elucidated with Respect to Bcl-2, Caspase, and Cyclin-Dependent Kinase Activities. Mol. Cell. Biol. 1998, 18, 2912–2922. [Google Scholar] [CrossRef][Green Version]
- Saraste, A. Morphologic and Biochemical Hallmarks of Apoptosis. Cardiovasc. Res. 2000, 45, 528–537. [Google Scholar] [CrossRef]
- Guicciardi, M.E.; Gores, G.J. Life and Death by Death Receptors. FASEB J. 2009, 23, 1625–1637. [Google Scholar] [CrossRef]
- Hillert-Richter, L.K.; Lavrik, I.N. Measuring Composition of CD95 Death-Inducing Signaling Complex and Processing of Procaspase-8 in This Complex. JoVE 2021, 174, 62842. [Google Scholar] [CrossRef]
- Shimizu, Y.; Taraborrelli, L.; Walczak, H. Linear Ubiquitination in Immunity. Immunol. Rev. 2015, 266, 190–207. [Google Scholar] [CrossRef]
- Micheau, O.; Tschopp, J. Induction of TNF Receptor I-Mediated Apoptosis via Two Sequential Signaling Complexes. Cell 2003, 114, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Micheau, O.; Shirley, S.; Dufour, F. Death Receptors as Targets in Cancer. Br. J. Pharmacol. 2013, 169, 1723–1744. [Google Scholar] [CrossRef]
- Guerrache, A.; Micheau, O. TNF-Related Apoptosis-Inducing Ligand: Non-Apoptotic Signalling. Cells 2024, 13, 521. [Google Scholar] [CrossRef]
- Huang, K.; Zhang, J.; O’Neill, K.L.; Gurumurthy, C.B.; Quadros, R.M.; Tu, Y.; Luo, X. Cleavage by Caspase 8 and Mitochondrial Membrane Association Activate the BH3-Only Protein Bid during TRAIL-Induced Apoptosis. J. Biol. Chem. 2016, 291, 11843–11851. [Google Scholar] [CrossRef]
- Garrido, C.; Galluzzi, L.; Brunet, M.; Puig, P.E.; Didelot, C.; Kroemer, G. Mechanisms of Cytochrome c Release from Mitochondria. Cell Death Differ. 2006, 13, 1423–1433. [Google Scholar] [CrossRef]
- Beaudouin, J.; Liesche, C.; Aschenbrenner, S.; Hörner, M.; Eils, R. Caspase-8 Cleaves Its Substrates from the Plasma Membrane upon CD95-Induced Apoptosis. Cell Death Differ. 2013, 20, 599–610. [Google Scholar] [CrossRef]
- Liao, W.-T.; Chang, K.-L.; Yu, C.-L.; Chen, G.-S.; Chang, L.W.; Yu, H.-S. Arsenic Induces Human Keratinocyte Apoptosis by the FAS/FAS Ligand Pathway, Which Correlates with Alterations in Nuclear Factor-κB and Activator Protein-1 Activity. J. Investig. Dermatol. 2004, 122, 125–129. [Google Scholar] [CrossRef]
- Chaitanya, G.V.; Alexander, J.S.; Babu, P.P. PARP-1 Cleavage Fragments: Signatures of Cell-Death Proteases in Neurodegeneration. Cell Commun. Signal. 2010, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Smulson, M.E.; Pang, D.; Jung, M.; Dimtchev, A.; Chasovskikh, S.; Spoonde, A.; Simbulan-Rosenthal, C.; Rosenthal, D.; Yakovlev, A.; Dritschilo, A. Irreversible Binding of Poly(ADP)Ribose Polymerase Cleavage Product to DNA Ends Revealed by Atomic Force Microscopy: Possible Role in Apoptosis. Cancer Res. 1998, 58, 3495–3498. [Google Scholar] [PubMed]
- Whelan, R.S.; Kaplinskiy, V.; Kitsis, R.N. Cell Death in the Pathogenesis of Heart Disease: Mechanisms and Significance. Annu. Rev. Physiol. 2010, 72, 19–44. [Google Scholar] [CrossRef]
- Renault, T.T.; Teijido, O.; Antonsson, B.; Dejean, L.M.; Manon, S. Regulation of Bax Mitochondrial Localization by Bcl-2 and Bcl-xL: Keep Your Friends Close but Your Enemies Closer. Int. J. Biochem. Cell Biol. 2013, 45, 64–67. [Google Scholar] [CrossRef]
- Dlugosz, P.J.; Billen, L.P.; Annis, M.G.; Zhu, W.; Zhang, Z.; Lin, J.; Leber, B.; Andrews, D.W. Bcl-2 Changes Conformation to Inhibit Bax Oligomerization. EMBO J. 2006, 25, 2287–2296. [Google Scholar] [CrossRef]
- Bennett, M.R. Apoptosis in the cardiovascular system. Heart 2002, 87, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Kantari, C.; Walczak, H. Caspase-8 and Bid: Caught in the Act between Death Receptors and Mitochondria. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2011, 1813, 558–563. [Google Scholar] [CrossRef]
- Scaffidi, C.; Schmitz, I.; Zha, J.; Korsmeyer, S.J.; Krammer, P.H.; Peter, M.E. Differential Modulation of Apoptosis Sensitivity in CD95 Type I and Type II Cells. J. Biol. Chem. 1999, 274, 22532–22538. [Google Scholar] [CrossRef] [PubMed]
- Krammer, P.H. CD95′s Deadly Mission in the Immune System. Nature 2000, 407, 789–795. [Google Scholar] [CrossRef]
- Barnhart, B.C.; Alappat, E.C.; Peter, M.E. The CD95 Type I/Type II Model. Semin. Immunol. 2003, 15, 185–193. [Google Scholar] [CrossRef]
- Fussenegger, M.; Bailey, J.E.; Varner, J. A Mathematical Model of Caspase Function in Apoptosis. Nat. Biotechnol. 2000, 18, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Stucki, J.W.; Simon, H.-U. Mathematical Modeling of the Regulation of Caspase-3 Activation and Degradation. J. Theor. Biol. 2005, 234, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Harrington, H.A.; Ho, K.L.; Ghosh, S.; Tung, K. Construction and Analysis of a Modular Model of Caspase Activation in Apoptosis. Theor. Biol. Med. Model. 2008, 5, 26. [Google Scholar] [CrossRef]
- Hong, J.-Y.; Kim, G.-H.; Kim, J.-W.; Kwon, S.-S.; Sato, E.F.; Cho, K.-H.; Shim, E.B. Computational Modeling of Apoptotic Signaling Pathways Induced by Cisplatin. BMC Syst. Biol. 2012, 6, 122. [Google Scholar] [CrossRef]
- Hendrata, M.; Sudiono, J. A Computational Model for Investigating Tumor Apoptosis Induced by Mesenchymal Stem Cell-Derived Secretome. Comput. Math. Methods Med. 2016, 2016, 4910603. [Google Scholar] [CrossRef]
- Bentele, M.; Lavrik, I.; Ulrich, M.; Stößer, S.; Heermann, D.W.; Kalthoff, H.; Krammer, P.H.; Eils, R. Mathematical Modeling Reveals Threshold Mechanism in CD95-Induced Apoptosis. J. Cell Biol. 2004, 166, 839–851. [Google Scholar] [CrossRef]
- Hua, F.; Cornejo, M.G.; Cardone, M.H.; Stokes, C.L.; Lauffenburger, D.A. Effects of Bcl-2 Levels on Fas Signaling-Induced Caspase-3 Activation: Molecular Genetic Tests of Computational Model Predictions. J. Immunol. 2005, 175, 985–995. [Google Scholar] [CrossRef]
- Hua, F.; Hautaniemi, S.; Yokoo, R.; Lauffenburger, D.A. Integrated Mechanistic and Data-Driven Modelling for Multivariate Analysis of Signalling Pathways. J. R. Soc. Interface 2006, 3, 515–526. [Google Scholar] [CrossRef]
- Okazaki, N.; Asano, R.; Kinoshita, T.; Chuman, H. Simple Computational Models of Type I/Type II Cells in Fas Signaling-Induced Apoptosis. J. Theor. Biol. 2008, 250, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Neumann, L.; Pforr, C.; Beaudouin, J.; Pappa, A.; Fricker, N.; Krammer, P.H.; Lavrik, I.N.; Eils, R. Dynamics within the CD95 Death-inducing Signaling Complex Decide Life and Death of Cells. Mol. Syst. Biol. 2010, 6, 352. [Google Scholar] [CrossRef]
- Fricker, N.; Beaudouin, J.; Richter, P.; Eils, R.; Krammer, P.H.; Lavrik, I.N. Model-Based Dissection of CD95 Signaling Dynamics Reveals Both a pro- and Antiapoptotic Role of c-FLIPL. J. Cell Biol. 2010, 190, 377–389. [Google Scholar] [CrossRef]
- Kutumova, E.; Zinovyev, A.; Sharipov, R.; Kolpakov, F. Model Composition through Model Reduction: A Combined Model of CD95 and NF-κB Signaling Pathways. BMC Syst. Biol. 2013, 7, 13. [Google Scholar] [CrossRef]
- Kutumova, E.; Kiselev, I.; Sharipov, R.; Lifshits, G.; Kolpakov, F. Thoroughly Calibrated Modular Agent-Based Model of the Human Cardiovascular and Renal Systems for Blood Pressure Regulation in Health and Disease. Front. Physiol. 2021, 12, 746300. [Google Scholar] [CrossRef]
- Kutumova, E.; Kiselev, I.; Sharipov, R.; Lifshits, G.; Kolpakov, F. Mathematical Modeling of Antihypertensive Therapy. Front. Physiol. 2022, 13, 1070115. [Google Scholar] [CrossRef] [PubMed]
- Kutumova, E.; Kovaleva, A.; Sharipov, R.; Lifshits, G.; Kolpakov, F. Mathematical Modelling of the Influence of ACE I/D Polymorphism on Blood Pressure and Antihypertensive Therapy. Heliyon 2024, 10, e29988. [Google Scholar] [CrossRef] [PubMed]
- Kutumova, E.; Kiselev, I.; Kolpakov, F. A Computational Model of Age-dependent Cardiomyocyte Apoptosis. J. Physiol. 2025, JP288853. [Google Scholar] [CrossRef]
- Wu, Q.; Finley, S.D. Predictive Model Identifies Strategies to Enhance TSP1-Mediated Apoptosis Signaling. Cell Commun. Signal. 2017, 15, 53. [Google Scholar] [CrossRef]
- Buchbinder, J.H.; Pischel, D.; Sundmacher, K.; Flassig, R.J.; Lavrik, I.N. Quantitative Single Cell Analysis Uncovers the Life/Death Decision in CD95 Network. PLoS Comput. Biol. 2018, 14, e1006368. [Google Scholar] [CrossRef]
- Hillert, L.K.; Ivanisenko, N.V.; Busse, D.; Espe, J.; König, C.; Peltek, S.E.; Kolchanov, N.A.; Ivanisenko, V.A.; Lavrik, I.N. Dissecting DISC Regulation via Pharmacological Targeting of Caspase-8/c-FLIPL Heterodimer. Cell Death Differ. 2020, 27, 2117–2130. [Google Scholar] [CrossRef] [PubMed]
- Ivanisenko, N.V.; Lavrik, I.N. Mathematical Modeling Reveals the Importance of the DED Filament Composition in the Effects of Small Molecules Targeting Caspase-8/c-FLIPL Heterodimer. Biochem. Mosc. 2020, 85, 1134–1144. [Google Scholar] [CrossRef] [PubMed]
- Eissing, T.; Conzelmann, H.; Gilles, E.D.; Allgöwer, F.; Bullinger, E.; Scheurich, P. Bistability Analyses of a Caspase Activation Model for Receptor-Induced Apoptosis. J. Biol. Chem. 2004, 279, 36892–36897. [Google Scholar] [CrossRef]
- Mangrum, D.S.; Finley, S.D. Modeling the Heterogeneous Apoptotic Response of Caspase-Mediated Signaling in Tumor Cells. J. Theor. Biol. 2024, 590, 111857. [Google Scholar] [CrossRef]
- Albeck, J.G.; Burke, J.M.; Spencer, S.L.; Lauffenburger, D.A.; Sorger, P.K. Modeling a Snap-Action, Variable-Delay Switch Controlling Extrinsic Cell Death. PLoS Biol. 2008, 6, e299. [Google Scholar] [CrossRef]
- Albeck, J.G.; Burke, J.M.; Aldridge, B.B.; Zhang, M.; Lauffenburger, D.A.; Sorger, P.K. Quantitative Analysis of Pathways Controlling Extrinsic Apoptosis in Single Cells. Mol. Cell 2008, 30, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wu, M.; Chen, Q.; Sun, Z. Investigation into the Regulation Mechanisms of TRAIL Apoptosis Pathway by Mathematical Modeling. ABBS 2010, 42, 98–108. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Laussmann, M.A.; Passante, E.; Hellwig, C.T.; Tomiczek, B.; Flanagan, L.; Prehn, J.H.M.; Huber, H.J.; Rehm, M. Proteasome Inhibition Can Impair Caspase-8 Activation upon Submaximal Stimulation of Apoptotic Tumor Necrosis Factor-Related Apoptosis Inducing Ligand (TRAIL) Signaling. J. Biol. Chem. 2012, 287, 14402–14411. [Google Scholar] [CrossRef]
- Anderson, M.W.; Moss, J.J.; Szalai, R.; Lane, J.D. Mathematical Modeling Highlights the Complex Role of AKT in TRAIL-Induced Apoptosis of Colorectal Carcinoma Cells. iScience 2019, 12, 182–193. [Google Scholar] [CrossRef]
- Cho, K.-H.; Shin, S.-Y.; Lee, H.-W.; Wolkenhauer, O. Investigations Into the Analysis and Modeling of the TNFα-Mediated NF-κB-Signaling Pathway. Genome Res. 2003, 13, 2413–2422. [Google Scholar] [CrossRef]
- Rangamani, P.; Sirovich, L. Survival and Apoptotic Pathways Initiated by TNF-α: Modeling and Predictions. Biotech Bioeng. 2007, 97, 1216–1229. [Google Scholar] [CrossRef]
- Koh, G.; Lee, D.-Y. Mathematical Modeling and Sensitivity Analysis of the Integrated TNFα-Mediated Apoptotic Pathway for Identifying Key Regulators. Comput. Biol. Med. 2011, 41, 512–528. [Google Scholar] [CrossRef]
- Schliemann, M.; Bullinger, E.; Borchers, S.; Allgöwer, F.; Findeisen, R.; Scheurich, P. Heterogeneity Reduces Sensitivity of Cell Death for TNF-Stimuli. BMC Syst. Biol. 2011, 5, 204. [Google Scholar] [CrossRef] [PubMed]
- Halder, S.; Chatterjee, S. Bistability Regulates TNFR2-Mediated Survival and Death of T-Regulatory Cells. J. Biol. Phys. 2023, 49, 95–119. [Google Scholar] [CrossRef] [PubMed]
- Bagci, E.Z.; Vodovotz, Y.; Billiar, T.R.; Ermentrout, G.B.; Bahar, I. Bistability in Apoptosis: Roles of Bax, Bcl-2, and Mitochondrial Permeability Transition Pores. Biophys. J. 2006, 90, 1546–1559. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, S.K.; Roy, A.M.; Baliga, M.S. Silymarin Induces Apoptosis Primarily through a P53-Dependent Pathway Involving Bcl-2/Bax, Cytochrome c Release, and Caspase Activation. Mol. Cancer Ther. 2005, 4, 207–216. [Google Scholar] [CrossRef]
- Bagci, E.Z.; Vodovotz, Y.; Billiar, T.R.; Ermentrout, B.; Bahar, I. Computational Insights on the Competing Effects of Nitric Oxide in Regulating Apoptosis. PLoS ONE 2008, 3, e2249. [Google Scholar] [CrossRef]
- Hamada, H.; Tashima, Y.; Kisaka, Y.; Iwamoto, K.; Hanai, T.; Eguchi, Y.; Okamoto, M. Sophisticated Framework between Cell Cycle Arrest and Apoptosis Induction Based on P53 Dynamics. PLoS ONE 2009, 4, e4795. [Google Scholar] [CrossRef]
- Zhang, T.; Brazhnik, P.; Tyson, J.J. Computational Analysis of Dynamical Responses to the Intrinsic Pathway of Programmed Cell Death. Biophys. J. 2009, 97, 415–434. [Google Scholar] [CrossRef]
- Paek, A.L.; Liu, J.C.; Loewer, A.; Forrester, W.C.; Lahav, G. Cell-to-Cell Variation in P53 Dynamics Leads to Fractional Killing. Cell 2016, 165, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Ballweg, R.; Paek, A.L.; Zhang, T. A Dynamical Framework for Complex Fractional Killing. Sci. Rep. 2017, 7, 8002. [Google Scholar] [CrossRef]
- McKenna, S.; García-Gutiérrez, L.; Matallanas, D.; Fey, D. BAX and SMAC Regulate Bistable Properties of the Apoptotic Caspase System. Sci. Rep. 2021, 11, 3272. [Google Scholar] [CrossRef] [PubMed]
- Rehm, M.; Huber, H.J.; Dussmann, H.; Prehn, J.H.M. Systems Analysis of Effector Caspase Activation and Its Control by X-Linked Inhibitor of Apoptosis Protein. EMBO J. 2006, 25, 4338–4349. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.; Lin, S.; Ugel, N.; Antoniotti, M.; Mishra, B. Mathematical Modeling of the Formation of Apoptosome in Intrinsic Pathway of Apoptosis. Syst. Synth. Biol. 2008, 2, 49–66. [Google Scholar] [CrossRef]
- Rodriguez, J.; Lazebnik, Y. Caspase-9 and APAF-1 Form an Active Holoenzyme. Genes Dev. 1999, 13, 3179–3184. [Google Scholar] [CrossRef]
- Legewie, S.; Blüthgen, N.; Herzel, H. Mathematical Modeling Identifies Inhibitors of Apoptosis as Mediators of Positive Feedback and Bistability. PLoS Comput. Biol. 2006, 2, e120. [Google Scholar] [CrossRef]
- Ooi, H.; Ma, L. Modeling Heterogeneous Responsiveness of Intrinsic Apoptosis Pathway. BMC Syst. Biol. 2013, 7, 65. [Google Scholar] [CrossRef]
- Burt, P.; Cornelis, R.; Geißler, G.; Hahne, S.; Radbruch, A.; Chang, H.-D.; Thurley, K. Data-Driven Mathematical Model of Apoptosis Regulation in Memory Plasma Cells. Cells 2022, 11, 1547. [Google Scholar] [CrossRef]
- Kutumova, E.O.; Kiselev, I.N.; Sharipov, R.N.; Lavrik, I.N.; Kolpakov, F.A. A Modular Model of the Apoptosis Machinery. In Advances in Systems Biology; Goryanin, I.I., Goryachev, A.B., Eds.; Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2012; Volume 736, pp. 235–245. ISBN 978-1-4419-7209-5. [Google Scholar]
- Liu, S.; Yao, S.; Yang, H.; Liu, S.; Wang, Y. Autophagy: Regulator of Cell Death. Cell Death Dis. 2023, 14, 648. [Google Scholar] [CrossRef]
- Khandia, R.; Dadar, M.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Iqbal, H.M.N.; Singh, K.P.; Joshi, S.K.; et al. A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells 2019, 8, 674. [Google Scholar] [CrossRef]
- Deter, R.L. Analog Modeling of Glucagon-Induced Autophagy in Rat Liver. Exp. Cell Res. 1975, 94, 122–126. [Google Scholar] [CrossRef]
- Han, K.; Kim, J.; Choi, M. Autophagy Mediates Phase Transitions from Cell Death to Life. Heliyon 2015, 1, e00027. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Kim, S.H.; Choi, M. Computational Modeling of the Effects of Autophagy on Amyloid-β Peptide Levels. Theor. Biol Med. Model. 2020, 17, 2. [Google Scholar] [CrossRef]
- Dalle Pezze, P.; Ruf, S.; Sonntag, A.G.; Langelaar-Makkinje, M.; Hall, P.; Heberle, A.M.; Razquin Navas, P.; Van Eunen, K.; Tölle, R.C.; Schwarz, J.J.; et al. A Systems Study Reveals Concurrent Activation of AMPK and mTOR by Amino Acids. Nat. Commun. 2016, 7, 13254. [Google Scholar] [CrossRef] [PubMed]
- Ichimiya, T.; Yamakawa, T.; Hirano, T.; Yokoyama, Y.; Hayashi, Y.; Hirayama, D.; Wagatsuma, K.; Itoi, T.; Nakase, H. Autophagy and Autophagy-Related Diseases: A Review. Int. J. Mol. Sci. 2020, 21, 8974. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Goronzy, J.J.; Weyand, C.M. Autophagy in Autoimmune Disease. J. Mol. Med. 2015, 93, 707–717. [Google Scholar] [CrossRef]
- Giovedì, S.; Ravanelli, M.M.; Parisi, B.; Bettegazzi, B.; Guarnieri, F.C. Dysfunctional Autophagy and Endolysosomal System in Neurodegenerative Diseases: Relevance and Therapeutic Options. Front. Cell Neurosci. 2020, 14, 602116. [Google Scholar] [CrossRef]
- Jiang, P.; Mizushima, N. Autophagy and Human Diseases. Cell Res. 2014, 24, 69–79. [Google Scholar] [CrossRef]
- Cheng, Y.; Ren, X.; Hait, W.N.; Yang, J.-M. Therapeutic Targeting of Autophagy in Disease: Biology and Pharmacology. Pharmacol. Rev. 2013, 65, 1162–1197. [Google Scholar] [CrossRef]
- Yuan, W.; Fang, W.; Zhang, R.; Lyu, H.; Xiao, S.; Guo, D.; Ali, D.W.; Michalak, M.; Chen, X.-Z.; Zhou, C.; et al. Therapeutic Strategies Targeting AMPK-Dependent Autophagy in Cancer Cells. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2023, 1870, 119537. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Yan, L.; Liao, N.; Wu, W.-Q.; Shi, J.-L. A Review of ULK1-Mediated Autophagy in Drug Resistance of Cancer. Cancers 2020, 12, 352. [Google Scholar] [CrossRef]
- Shirin, A.; Klickstein, I.S.; Feng, S.; Lin, Y.T.; Hlavacek, W.S.; Sorrentino, F. Prediction of Optimal Drug Schedules for Controlling Autophagy. Sci. Rep. 2019, 9, 1428. [Google Scholar] [CrossRef] [PubMed]
- Dalle Pezze, P.; Karanasios, E.; Kandia, V.; Manifava, M.; Walker, S.A.; Gambardella Le Novère, N.; Ktistakis, N.T. ATG13 Dynamics in Nonselective Autophagy and Mitophagy: Insights from Live Imaging Studies and Mathematical Modeling. Autophagy 2021, 17, 1131–1141. [Google Scholar] [CrossRef]
- Sadria, M.; Layton, A.T. Interactions among mTORC, AMPK and SIRT: A Computational Model for Cell Energy Balance and Metabolism. Cell Commun. Signal. 2021, 19, 57. [Google Scholar] [CrossRef] [PubMed]
- Sadria, M.; Seo, D.; Layton, A.T. The Mixed Blessing of AMPK Signaling in Cancer Treatments. BMC Cancer 2022, 22, 105. [Google Scholar] [CrossRef]
- Mehta, K.; Guo, T.; Wallis, R.S.; Van Der Graaf, P.H.; Van Hasselt, J.G.C. Quantitative Systems Pharmacology Modeling Framework of Autophagy in Tuberculosis: Application to Adjunctive Metformin Host-Directed Therapy. Antimicrob. Agents Chemother. 2022, 66, e00366-22. [Google Scholar] [CrossRef]
- Tavassoly, I.; Parmar, J.; Shajahan-Haq, A.; Clarke, R.; Baumann, W.; Tyson, J. Dynamic Modeling of the Interaction Between Autophagy and Apoptosis in Mammalian Cells. CPT Pharmacom. Syst. Pharma. 2015, 4, 263–272. [Google Scholar] [CrossRef]
- Cook, K.L.; Clarke, P.A.G.; Parmar, J.; Hu, R.; Schwartz-Roberts, J.L.; Abu-Asab, M.; Wärri, A.; Baumann, W.T.; Clarke, R. Knockdown of Estrogen Receptor-α Induces Autophagy and Inhibits Antiestrogen-mediated Unfolded Protein Response Activation, Promoting ROS-induced Breast Cancer Cell Death. FASEB J. 2014, 28, 3891–3905. [Google Scholar] [CrossRef]
- Levine, B.; Sinha, S.C.; Kroemer, G. Bcl-2 Family Members: Dual Regulators of Apoptosis and Autophagy. Autophagy 2008, 4, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yang, Y.; Xing, D. Bcl-2 and Bcl-xL Play Important Roles in the Crosstalk between Autophagy and Apoptosis. FEBS J. 2011, 278, 403–413. [Google Scholar] [CrossRef]
- Kapuy, O.; Vinod, P.K.; Mandl, J.; Bánhegyi, G. A Cellular Stress-Directed Bistable Switch Controls the Crosstalk between Autophagy and Apoptosis. Mol. BioSyst. 2013, 9, 296–306. [Google Scholar] [CrossRef]
- Holczer, M.; Hajdú, B.; Lőrincz, T.; Szarka, A.; Bánhegyi, G.; Kapuy, O. A Double Negative Feedback Loop between mTORC1 and AMPK Kinases Guarantees Precise Autophagy Induction upon Cellular Stress. Int. J. Mol. Sci. 2019, 20, 5543. [Google Scholar] [CrossRef]
- Szymańska, P.; Martin, K.R.; MacKeigan, J.P.; Hlavacek, W.S.; Lipniacki, T. Computational Analysis of an Autophagy/Translation Switch Based on Mutual Inhibition of MTORC1 and ULK1. PLoS ONE 2015, 10, e0116550. [Google Scholar] [CrossRef]
- Holczer, M.; Hajdú, B.; Lőrincz, T.; Szarka, A.; Bánhegyi, G.; Kapuy, O. Fine-Tuning of AMPK–ULK1–mTORC1 Regulatory Triangle Is Crucial for Autophagy Oscillation. Sci. Rep. 2020, 10, 17803. [Google Scholar] [CrossRef]
- Kapuy, O.; Holczer, M.; Márton, M.; Korcsmáros, T. Autophagy-Dependent Survival Is Controlled with a Unique Regulatory Network upon Various Cellular Stress Events. Cell Death Dis. 2021, 12, 309. [Google Scholar] [CrossRef]
- Hajdú, B.; Holczer, M.; Horváth, G.; Szederkényi, G.; Kapuy, O. Fine-Tuning of mTORC1-ULK1-PP2A Regulatory Triangle Is Crucial for Robust Autophagic Response upon Cellular Stress. Biomolecules 2022, 12, 1587. [Google Scholar] [CrossRef]
- Hajdú, B.; Csabai, L.; Márton, M.; Holczer, M.; Korcsmáros, T.; Kapuy, O. Oscillation of Autophagy Induction under Cellular Stress and What Lies behind It, a Systems Biology Study. Int. J. Mol. Sci. 2023, 24, 7671. [Google Scholar] [CrossRef] [PubMed]
- Holczer, M.; Besze, B.; Lehel, A.; Kapuy, O. The Dual Role of Sulforaphane-Induced Cellular Stress—A Systems Biological Study. Int. J. Mol. Sci. 2024, 25, 1220. [Google Scholar] [CrossRef] [PubMed]
- Kapuy, O.; Holczer, M.; Csabai, L.; Korcsmáros, T. Oscillatory Autophagy Induction Is Enabled by an Updated AMPK-ULK1 Regulatory Wiring. PLoS ONE 2024, 19, e0313302. [Google Scholar] [CrossRef] [PubMed]
- Kapuy, O.; Papp, D.; Vellai, T.; Bánhegyi, G.; Korcsmáros, T. Systems-Level Feedbacks of NRF2 Controlling Autophagy upon Oxidative Stress Response. Antioxidants 2018, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Márton, M.; Kurucz, A.; Lizák, B.; Margittai, É.; Bánhegyi, G.; Kapuy, O. A Systems Biological View of Life-and-Death Decision with Respect to Endoplasmic Reticulum Stress—The Role of PERK Pathway. Int. J. Mol. Sci. 2017, 18, 58. [Google Scholar] [CrossRef] [PubMed]
- Kapuy, O.; Márton, M.; Bánhegyi, G.; Vinod, P.K. Multiple System-level Feedback Loops Control Life-and-death Decisions in Endoplasmic Reticulum Stress. FEBS Lett. 2020, 594, 1112–1123. [Google Scholar] [CrossRef]
- Márton, M.; Bánhegyi, G.; Gyöngyösi, N.; Kálmán, E.É.; Pettkó-Szandtner, A.; Káldi, K.; Kapuy, O. A Systems Biological Analysis of the ATF4-GADD34-CHOP Regulatory Triangle upon Endoplasmic Reticulum Stress. FEBS Open Bio. 2022, 12, 2065–2082. [Google Scholar] [CrossRef]
- Kapuy, O.; Korcsmáros, T. Chloroquine and COVID-19—A Systems Biology Model Uncovers the Drug’s Detrimental Effect on Autophagy and Explains Its Failure. PLoS ONE 2022, 17, e0266337. [Google Scholar] [CrossRef]
- Kapuy, O.; Makk-Merczel, K.; Szarka, A. Therapeutic Approach of KRAS Mutant Tumours by the Combination of Pharmacologic Ascorbate and Chloroquine. Biomolecules 2021, 11, 652. [Google Scholar] [CrossRef]
- Kapuy, O.; Vinod, P.K.; Bánhegyi, G. mTOR Inhibition Increases Cell Viability via Autophagy Induction during Endoplasmic Reticulum Stress—An Experimental and Modeling Study. FEBS Open Bio. 2014, 4, 704–713. [Google Scholar] [CrossRef]
- Holczer, M.; Márton, M.; Kurucz, A.; Bánhegyi, G.; Kapuy, O. A Comprehensive Systems Biological Study of Autophagy-Apoptosis Crosstalk during Endoplasmic Reticulum Stress. BioMed Res. Int. 2015, 2015, 319589. [Google Scholar] [CrossRef]
- Li, X.; Lyu, Y.; Li, J.; Wang, X. AMBRA1 and Its Role as a Target for Anticancer Therapy. Front. Oncol. 2022, 12, 946086. [Google Scholar] [CrossRef]
- Yang, B.; Liu, Q.; Bi, Y. Autophagy and Apoptosis Are Regulated by Stress on Bcl2 by AMBRA1 in the Endoplasmic Reticulum and Mitochondria. Theor. Biol. Med. Model. 2019, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Yang, Z. Deterministic and Stochastic Approaches to a Minimal Model for the Transition from Autophagy to Apoptosis. Math. Biosci. Eng. 2024, 21, 3207–3228. [Google Scholar] [CrossRef]
- Yang, B.; Yang, Z.; Hao, L. Dynamics of a Model for the Degradation Mechanism of Aggregated α-Synuclein in Parkinson’s Disease. Front. Comput. Neurosci. 2023, 17, 1068150. [Google Scholar] [CrossRef]
- Sukumaran, P.; Nascimento Da Conceicao, V.; Sun, Y.; Ahamad, N.; Saraiva, L.R.; Selvaraj, S.; Singh, B.B. Calcium Signaling Regulates Autophagy and Apoptosis. Cells 2021, 10, 2125. [Google Scholar] [CrossRef]
- Decuypere, J.-P.; Bultynck, G.; Parys, J.B. A Dual Role for Ca2+ in Autophagy Regulation. Cell Calcium 2011, 50, 242–250. [Google Scholar] [CrossRef]
- Hajdú, B.; Kapuy, O.; Nagy, T. Basal State Calibration of a Chemical Reaction Network Model for Autophagy. Int. J. Mol. Sci. 2024, 25, 11316. [Google Scholar] [CrossRef]
- Ge, Z.; Wang, R. Fate Decisions Mediated by Crosstalk of Autophagy and Apoptosis in Mammalian Cells. J. Biol. Phys. 2020, 46, 133–149. [Google Scholar] [CrossRef]
- Sarmah, D.T.; Bairagi, N.; Chatterjee, S. The Interplay between DNA Damage and Autophagy in Lung Cancer: A Mathematical Study. Biosystems 2021, 206, 104443. [Google Scholar] [CrossRef] [PubMed]
- Schwartz-Roberts, J.L.; Cook, K.L.; Chen, C.; Shajahan-Haq, A.N.; Axelrod, M.; Wärri, A.; Riggins, R.B.; Jin, L.; Haddad, B.R.; Kallakury, B.V.; et al. Interferon Regulatory Factor-1 Signaling Regulates the Switch between Autophagy and Apoptosis to Determine Breast Cancer Cell Fate. Cancer Res. 2015, 75, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Pavel, M.; Park, S.J.; Frake, R.A.; Son, S.M.; Manni, M.M.; Bento, C.F.; Renna, M.; Ricketts, T.; Menzies, F.M.; Tanasa, R.; et al. α-Catenin Levels Determine Direction of YAP/TAZ Response to Autophagy Perturbation. Nat. Commun. 2021, 12, 1703. [Google Scholar] [CrossRef]
- Zhou, Q.; Meng, Y.; Li, D.; Yao, L.; Le, J.; Liu, Y.; Sun, Y.; Zeng, F.; Chen, X.; Deng, G. Ferroptosis in Cancer: From Molecular Mechanisms to Therapeutic Strategies. Signal Transduct. Target. Ther. 2024, 9, 55. [Google Scholar] [CrossRef]
- Berndt, C.; Alborzinia, H.; Amen, V.S.; Ayton, S.; Barayeu, U.; Bartelt, A.; Bayir, H.; Bebber, C.M.; Birsoy, K.; Böttcher, J.P.; et al. Ferroptosis in Health and Disease. Redox Biol. 2024, 75, 103211. [Google Scholar] [CrossRef]
- Dixon, S.J.; Olzmann, J.A. The Cell Biology of Ferroptosis. Nat. Rev. Mol. Cell Biol. 2024, 25, 424–442. [Google Scholar] [CrossRef]
- Jin, X.; Tang, J.; Qiu, X.; Nie, X.; Ou, S.; Wu, G.; Zhang, R.; Zhu, J. Ferroptosis: Emerging Mechanisms, Biological Function, and Therapeutic Potential in Cancer and Inflammation. Cell Death Discov. 2024, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Zhang, X.; Duan, D.; Zhao, L. Organelle-Specific Mechanisms in Crosstalk between Apoptosis and Ferroptosis. Oxidative Med. Cell. Longev. 2023, 2023, 3400147. [Google Scholar] [CrossRef]
- Eskander, G.; Abdelhamid, S.G.; Wahdan, S.A.; Radwan, S.M. Insights on the Crosstalk among Different Cell Death Mechanisms. Cell Death Discov. 2025, 11, 56. [Google Scholar] [CrossRef]
- Qiu, Y.; Hüther, J.A.; Wank, B.; Rath, A.; Tykwe, R.; Aldrovandi, M.; Henkelmann, B.; Mergner, J.; Nakamura, T.; Laschat, S.; et al. Interplay of Ferroptotic and Apoptotic Cell Death and Its Modulation by BH3-Mimetics. Cell Death Differ. 2025, 32, 1970–1985. [Google Scholar] [CrossRef]
- Kagan, V.E.; Mao, G.; Qu, F.; Angeli, J.P.F.; Doll, S.; Croix, C.S.; Dar, H.H.; Liu, B.; Tyurin, V.A.; Ritov, V.B.; et al. Oxidized Arachidonic and Adrenic PEs Navigate Cells to Ferroptosis. Nat. Chem. Biol. 2017, 13, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Kapralov, A.A.; Yang, Q.; Dar, H.H.; Tyurina, Y.Y.; Anthonymuthu, T.S.; Kim, R.; St. Croix, C.M.; Mikulska-Ruminska, K.; Liu, B.; Shrivastava, I.H.; et al. Redox Lipid Reprogramming Commands Susceptibility of Macrophages and Microglia to Ferroptotic Death. Nat. Chem. Biol. 2020, 16, 278–290. [Google Scholar] [CrossRef]
- Arbatskiy, M.; Balandin, D.; Akberdin, I.; Churov, A. A Systems Biology Approach Towards a Comprehensive Understanding of Ferroptosis. Int. J. Mol. Sci. 2024, 25, 11782. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Yan, Y.; Yang, C.; Cai, H. Immunogenic Cell Death and Metabolic Reprogramming in Cancer: Mechanisms, Synergies, and Innovative Therapeutic Strategies. Biomedicines 2025, 13, 950. [Google Scholar] [CrossRef]
- Arimoto, K.; Miyauchi, S.; Liu, M.; Zhang, D.-E. Emerging Role of Immunogenic Cell Death in Cancer Immunotherapy. Front. Immunol. 2024, 15, 1390263. [Google Scholar] [CrossRef]
- Galluzzi, L.; Guilbaud, E.; Schmidt, D.; Kroemer, G.; Marincola, F.M. Targeting Immunogenic Cell Stress and Death for Cancer Therapy. Nat. Rev. Drug. Discov. 2024, 23, 445–460. [Google Scholar] [CrossRef]
- Rodrigues, M.C.; Morais, J.A.V.; Ganassin, R.; Oliveira, G.R.T.; Costa, F.C.; Morais, A.A.C.; Silveira, A.P.; Silva, V.C.M.; Longo, J.P.F.; Muehlmann, L.A. An Overview on Immunogenic Cell Death in Cancer Biology and Therapy. Pharmaceutics 2022, 14, 1564. [Google Scholar] [CrossRef]
- Ahmed, A.; Tait, S.W.G. Targeting Immunogenic Cell Death in Cancer. Mol. Oncol. 2020, 14, 2994–3006. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Liu, Y.; Chen, D.; Sun, Y.; Li, D.; Meng, Y.; Zhou, Q.; Zeng, F.; Deng, G.; Chen, X. Targeting Regulated Cell Death: Apoptosis, Necroptosis, Pyroptosis, Ferroptosis, and Cuproptosis in Anticancer Immunity. J. Transl. Intern. Med. 2025, 13, 10–32. [Google Scholar] [CrossRef] [PubMed]
- Vandenabeele, P.; Vandecasteele, K.; Bachert, C.; Krysko, O.; Krysko, D.V. Immunogenic Apoptotic Cell Death and Anticancer Immunity. In Apoptosis in Cancer Pathogenesis and Anti-Cancer Therapy; Gregory, C.D., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2016; Volume 930, pp. 133–149. ISBN 978-3-319-39404-6. [Google Scholar]
- Montico, B.; Nigro, A.; Casolaro, V.; Dal Col, J. Immunogenic Apoptosis as a Novel Tool for Anticancer Vaccine Development. Int. J. Mol. Sci. 2018, 19, 594. [Google Scholar] [CrossRef] [PubMed]
- Hänggi, K.; Ruffell, B. Cell Death, Therapeutics, and the Immune Response in Cancer. Trends Cancer 2023, 9, 381–396. [Google Scholar] [CrossRef]
- DeLisi, C.; Rescigno, A. Immune Surveillance and Neoplasia—1 a Minimal Mathematical Model. Bull. Math. Biol. 1977, 39, 201–221. [Google Scholar] [CrossRef]
- Rescigno, A.; DeLisi, C. Immune Surveillance and Neoplasia—II A Two-Stage Mathematical Model. Bull. Math. Biol. 1977, 39, 487–497. [Google Scholar] [CrossRef]
- Deichman, G.I.; Kluchareva, T.E.; Kashkina, L.M.; Matveeva, V.A. Reproducibility and Relation to Specific and Non-specific Anti-tumor Resistance of the Tumor “Sneaking through” Phenomenon. Int. J. Cancer 1979, 23, 571–584. [Google Scholar] [CrossRef]
- Gatenby, P.A.; Basten, A.; Creswick, P. “Sneaking through”: A T-Cell-Dependent Phenomenon. Br. J. Cancer 1981, 44, 753–756. [Google Scholar] [CrossRef][Green Version]
- Grossman, Z.; Berke, G. Tumor Escape from Immune Elimination. J. Theor. Biol. 1980, 83, 267–296. [Google Scholar] [CrossRef]
- De Boer, R.J.; Hogeweg, P. Tumor Escape from Immune Elimination: Simplified Precursor Bound Cytotoxicity Models. J. Theor. Biol. 1985, 113, 719–736. [Google Scholar] [CrossRef][Green Version]
- De Boer, R.J.; Hogeweg, P.; Dullens, H.F.; De Weger, R.A.; Den Otter, W. Macrophage T Lymphocyte Interactions in the Anti-Tumor Immune Response: A Mathematical Model. J. Immunol. 1985, 134, 2748–2758. [Google Scholar] [CrossRef]
- De Boer, R.J.; Hogeweg, P. Interactions between Macrophages and T-Lymphocytes: Tumor Sneaking through Intrinsic to Helper T Cell Dynamics. J. Theor. Biol. 1986, 120, 331–351. [Google Scholar] [CrossRef]
- Kuznetsov, V.A.; Makalkin, I.A.; Taylor, M.A.; Perelson, A.S. Nonlinear Dynamics of Immunogenic Tumors: Parameter Estimation and Global Bifurcation Analysis. Bull. Math. Biol. 1994, 56, 295–321. [Google Scholar] [CrossRef]
- El Wajeh, M.; Jung, F.; Bongartz, D.; Kappatou, C.D.; Ghaffari Laleh, N.; Mitsos, A.; Kather, J.N. Can the Kuznetsov Model Replicate and Predict Cancer Growth in Humans? Bull. Math. Biol. 2022, 84, 130. [Google Scholar] [CrossRef]
- Moore, H.; Li, N.K. A Mathematical Model for Chronic Myelogenous Leukemia (CML) and T Cell Interaction. J. Theor. Biol. 2004, 227, 513–523. [Google Scholar] [CrossRef]
- De Pillis, L.G.; Radunskaya, A.E.; Wiseman, C.L. A Validated Mathematical Model of Cell-Mediated Immune Response to Tumor Growth. Cancer Res. 2005, 65, 7950–7958. [Google Scholar] [CrossRef]
- Adam, J.A. Effects of Vascularization on Lymphocyte/Tumor Cell Dynamics: Qualitative Features. Math. Comput. Model. 1996, 23, 1–10. [Google Scholar] [CrossRef][Green Version]
- Hatzikirou, H.; Alfonso, J.C.L.; Mühle, S.; Stern, C.; Weiss, S.; Meyer-Hermann, M. Cancer Therapeutic Potential of Combinatorial Immuno- and Vasomodulatory Interventions. J. R. Soc. Interface 2015, 12, 20150439. [Google Scholar] [CrossRef]
- Kirshtein, A.; Akbarinejad, S.; Hao, W.; Le, T.; Su, S.; Aronow, R.A.; Shahriyari, L. Data Driven Mathematical Model of Colon Cancer Progression. J. Clin. Med. 2020, 9, 3947. [Google Scholar] [CrossRef]
- Sofia, D.; Mohammad Mirzaei, N.; Shahriyari, L. Patient-Specific Mathematical Model of the Clear Cell Renal Cell Carcinoma Microenvironment. J. Pers. Med. 2022, 12, 1681. [Google Scholar] [CrossRef]
- Kuznetsov, V.A.; Knott, G.D. Modeling Tumor Regrowth and Immunotherapy. Math. Comput. Model. 2001, 33, 1275–1287. [Google Scholar] [CrossRef]
- Poleszczuk, J.T.; Luddy, K.A.; Prokopiou, S.; Robertson-Tessi, M.; Moros, E.G.; Fishman, M.; Djeu, J.Y.; Finkelstein, S.E.; Enderling, H. Abscopal Benefits of Localized Radiotherapy Depend on Activated T-Cell Trafficking and Distribution between Metastatic Lesions. Cancer Res. 2016, 76, 1009–1018. [Google Scholar] [CrossRef]
- Kosinsky, Y.; Dovedi, S.J.; Peskov, K.; Voronova, V.; Chu, L.; Tomkinson, H.; Al-Huniti, N.; Stanski, D.R.; Helmlinger, G. Radiation and PD-(L)1 Treatment Combinations: Immune Response and Dose Optimization via a Predictive Systems Model. J. Immunother. Cancer 2018, 6, 17. [Google Scholar] [CrossRef]
- Poleszczuk, J.; Enderling, H. The Optimal Radiation Dose to Induce Robust Systemic Anti-Tumor Immunity. Int. J. Mol. Sci. 2018, 19, 3377. [Google Scholar] [CrossRef]
- Nikolopoulou, E.; Johnson, L.; Harris, D.; Nagy, J.; Stites, E.; Kuang, Y. Tumour-Immune Dynamics with an Immune Checkpoint Inhibitor. LiB 2018, 5, 137–159. [Google Scholar] [CrossRef]
- Byun, J.H.; Yoon, I.-S.; Jeong, Y.D.; Kim, S.; Jung, I.H. A Tumor-Immune Interaction Model for Synergistic Combinations of Anti PD-L1 and Ionizing Irradiation Treatment. Pharmaceutics 2020, 12, 830. [Google Scholar] [CrossRef]
- Storey, K.M.; Lawler, S.E.; Jackson, T.L. Modeling Oncolytic Viral Therapy, Immune Checkpoint Inhibition, and the Complex Dynamics of Innate and Adaptive Immunity in Glioblastoma Treatment. Front. Physiol. 2020, 11, 151. [Google Scholar] [CrossRef]
- Okuneye, K.; Bergman, D.; Bloodworth, J.C.; Pearson, A.T.; Sweis, R.F.; Jackson, T.L. A Validated Mathematical Model of FGFR3-mediated Tumor Growth Reveals Pathways to Harness the Benefits of Combination Targeted Therapy and Immunotherapy in Bladder Cancer. Comput. Syst. Oncol. 2021, 1, e1019. [Google Scholar] [CrossRef]
- Sung, W.; Hong, T.S.; Poznansky, M.C.; Paganetti, H.; Grassberger, C. Mathematical Modeling to Simulate the Effect of Adding Radiation Therapy to Immunotherapy and Application to Hepatocellular Carcinoma. Int. J. Radiat. Oncol. *Biol. *Phys. 2022, 112, 1055–1062. [Google Scholar] [CrossRef]
- Kim, Y.; Choe, B.-Y.; Suh, T.S.; Sung, W. A Mathematical Model for Predicting Patient Responses to Combined Radiotherapy with CTLA-4 Immune Checkpoint Inhibitors. Cells 2023, 12, 1305. [Google Scholar] [CrossRef]
- Wang, Y.; Bergman, D.R.; Trujillo, E.; Pearson, A.T.; Sweis, R.F.; Jackson, T.L. Mathematical Model Predicts Tumor Control Patterns Induced by Fast and Slow Cytotoxic T Lymphocyte Killing Mechanisms. Sci. Rep. 2023, 13, 22541. [Google Scholar] [CrossRef]
- Yu, J.-L.; Jang, S.R.-J.; Liu, K.-Y. Exploring the Interactions of Oncolytic Viral Therapy and Immunotherapy of Anti-CTLA-4 for Malignant Melanoma Mice Model. Cells 2023, 12, 507. [Google Scholar] [CrossRef]
- Yao, Y.; Chen, Y.F.; Zhang, Q. Optimized Patient-Specific Immune Checkpoint Inhibitor Therapies for Cancer Treatment Based on Tumor Immune Microenvironment Modeling. Brief. Bioinform. 2024, 25, bbae547. [Google Scholar] [CrossRef]
- De Pillis, L.G.; Gu, W.; Radunskaya, A.E. Mixed Immunotherapy and Chemotherapy of Tumors: Modeling, Applications and Biological Interpretations. J. Theor. Biol. 2006, 238, 841–862. [Google Scholar] [CrossRef]
- Leon, K.; Garcia, K.; Carneiro, J.; Lage, A. How Regulatory CD25+CD4+ T Cells Impinge on Tumor Immunobiology: The Differential Response of Tumors to Therapies. J. Immunol. 2007, 179, 5659–5668. [Google Scholar] [CrossRef]
- Eftimie, R.; Dushoff, J.; Bridle, B.W.; Bramson, J.L.; Earn, D.J.D. Multi-Stability and Multi-Instability Phenomena in a Mathematical Model of Tumor-Immune-Virus Interactions. Bull. Math. Biol. 2011, 73, 2932–2961. [Google Scholar] [CrossRef]
- Robertson-Tessi, M.; El-Kareh, A.; Goriely, A. A Mathematical Model of Tumor–Immune Interactions. J. Theor. Biol. 2012, 294, 56–73. [Google Scholar] [CrossRef]
- Kirschner, D.; Panetta, J.C. Modeling Immunotherapy of the Tumor-Immune Interaction. J. Math. Biol. 1998, 37, 235–252. [Google Scholar] [CrossRef]
- Rodrigues, D.S.; Mancera, P.F.A.; Carvalho, T.; Gonçalves, L.F. A Mathematical Model for Chemoimmunotherapy of Chronic Lymphocytic Leukemia. Appl. Math. Comput. 2019, 349, 118–133. [Google Scholar] [CrossRef]
- Sotolongo-Costa, O.; Morales Molina, L.; Rodríguez Perez, D.; Antoranz, J.C.; Chacón Reyes, M. Behavior of Tumors under Nonstationary Therapy. Phys. D Nonlinear Phenom. 2003, 178, 242–253. [Google Scholar] [CrossRef]
- Bunimovich-Mendrazitsky, S.; Shochat, E.; Stone, L. Mathematical Model of BCG Immunotherapy in Superficial Bladder Cancer. Bull. Math. Biol. 2007, 69, 1847–1870. [Google Scholar] [CrossRef]
- Rodriguez-Perez, D.; Sotolongo-Grau, O.; Espinosa Riquelme, R.; Sotolongo-Costa, O.; Santos Miranda, J.A.; Antoranz, J.C. Assessment of Cancer Immunotherapy Outcome in Terms of the Immune Response Time Features. Math. Med. Biol. 2007, 24, 287–300. [Google Scholar] [CrossRef]
- Shaikhet, L.; Bunimovich-Mendrazitsky, S. Stability Analysis of Delayed Immune Response BCG Infection in Bladder Cancer Treatment Model by Stochastic Perturbations. Comput. Math. Methods Med. 2018, 2018, 9653873. [Google Scholar] [CrossRef]
- Wodarz, D. Viruses as Antitumor Weapons: Defining Conditions for Tumor Remission. Cancer Res. 2001, 61, 3501–3507. [Google Scholar]
- Senekal, N.S.; Mahasa, K.J.; Eladdadi, A.; De Pillis, L.; Ouifki, R. Natural Killer Cells Recruitment in Oncolytic Virotherapy: A Mathematical Model. Bull. Math. Biol. 2021, 83, 75. [Google Scholar] [CrossRef]
- Sotolongo-Grau, O.; Rodriguez-Perez, D.; Santos-Miranda, J.A.; Sotolongo-Costa, O.; Antoranz, J.C. Immune System-Tumour Efficiency Ratio as a New Oncological Index for Radiotherapy Treatment Optimization. Math. Med. Biol. 2009, 26, 297–307. [Google Scholar] [CrossRef]
- Walker, R.; Poleszczuk, J.; Pilon-Thomas, S.; Kim, S.; Anderson, A.A.R.A.; Czerniecki, B.J.; Harrison, L.B.; Moros, E.G.; Enderling, H. Immune Interconnectivity of Anatomically Distant Tumors as a Potential Mediator of Systemic Responses to Local Therapy. Sci. Rep. 2018, 8, 9474. [Google Scholar] [CrossRef]
- López Alfonso, J.C.; Poleszczuk, J.; Walker, R.; Kim, S.; Pilon-Thomas, S.; Conejo-Garcia, J.J.; Soliman, H.; Czerniecki, B.; Harrison, L.B.; Enderling, H. Immunologic Consequences of Sequencing Cancer Radiotherapy and Surgery. JCO Clin. Cancer Inform. 2019, 3, 1–16. [Google Scholar] [CrossRef]
- Alfonso, J.C.L.; Papaxenopoulou, L.A.; Mascheroni, P.; Meyer-Hermann, M.; Hatzikirou, H. On the Immunological Consequences of Conventionally Fractionated Radiotherapy. iScience 2020, 23, 100897. [Google Scholar] [CrossRef]
- Montaseri, G.; Alfonso, J.C.L.; Hatzikirou, H.; Meyer-Hermann, M. A Minimal Modeling Framework of Radiation and Immune System Synergy to Assist Radiotherapy Planning. J. Theor. Biol. 2020, 486, 110099. [Google Scholar] [CrossRef]
- Sung, W.; Grassberger, C.; McNamara, A.L.; Basler, L.; Ehrbar, S.; Tanadini-Lang, S.; Hong, T.S.; Paganetti, H. A Tumor-Immune Interaction Model for Hepatocellular Carcinoma Based on Measured Lymphocyte Counts in Patients Undergoing Radiotherapy. Radiother. Oncol. 2020, 151, 73–81. [Google Scholar] [CrossRef]
- Forjanič, T.; Miklavčič, D. Mathematical Model of Tumor Volume Dynamics in Mice Treated with Electrochemotherapy. Med. Biol. Eng. Comput. 2017, 55, 1085–1096. [Google Scholar] [CrossRef]
- Yosef, M.; Bunimovich-Mendrazitsky, S. Mathematical Model of MMC Chemotherapy for Non-Invasive Bladder Cancer Treatment. Front. Oncol. 2024, 14, 1352065. [Google Scholar] [CrossRef]
- Crosby, D.; Bhatia, S.; Brindle, K.M.; Coussens, L.M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald, R.C.; Gambhir, S.S.; Kuhn, P.; et al. Early Detection of Cancer. Science 2022, 375, eaay9040. [Google Scholar] [CrossRef]
- Galluzzi, L.; Kepp, O.; Chan, F.K.-M.; Kroemer, G. Necroptosis: Mechanisms and Relevance to Disease. Annu. Rev. Pathol. Mech. Dis. 2017, 12, 103–130. [Google Scholar] [CrossRef]
- Dhuriya, Y.K.; Sharma, D. Necroptosis: A Regulated Inflammatory Mode of Cell Death. J. Neuro Inflamm. 2018, 15, 199. [Google Scholar] [CrossRef]
- Yao, K.; Shi, Z.; Zhao, F.; Tan, C.; Zhang, Y.; Fan, H.; Wang, Y.; Li, X.; Kong, J.; Wang, Q.; et al. RIPK1 in Necroptosis and Recent Progress in Related Pharmaceutics. Front. Immunol. 2025, 16, 1480027. [Google Scholar] [CrossRef]
- Salvesen, G.S.; Walsh, C.M. Functions of Caspase 8: The Identified and the Mysterious. Semin. Immunol. 2014, 26, 246–252. [Google Scholar] [CrossRef]
- Yuan, J.; Najafov, A.; Py, B.F. Roles of Caspases in Necrotic Cell Death. Cell 2016, 167, 1693–1704. [Google Scholar] [CrossRef]
- Ye, K.; Chen, Z.; Xu, Y. The Double-Edged Functions of Necroptosis. Cell Death Dis. 2023, 14, 163. [Google Scholar] [CrossRef]
- Oberst, A.; Dillon, C.P.; Weinlich, R.; McCormick, L.L.; Fitzgerald, P.; Pop, C.; Hakem, R.; Salvesen, G.S.; Green, D.R. Catalytic Activity of the Caspase-8–FLIPL Complex Inhibits RIPK3-Dependent Necrosis. Nature 2011, 471, 363–367. [Google Scholar] [CrossRef]
- Oliver Metzig, M.; Tang, Y.; Mitchell, S.; Taylor, B.; Foreman, R.; Wollman, R.; Hoffmann, A. An Incoherent Feedforward Loop Interprets NFκB/RelA Dynamics to Determine TNF-induced Necroptosis Decisions. Mol. Syst. Biol. 2020, 16, e9677. [Google Scholar] [CrossRef]
- Xu, F.; Yin, Z.; Zhu, L.; Jin, J.; He, Q.; Li, X.; Shuai, J. Oscillations Governed by the Incoherent Dynamics in Necroptotic Signaling. Front. Phys. 2021, 9, 726638. [Google Scholar] [CrossRef]
- Ildefonso, G.V.; Oliver Metzig, M.; Hoffmann, A.; Harris, L.A.; Lopez, C.F. A Biochemical Necroptosis Model Explains Cell-Type-Specific Responses to Cell Death Cues. Biophys. J. 2023, 122, 817–834. [Google Scholar] [CrossRef]
- Lee, D.; De Los Reyes, V.A.A.; Kim, Y. Optimal Strategies of Oncolytic Virus-Bortezomib Therapy via the Apoptotic, Necroptotic, and Oncolysis Signaling Network. Math. Biosci. Eng. 2024, 21, 3876–3909. [Google Scholar] [CrossRef]
- Jorgensen, I.; Miao, E.A. Pyroptotic Cell Death Defends against Intracellular Pathogens. Immunol. Rev. 2015, 265, 130–142. [Google Scholar] [CrossRef]
- Hou, J.; Hsu, J.-M.; Hung, M.-C. Molecular Mechanisms and Functions of Pyroptosis in Inflammation and Antitumor Immunity. Mol. Cell 2021, 81, 4579–4590. [Google Scholar] [CrossRef]
- Tan, Y.; Chen, Q.; Li, X.; Zeng, Z.; Xiong, W.; Li, G.; Li, X.; Yang, J.; Xiang, B.; Yi, M. Pyroptosis: A New Paradigm of Cell Death for Fighting against Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 153. [Google Scholar] [CrossRef]
- Yu, P.; Zhang, X.; Liu, N.; Tang, L.; Peng, C.; Chen, X. Pyroptosis: Mechanisms and Diseases. Signal Transduct. Target Ther. 2021, 6, 128. [Google Scholar] [CrossRef]
- Wei, S.; Feng, M.; Zhang, S. Molecular Characteristics of Cell Pyroptosis and Its Inhibitors: A Review of Activation, Regulation, and Inhibitors. Int. J. Mol. Sci. 2022, 23, 16115. [Google Scholar] [CrossRef] [PubMed]
- Chai, R.; Li, Y.; Shui, L.; Ni, L.; Zhang, A. The Role of Pyroptosis in Inflammatory Diseases. Front. Cell Dev. Biol. 2023, 11, 1173235. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Pan, R.; Ouyang, Y.; Gu, W.; Xiao, T.; Yang, H.; Tang, L.; Wang, H.; Xiang, B.; Chen, P. Pyroptosis in Health and Disease: Mechanisms, Regulation and Clinical Perspective. Signal Transduct. Target. Ther. 2024, 9, 245. [Google Scholar] [CrossRef]
- Broz, P. Pyroptosis: Molecular Mechanisms and Roles in Disease. Cell Res. 2025, 35, 334–344. [Google Scholar] [CrossRef]
- Jiang, M.; Qi, L.; Li, L.; Li, Y. The Caspase-3/GSDME Signal Pathway as a Switch between Apoptosis and Pyroptosis in Cancer. Cell Death Discov. 2020, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.A.; Thapa, R.; Afzal, O.; Agrawal, N.; Almalki, W.H.; Kazmi, I.; Alzarea, S.I.; Altamimi, A.S.A.; Prasher, P.; Singh, S.K.; et al. The Pyroptotic Role of Caspase-3/GSDME Signalling Pathway among Various Cancer: A Review. Int. J. Biol. Macromol. 2023, 242, 124832. [Google Scholar] [CrossRef]
- Cookson, B.T.; Brennan, M.A. Pro-Inflammatory Programmed Cell Death. Trends Microbiol. 2001, 9, 113–114. [Google Scholar] [CrossRef]
- Ma, D.; Yang, B.; Guan, B.; Song, L.; Liu, Q.; Fan, Y.; Zhao, L.; Wang, T.; Zhang, Z.; Gao, Z.; et al. A Bibliometric Analysis of Pyroptosis From 2001 to 2021. Front. Immunol. 2021, 12, 731933. [Google Scholar] [CrossRef]
- Hamis, S.J.; Macfarlane, F.R. A Single-Cell Mathematical Model of SARS-CoV-2 Induced Pyroptosis and the Effects of Anti-Inflammatory Intervention. AIMS Math. 2021, 6, 6050–6086. [Google Scholar] [CrossRef]
- Zhu, L.; Li, X.; Xu, F.; Yin, Z.; Jin, J.; Liu, Z.; Qi, H.; Shuai, J. Network Modeling-Based Identification of the Switching Targets between Pyroptosis and Secondary Pyroptosis. Chaos Solitons Fractals 2022, 155, 111724. [Google Scholar] [CrossRef]
- Dai, J.; Wang, Y.; Wang, H.; Gao, Z.; Wang, Y.; Fang, M.; Shi, S.; Zhang, P.; Wang, H.; Su, Y.; et al. Toll-Like Receptor Signaling in Severe Acute Respiratory Syndrome Coronavirus 2-Induced Innate Immune Responses and the Potential Application Value of Toll-Like Receptor Immunomodulators in Patients with Coronavirus Disease 2019. Front. Microbiol. 2022, 13, 948770. [Google Scholar] [CrossRef]
- Barker, B.R.; Taxman, D.J.; Ting, J.P.-Y. Cross-Regulation between the IL-1β/IL-18 Processing Inflammasome and Other Inflammatory Cytokines. Curr. Opin. Immunol. 2011, 23, 591–597. [Google Scholar] [CrossRef] [PubMed]
- Van De Veerdonk, F.L.; Netea, M.G.; Dinarello, C.A.; Joosten, L.A.B. Inflammasome Activation and IL-1β and IL-18 Processing during Infection. Trends Immunol. 2011, 32, 110–116. [Google Scholar] [CrossRef] [PubMed]
- Dinarello, C.A. Overview of the IL-1 Family in Innate Inflammation and Acquired Immunity. Immunol. Rev. 2018, 281, 8–27. [Google Scholar] [CrossRef]
- Declercq, J.; De Leeuw, E.; Lambrecht, B.N. Inflammasomes and IL-1 Family Cytokines in SARS-CoV-2 Infection: From Prognostic Marker to Therapeutic Agent. Cytokine 2022, 157, 155934. [Google Scholar] [CrossRef] [PubMed]
- Ihim, S.A.; Abubakar, S.D.; Zian, Z.; Sasaki, T.; Saffarioun, M.; Maleknia, S.; Azizi, G. Interleukin-18 Cytokine in Immunity, Inflammation, and Autoimmunity: Biological Role in Induction, Regulation, and Treatment. Front. Immunol. 2022, 13, 919973. [Google Scholar] [CrossRef]
- Glont, M.; Nguyen, T.V.N.; Graesslin, M.; Hälke, R.; Ali, R.; Schramm, J.; Wimalaratne, S.M.; Kothamachu, V.B.; Rodriguez, N.; Swat, M.J.; et al. BioModels: Expanding Horizons to Include More Modelling Approaches and Formats. Nucleic Acids Res. 2018, 46, D1248–D1253. [Google Scholar] [CrossRef]
- Malik-Sheriff, R.S.; Glont, M.; Nguyen, T.V.N.; Tiwari, K.; Roberts, M.G.; Xavier, A.; Vu, M.T.; Men, J.; Maire, M.; Kananathan, S.; et al. BioModels—15 Years of Sharing Computational Models in Life Science. Nucleic Acids Res. 2019, 48, D407–D415. [Google Scholar] [CrossRef] [PubMed]
- Kolpakov, F.; Akberdin, I.; Kiselev, I.; Kolmykov, S.; Kondrakhin, Y.; Kulyashov, M.; Kutumova, E.; Pintus, S.; Ryabova, A.; Sharipov, R.; et al. BioUML—Towards a Universal Research Platform. Nucleic Acids Res. 2022, 50, W124–W131. [Google Scholar] [CrossRef] [PubMed]
- Novère, N.L.; Hucka, M.; Mi, H.; Moodie, S.; Schreiber, F.; Sorokin, A.; Demir, E.; Wegner, K.; Aladjem, M.I.; Wimalaratne, S.M.; et al. The Systems Biology Graphical Notation. Nat. Biotechnol. 2009, 27, 735–741. [Google Scholar] [CrossRef] [PubMed]
- Hoang, M.T.; Ehrhardt, M. Differential Equation Models for Infectious Diseases: Mathematical Modeling, Qualitative Analysis, Numerical Methods and Applications. SeMA 2025. [Google Scholar] [CrossRef]
- Yin, A.; Moes, D.J.A.R.; Van Hasselt, J.G.C.; Swen, J.J.; Guchelaar, H. A Review of Mathematical Models for Tumor Dynamics and Treatment Resistance Evolution of Solid Tumors. CPT Pharmacom. Syst. Pharma. 2019, 8, 720–737. [Google Scholar] [CrossRef]
- Spencer, S.L.; Berryman, M.J.; García, J.A.; Abbott, D. An Ordinary Differential Equation Model for the Multistep Transformation to Cancer. J. Theor. Biol. 2004, 231, 515–524. [Google Scholar] [CrossRef]
- Lee, J.; Kim, E. Ordinary Differential Equation Model of Cancer-Associated Fibroblast Heterogeneity Predicts Treatment Outcomes. Npj Syst. Biol. Appl. 2025, 11, 96. [Google Scholar] [CrossRef]
- Guyton, A.C.; Coleman, T.G.; Granger, H.J. Circulation: Overall Regulation. Annu. Rev. Physiol. 1972, 34, 13–44. [Google Scholar] [CrossRef]
- Bhattacharya-Ghosh, B.; Bozkurt, S.; Rutten, M.C.M.; Van De Vosse, F.N.; Díaz-Zuccarini, V. An in Silico Case Study of Idiopathic Dilated Cardiomyopathy via a Multi-Scale Model of the Cardiovascular System. Comput. Biol. Med. 2014, 53, 141–153. [Google Scholar] [CrossRef]
- Hallow, K.M.; Lo, A.; Beh, J.; Rodrigo, M.; Ermakov, S.; Friedman, S.; De Leon, H.; Sarkar, A.; Xiong, Y.; Sarangapani, R.; et al. A Model-Based Approach to Investigating the Pathophysiological Mechanisms of Hypertension and Response to Antihypertensive Therapies: Extending the Guyton Model. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2014, 306, R647–R662. [Google Scholar] [CrossRef]
- Kurian, V.; Gee, M.; Farrington, S.; Yang, E.; Okossi, A.; Chen, L.; Beris, A.N. Systems Engineering Approach to Modeling and Analysis of Chronic Obstructive Pulmonary Disease Part II: Extension for Variable Metabolic Rates. ACS Omega 2024, 9, 494–508. [Google Scholar] [CrossRef]
- Siewe, N.; Friedman, A. A Mathematical Model of Obesity-Induced Type 2 Diabetes and Efficacy of Anti-Diabetic Weight Reducing Drug. J. Theor. Biol. 2024, 581, 111756. [Google Scholar] [CrossRef]
- Aliffi, G.E.; Nastasi, G.; Romano, V.; Pitocco, D.; Rizzi, A.; Moore, E.J.; De Gaetano, A. A System of ODEs for Representing Trends of CGM Signals. J. Math. Ind. 2024, 14, 23. [Google Scholar] [CrossRef]
- Ugolkov, Y.; Nikitich, A.; Leon, C.; Helmlinger, G.; Peskov, K.; Sokolov, V.; Volkova, A. Mathematical Modeling in Autoimmune Diseases: From Theory to Clinical Application. Front. Immunol. 2024, 15, 1371620. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Chen, K.; Chen, W.; Yao, Y.; Ni, S.; Ye, M.; Zhuang, G.; Hu, M.; Gao, J.; Gao, C.; et al. Paradoxical Mitophagy Regulation by PINK1 and TUFm. Mol. Cell 2020, 80, 607–620.e12. [Google Scholar] [CrossRef] [PubMed]
- Goglia, I.; Węglarz-Tomczak, E.; Gioia, C.; Liu, Y.; Virtuoso, A.; Bonanomi, M.; Gaglio, D.; Salmistraro, N.; De Luca, C.; Papa, M.; et al. Fusion–Fission–Mitophagy Cycling and Metabolic Reprogramming Coordinate Nerve Growth Factor (NGF)-dependent Neuronal Differentiation. FEBS J. 2024, 291, 2811–2835. [Google Scholar] [CrossRef]
- Fu, C.; Jiang, A. Dendritic Cells and CD8 T Cell Immunity in Tumor Microenvironment. Front. Immunol. 2018, 9, 3059. [Google Scholar] [CrossRef]
- Garris, C.S.; Luke, J.J. Dendritic Cells, the T-Cell-Inflamed Tumor Microenvironment, and Immunotherapy Treatment Response. Clin. Cancer Res. 2020, 26, 3901–3907. [Google Scholar] [CrossRef]
- Heras-Murillo, I.; Adán-Barrientos, I.; Galán, M.; Wculek, S.K.; Sancho, D. Dendritic Cells as Orchestrators of Anticancer Immunity and Immunotherapy. Nat. Rev. Clin. Oncol. 2024, 21, 257–277. [Google Scholar] [CrossRef]
- Peng, F.; Liao, M.; Qin, R.; Zhu, S.; Peng, C.; Fu, L.; Chen, Y.; Han, B. Regulated Cell Death (RCD) in Cancer: Key Pathways and Targeted Therapies. Signal Transduct. Target. Ther. 2022, 7, 286. [Google Scholar] [CrossRef]
- Zhang, Y.; Yi, S.; Luan, M. Advances in Non-Apoptotic Regulated Cell Death: Implications for Malignant Tumor Treatment. Front. Oncol. 2025, 15, 1519119. [Google Scholar] [CrossRef]
- Neal, M.L.; Cooling, M.T.; Smith, L.P.; Thompson, C.T.; Sauro, H.M.; Carlson, B.E.; Cook, D.L.; Gennari, J.H. A Reappraisal of How to Build Modular, Reusable Models of Biological Systems. PLoS Comput. Biol. 2014, 10, e1003849. [Google Scholar] [CrossRef]
- Soheilypour, M.; Mofrad, M.R.K. Agent-Based Modeling in Molecular Systems Biology. BioEssays 2018, 40, 1800020. [Google Scholar] [CrossRef]
- Macal, C.M.; North, M.J. Tutorial on Agent-Based Modelling and Simulation. J. Simul. 2010, 4, 151–162. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Berghe, T.V.; Vandenabeele, P.; Kroemer, G. The Molecular Machinery of Regulated Cell Death. Cell Res. 2019, 29, 347–364. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues De Souza, E.; Almeida Cordeiro Nogueira, H.; Da Silva Francisco Junior, R.; Garcia, A.B.; Medina-Acosta, E. Integrated Multi-Optosis Model for Pan-Cancer Candidate Biomarker and Therapy Target Discovery. Front. Bioinform. 2025, 5, 1630518. [Google Scholar] [CrossRef]
- Liu, J.; Kuang, F.; Kang, R.; Tang, D. Alkaliptosis: A New Weapon for Cancer Therapy. Cancer Gene Ther. 2020, 27, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Kang, R.; Liu, J.; Tang, D. Mechanisms of Alkaliptosis. Front. Cell Dev. Biol. 2023, 11, 1213995. [Google Scholar] [CrossRef] [PubMed]
- Pallichankandy, S.; Thayyullathil, F.; Cheratta, A.R.; Subburayan, K.; Alakkal, A.; Sultana, M.; Drou, N.; Arshad, M.; Tariq, S.; Galadari, S. Targeting Oxeiptosis-Mediated Tumor Suppression: A Novel Approach to Treat Colorectal Cancers by Sanguinarine. Cell Death Discov. 2023, 9, 94. [Google Scholar] [CrossRef]
- Bartoszewska, E.; Florek, K.; Zagórski, K.; Gachowska, M.; Wietrzyk, A.; Hutny, A.; Nowakowska-Toporowska, A.; Kulbacka, J. Methuosis, Alkaliptosis, and Oxeiptosis and Their Significance in Anticancer Therapy. Cells 2024, 13, 2095. [Google Scholar] [CrossRef]
- Chen, K.-Q.; Wang, S.-Z.; Lei, H.-B.; Liu, X. Mini-Review: Research and Progress of Oxeiptosis in Diseases. Front. Cell Dev. Biol. 2024, 12, 1428250. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Zhang, J.; Yang, Y.; Fleishman, J.S.; Wang, Y.; Wang, J.; Chen, J.; Li, Y.; Wang, H. Cuproptosis: A Novel Therapeutic Target for Overcoming Cancer Drug Resistance. Drug Resist. Updates 2024, 72, 101018. [Google Scholar] [CrossRef]
- Bhuvaneshwari, K.; Harithpriya, K.; Ganesan, K.; Xu, B.; Ramkumar, K.M. Role of Oxeiptosis in Disease Mechanisms and Therapeutic Opportunities. Apoptosis 2025, 30, 1182–1201. [Google Scholar] [CrossRef]
- Chen, R.; You, J.; Weng, S.; Zhao, T. Disulfidptosis Mechanisms and Therapeutic Implications in Cancer Metabolic Reprogramming and Future Perspectives. Discov. Oncol. 2025, 16, 1814. [Google Scholar] [CrossRef]
- Gan, B. Redox-Driven Cell Death by Disulfidptosis and Its Therapeutic Potential. Nat. Rev. Mol. Cell Biol. 2025, 26, 727–729. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, D.; Yao, L.; Sun, Y.; Li, D.; Le, J.; Dian, Y.; Zeng, F.; Chen, X.; Deng, G. The Molecular Mechanism and Therapeutic Landscape of Copper and Cuproptosis in Cancer. Signal Transduct. Target. Ther. 2025, 10, 149. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Xin, H.; Liu, Y.; Han, Y. Cuproptosis as a Therapeutic Target in Cancer: A Systematic Review and Bibliometric Analysis of the Research Landscape. Front. Oncol. 2025, 15, 1566986. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, S.; Wu, Y.; Zhang, P.; Yu, Y.; Chen, X.; Yu, L.; Yang, X.; Li, H.; Wu, C.; et al. Molecular Signatures of Disulfidptosis: Interplay with Programmed Cell Death Pathways and Therapeutic Implications in Oncology. Cell Mol. Biol. Lett. 2025, 30, 66. [Google Scholar] [CrossRef] [PubMed]
- Zhen, W.; Zhao, T.; Chen, X.; Zhang, J. Unlocking the Potential of Disulfidptosis: Nanotechnology-Driven Strategies for Advanced Cancer Therapy. Small 2025, 21, 2500880. [Google Scholar] [CrossRef]
- Yu, J.S.; Bagheri, N. Agent-Based Models Predict Emergent Behavior of Heterogeneous Cell Populations in Dynamic Microenvironments. Front. Bioeng. Biotechnol. 2020, 8, 249. [Google Scholar] [CrossRef]
| Number of Models | Most Used Modeling Formats | Public Availability | BioModels | ||||
|---|---|---|---|---|---|---|---|
| MATLAB | SBML | XPPAUT | Number of Models | Percentage of Total | |||
| Apoptosis | 40 | 14 | 12 | 2 | 20 | 50% | 9 |
| Autophagy | 20 | 2 | 2 | 8 | 14 | 70% | 1 |
| ICD | 46 | 18 | 13 | 0 | 17 | 37% | 13 |
| Ferroptosis | 3 | 1 | 0 | 0 | 1 | 33% | 0 |
| Necroptosis | 3 | 1 | 0 | 0 | 2 | 66% | 0 |
| Pyroptosis | 2 | 1 | 0 | 0 | 1 | 50% | 0 |
| Apoptosis and autophagy | 19 | 4 | 0 | 12 | 13 | 68% | 0 |
| Apoptosis and necroptosis | 2 | 2 | 0 | 0 | 0 | 0% | 0 |
| Apoptosis and pyroptosis | 2 | 2 | 0 | 0 | 1 | 50% | 0 |
| Total | 137 | 45 | 27 | 22 | 69 | 50% | 23 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kutumova, E.; Akberdin, I.; Lavrik, I.; Kolpakov, F. Mathematical Modeling of Cell Death and Survival: Toward an Integrated Computational Framework for Multi-Decision Regulatory Dynamics. Cells 2025, 14, 1792. https://doi.org/10.3390/cells14221792
Kutumova E, Akberdin I, Lavrik I, Kolpakov F. Mathematical Modeling of Cell Death and Survival: Toward an Integrated Computational Framework for Multi-Decision Regulatory Dynamics. Cells. 2025; 14(22):1792. https://doi.org/10.3390/cells14221792
Chicago/Turabian StyleKutumova, Elena, Ilya Akberdin, Inna Lavrik, and Fedor Kolpakov. 2025. "Mathematical Modeling of Cell Death and Survival: Toward an Integrated Computational Framework for Multi-Decision Regulatory Dynamics" Cells 14, no. 22: 1792. https://doi.org/10.3390/cells14221792
APA StyleKutumova, E., Akberdin, I., Lavrik, I., & Kolpakov, F. (2025). Mathematical Modeling of Cell Death and Survival: Toward an Integrated Computational Framework for Multi-Decision Regulatory Dynamics. Cells, 14(22), 1792. https://doi.org/10.3390/cells14221792








