Neuronal Enriched Extracellular Vesicle miR-122-5p as a Potential Biomarker for Alzheimer’s Disease
Highlights
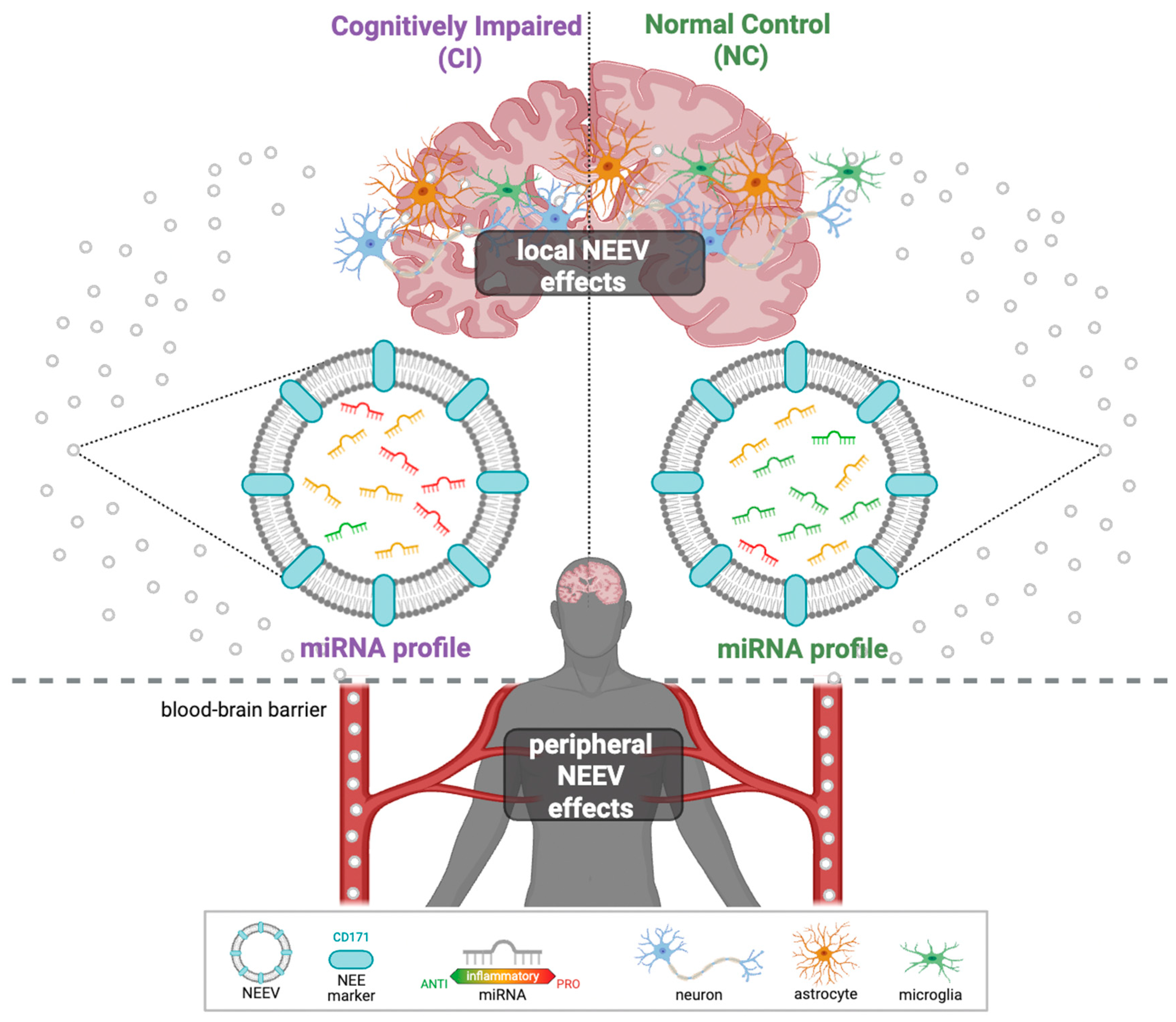
- hsa-miR-122-5p was significantly underrepresented in cognitively impaired individuals in both Mexican American (MA) and Non-Hispanic White (NHW) groups.
- Population-specific NEEV miRNAs (MAs: hsa-miR-26a-5p, hsa-let-7f-5p, and hsa-miR-139-5p, NHWs: hsa-miR-133a-3p, hsa-miR-125b-5p, and hsa-miR-100-5p) were identified as potential blood-based biomarkers for Alzheimer’s disease (AD) and related cognitive impairment.
- Differentially expressed miRNAs were associated with APOE genotype, age, and metabolic burden, and their predicted targets are enriched in NF-κB-inflammatory pathways.
- Dysregulated miRNA networks may provide a mechanistic link between comorbidity burden and AD-related neuroinflammation, supporting their utility in AD precision medicine.
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Total EV Isolation
2.3. NEEV Enrichment
2.4. NEEV Characterization
2.5. RNA Extraction
2.6. miRNA Library Preparation and Sequencing
2.7. Data Analysis
2.7.1. Differential miRNA Expression Analysis
2.7.2. Covariate Modeling
2.7.3. Pathway Analysis and Target Genes
3. Results
3.1. EV Characterization
3.2. Differential Expression of miRNAs in NEEVs
3.2.1. Quality Control
3.2.2. Representative miRNAs in NEEVs
3.2.3. Differential Expression in Cognitive Impairment (CI)
Cross-Population Patterns
- hsa-miR-122-5p was consistently underrepresented in CI across MAs and NHWs (Table 2), a pattern that remained significant after adjusting for covariates (Figure 5, Table 2, Supplementary Document S2; Tables S1–S6). Males showed higher expression of hsa-miR-122-5p than females across populations, while no significant age association was observed in NHW (Supplementary Documents S3 and S4).
- hsa-let-7a-5p, hsa-let-7g-5p, and hsa-miR-122-5p were significantly DE in both MAs and NHWs when both the visits were analyzed (Supplementary Document S2; Table S6). hsa-miR-122-5p was underrepresented, and hsa-let-7a-5p was overrepresented in both population groups. hsa-let-7a-5p showed significant association with APOE4 index and the metabolic index in NHWs at Visit 2 but not in MAs. Notably, hsa-let-7g-5p was overrepresented in MAs but underrepresented in NHWs (Supplementary Document S1; Tables S2 and S3).
- miRNet linked these miRNAs to AD-, dementia-, and T2D-related genes (Supplementary Document S1; Figure S4).
Population-Specific Findings:
- MAs: hsa-miR-26a-5p, hsa-let-7f-5p, and hsa-miR-139-5p were consistently overrepresented in CI and linked to AD-, hypertension-, and diabetes-related genes (Figure 6, Table 3; Supplementary Document S2, Tables S8 and S10; Supplementary Document S1; Figure S5). Several showed associations with APOE4, metabolic index, age, and gender (Supplementary Document S3).
- NHWs: hsa-miR-133a-3p, hsa-miR-125b-5p and hsa-miR-100-5p were overrepresented in CI and linked to hypertension-related genes (Figure 6, Table 4; Supplementary Document S2, Tables S9 and S11; Supplementary Document S1; Figure S6). Many NHW-specific miRNAs were strongly associated with metabolic index, APOE4, and demographic variables (e.g., hsa-miR-133a-3p increased with APOE4 index and metabolic index; hsa-miR-30c-1-3p higher in males; hsa-miR-1268a increased with age) (Supplementary Document S4).
3.2.4. Longitudinal Changes
4. Discussion
4.1. Strengths
4.2. Limitations
4.3. Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DE | Differential Expression |
| AD | Alzheimer’s disease |
| CI | Cognitive Impairment |
| EV | Extracellular Vesicle |
| NEEV | Neuronal Enriched Extracellular Vesicle |
| MA | Mexican American |
| NHW | Non-Hispanic White |
References
- Mahaman, Y.A.R.; Embaye, K.S.; Huang, F.; Li, L.; Zhu, F.; Wang, J.Z.; Liu, R.; Feng, J.; Wang, X. Biomarkers used in Alzheimer’s disease diagnosis, treatment, and prevention. Ageing Res. Rev. 2022, 74, 101544. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.K.; Mehta, V.; Singh, T.G. Alzheimer’s Disorder: Epigenetic Connection and Associated Risk Factors. Curr. Neuropharmacol. 2020, 18, 740–753. [Google Scholar] [CrossRef] [PubMed]
- Tahami Monfared, A.A.; Byrnes, M.J.; White, L.A.; Zhang, Q. Alzheimer’s Disease: Epidemiology and Clinical Progression. Neurol. Ther. 2022, 11, 553–569. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Chung, J.Y. Pathobiolgy and Management of Alzheimer’s Disease. Chonnam Med. J. 2021, 57, 108. [Google Scholar] [CrossRef]
- Rasmussen, J.; Langerman, H. Alzheimer’s Disease—Why We Need Early Diagnosis. Degener. Neurol. Neuromuscul. Dis. 2019, 9, 123–130. [Google Scholar] [CrossRef]
- Barnett, J.H.; Lewis, L.; Blackwell, A.D.; Taylor, M. Early intervention in Alzheimer’s disease: A health economic study of the effects of diagnostic timing. BMC Neurol. 2014, 14, 101. [Google Scholar] [CrossRef]
- Frederiksen, K.S.; Arus, X.M.; Zetterberg, H.; Gauthier, S.; Boada, M.; Pytel, V.; Hahn-Pedersen, J.; Solís Tarazona, L.R.; Mattke, S. Focusing on earlier diagnosis of Alzheimer’s disease. Future Neurol. 2024, 19, 2337452. [Google Scholar] [CrossRef]
- O’Bryant, S.E.; Johnson, L.; Reisch, J.; Edwards, M.; Hall, J.; Barber, R.; Devous, M.D.; Royall, D.; Singh, M. Risk factors for mild cognitive impairment among Mexican Americans. Alzheimer’s Dement. 2013, 9, 622. [Google Scholar] [CrossRef]
- O’Bryant, S.E.; Zhang, F.; Petersen, M.; Hall, J.R.; Johnson, L.A.; Yaffe, K.; Braskie, M.; Vig, R.; Toga, A.W.; Rissman, R.A.; et al. Proteomic Profiles of Neurodegeneration Among Mexican Americans and Non-Hispanic Whites in the HABS-HD Study. J. Alzheimer’s Dis. 2022, 86, 1243–1254. [Google Scholar] [CrossRef]
- Tran, E.; Cabán, M.; Meng, A.; Wetmore, J.; Ottman, R.; Siegel, K. Beliefs About the Causes of Alzheimer’s Disease Among Latinos in New York City. J. Community Health 2025, 50, 10–22. [Google Scholar] [CrossRef]
- Quiroz, Y.T.; Solis, M.; Aranda, M.P.; Arbaje, A.I.; Arroyo-Miranda, M.; Cabrera, L.Y.; Carrasquillo, M.M.; Corrada, M.M.; Crivelli, L.; Diminich, E.D.; et al. Addressing the disparities in dementia risk, early detection and care in Latino populations: Highlights from the second Latinos & Alzheimer’s Symposium. Alzheimer’s Dement. 2022, 18, 1677–1686. [Google Scholar] [CrossRef]
- Reid, D.M.; Barber, R.C.; Thorpe, R.J.; Sun, J.; Zhou, Z.; Phillips, N.R. Mitochondrial DNA oxidative mutations are elevated in Mexican American women potentially implicating Alzheimer’s disease. NPJ Aging 2022, 8, 2. [Google Scholar] [CrossRef]
- O’Bryant, S.E.; Johnson, L.A.; Barber, R.C.; Braskie, M.N.; Christian, B.; Hall, J.R.; Hazra, N.; King, K.; Kothapalli, D.; Large, S.; et al. The Health & Aging Brain among Latino Elders (HABLE) study methods and participant characteristics. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2021, 13, e12202. [Google Scholar] [CrossRef]
- Zhang, Y.; Bi, K.; Zhou, L.; Wang, J.; Huang, L.; Sun, Y.; Peng, G.; Wu, W. Advances in Blood Biomarkers for Alzheimer’s Disease: Ultra-Sensitive Detection Technologies and Impact on Clinical Diagnosis. Degener. Neurol. Neuromuscul. Dis. 2024, 14, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Colvee-Martin, H.; Parra, J.R.; Gonzalez, G.A.; Barker, W.; Duara, R. Neuropathology, Neuroimaging, and Fluid Biomarkers in Alzheimer’s Disease. Diagnostics 2024, 14, 704. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R.; Andrews, J.S.; Beach, T.G.; Buracchio, T.; Dunn, B.; Graf, A.; Hansson, O.; Ho, C.; Jagust, W.; McDade, E.; et al. Revised criteria for diagnosis and staging of Alzheimer’s disease: Alzheimer’s Association Workgroup. Alzheimer’s Dement. 2024, 20, 5143–5169. [Google Scholar] [CrossRef]
- Nikolac Perkovic, M.; Videtic Paska, A.; Konjevod, M.; Kouter, K.; Svob Strac, D.; Nedic Erjavec, G.; Pivac, N. Epigenetics of Alzheimer’s Disease. Biomolecules 2021, 11, 195. [Google Scholar] [CrossRef]
- Abdelsalam, M.; Ahmed, M.; Osaid, Z.; Hamoudi, R.; Harati, R. Insights into Exosome Transport through the Blood-Brain Barrier and the Potential Therapeutical Applications in Brain Diseases. Pharmaceuticals 2023, 16, 571. [Google Scholar] [CrossRef]
- Mashouri, L.; Yousefi, H.; Aref, A.R.; Ahadi, A.M.; Molaei, F.; Alahari, S.K. Exosomes: Composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol. Cancer 2019, 18, 75. [Google Scholar] [CrossRef]
- Banack, S.A.; Dunlop, R.A.; Cox, P.A. An miRNA fingerprint using neural-enriched extracellular vesicles from blood plasma: Towards a biomarker for amyotrophic lateral sclerosis/motor neuron disease. Open Biol. 2020, 10, 200116. [Google Scholar] [CrossRef]
- Mustapic, M.; Eitan, E.; Werner, J.K., Jr.; Berkowitz, S.T.; Lazaropoulos, M.P.; Tran, J.; Goetzl, E.J.; Kapogiannis, D. Plasma Extracellular Vesicles Enriched for Neuronal Origin: A Potential Window into Brain Pathologic Processes. Front. Neurosci. 2017, 11, 278. [Google Scholar] [CrossRef]
- Hotta, N.; Tadokoro, T.; Henry, J.; Koga, D.; Kawata, K.; Ishida, H.; Oguma, Y.; Hirata, A.; Mitsuhashi, M.; Yoshitani, K. Monitoring of Post-Brain Injuries By Measuring Plasma Levels of Neuron-Derived Extracellular Vesicles. Biomark. Insights 2022, 17, 11772719221128145. [Google Scholar] [CrossRef]
- Durur, D.Y.; Tastan, B.; Ugur Tufekci, K.; Olcum, M.; Uzuner, H.; Karakülah, G.; Yener, G.; Genc, S. Alteration of miRNAs in Small Neuron-Derived Extracellular Vesicles of Alzheimer’s Disease Patients and the Effect of Extracellular Vesicles on Microglial Immune Responses. J. Mol. Neurosci. 2022, 72, 1182–1194. [Google Scholar] [CrossRef] [PubMed]
- Burrows, K.; Figueroa-Hall, L.K.; Stewart, J.L.; Alarbi, A.M.; Kuplicki, R.; Hannafon, B.N.; Tan, C.; Risbrough, V.B.; McKinney, B.A.; Ramesh, R.; et al. Exploring the role of neuronal-enriched extracellular vesicle miR-93 and interoception in major depressive disorder. Transl. Psychiatry 2024, 14, 199. [Google Scholar] [CrossRef] [PubMed]
- Pathak, G.A.; Zhou, Z.; Silzer, T.K.; Barber, R.C.; Phillips, N.R. Two-stage Bayesian GWAS of 9576 individuals identifies SNP regions that are targeted by miRNAs inversely expressed in Alzheimer’s and cancer. Alzheimer’s Dement. 2020, 16, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, Y.; Gao, Q.; Ping, D.; Wang, Y.; Wu, W.; Lin, X.; Fang, Y.; Zhang, J.; Shao, A. The role of exosomal microRNAs and oxidative stress in neurodegenerative diseases. Oxidative Med. Cell. Longev. 2020, 2020, 3232869. [Google Scholar] [CrossRef]
- Yang, E.; Wang, X.; Gong, Z.; Yu, M.; Wu, H.; Zhang, D. Exosome-mediated metabolic reprogramming: The emerging role in tumor microenvironment remodeling and its influence on cancer progression. Signal Transduct. Target. Ther. 2020, 5, 242. [Google Scholar] [CrossRef]
- Kaur, S.; Verma, H.; Dhiman, M.; Tell, G.; Gigli, G.L.; Janes, F.; Mantha, A.K. Brain Exosomes: Friend or Foe in Alzheimer’s Disease? Mol. Neurobiol. 2021, 58, 6610–6624. [Google Scholar] [CrossRef]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef]
- You, Y.; Zhang, Z.; Sultana, N.; Ericsson, M.; Martens, Y.A.; Sun, M.; Kanekiyo, T.; Ikezu, S.; Shaffer, S.A.; Ikezu, T. ATP1A3 as a target for isolating neuron-specific extracellular vesicles from human brain and biofluids. Sci. Adv. 2023, 9, eadi3647. [Google Scholar] [CrossRef]
- Fiandaca, M.S.; Kapogiannis, D.; Mapstone, M.; Boxer, A.; Eitan, E.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Federoff, H.J.; Miller, B.L.; et al. Identification of preclinical Alzheimer’s disease by a profile of pathogenic proteins in neurally derived blood exosomes: A case-control study. Alzheimer’s Dement. 2015, 11, 600. [Google Scholar] [CrossRef]
- Vukojevic, V.; Mastrandreas, P.; Arnold, A.; Peter, F.; Kolassa, I.-T.; Wilker, S.; Elbert, T.; De Quervain, D.J.F.; Papassotiropoulos, A.; Stetak, A. Evolutionary conserved role of neural cell adhesion molecule-1 in memory. Transl. Psychiatry 2020, 10, 217. [Google Scholar] [CrossRef]
- QIAGEN RNA-seq Analysis Portal 5.1 (QIAGEN, Aarhus, Denmark). Available online: https://rnaportal.qiagen.com (accessed on 20 January 2025).
- Tripathi, S.; Mathaiyan, J.; Kayal, S.; Nachiappa Ganesh, R. Identification of Differentially Expressed Mirna by Next Generation Sequencing in Locally Advanced Breast Cancer Patients of South Indian Origin. Asian Pac. J. Cancer Prev. 2022, 23, 2255–2261. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Rafsanjani, M.R. Analysing and identifying miRNAs from RNA-seq data using miRDeep2 tool in Galaxy, a practical guide. bioRxiv 2021. [Google Scholar] [CrossRef]
- Ruhela, V.; Gupta, A.; Sriram, K.; Ahuja, G.; Kaur, G.; Gupta, R. A Unified Computational Framework for a Robust, Reliable, and Reproducible Identification of Novel miRNAs From the RNA Sequencing Data. Front. Bioinform. 2022, 2, 842051. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y. The comparison of limma and DESeq2 in gene analysis. E3S Web Conf. 2021, 271, 03058. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020, 10, 20560. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Hoffman, G.E.; Roussos, P. Dream: Powerful differential expression analysis for repeated measures designs. Bioinformatics 2021, 37, 192–201. [Google Scholar] [CrossRef]
- Chang, L.; Zhou, G.; Soufan, O.; Xia, J. miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 2020, 48, W244–W251. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2019, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed]
- Boccardi, V.; Poli, G.; Cecchetti, R.; Bastiani, P.; Scamosci, M.; Febo, M.; Mazzon, E.; Bruscoli, S.; Brancorsini, S.; Mecocci, P. miRNAs and Alzheimer’s Disease: Exploring the Role of Inflammation and Vitamin E in an Old-Age Population. Nutrients 2023, 15, 634. [Google Scholar] [CrossRef]
- Hu, Y.; Peng, X.; Du, G.; Zhang, Z.; Zhai, Y.; Xiong, X.; Luo, X. MicroRNA-122-5p Inhibition Improves Inflammation and Oxidative Stress Damage in Dietary-Induced Non-alcoholic Fatty Liver Disease Through Targeting FOXO3. Front. Physiol. 2022, 13, 803445. [Google Scholar] [CrossRef]
- Faramin Lashkarian, M.; Hashemipour, N.; Niaraki, N.; Soghala, S.; Moradi, A.; Sarhangi, S.; Hatami, M.; Aghaei-Zarch, F.; Khosravifar, M.; Mohammadzadeh, A.; et al. MicroRNA-122 in human cancers: From mechanistic to clinical perspectives. Cancer Cell Int. 2023, 23, 29. [Google Scholar] [CrossRef]
- Salzmann, R.J.S.; Garbin, A.; Gaffo, E.; Elia, C.; Martire, G.; Bortoluzzi, S.; Tondo, A.; Muggeo, P.; Sala, A.; Pizzi, M.; et al. Extracellular Vesicle miR-122-5p as a Prognostic Biomarker in Pediatric Classical Hodgkin Lymphoma. Int. J. Mol. Sci. 2024, 25, 13243. [Google Scholar] [CrossRef]
- Zhu, Z.; Yin, J.; Li, D.C.; Mao, Z.Q. Role of microRNAs in the treatment of type 2 diabetes mellitus with Roux-en-Y gastric bypass. Braz. J. Med. Biol. Res. 2017, 50, e5817. [Google Scholar] [CrossRef]
- Hashemi, K.S.; Aliabadi, M.K.; Mehrara, A.; Talebi, E.; Hemmati, A.A.; Rezaeiye, R.D.; Ghanbary, M.J.; Motealleh, M.; Dayeri, B.; Alashti, S.K. A meta-analysis of microarray datasets to identify biological regulatory networks in Alzheimer’s disease. Front. Genet. 2023, 14, 1–13. [Google Scholar] [CrossRef]
- Chen, Y.-P.; Jin, X.; Kong, M.; Li, Y.-M. Pattern of microRNA expression associated with different stages of alcoholic liver disease in rat models. Mol. Med. Rep. 2014, 10, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Darvishi Talemi, M.; Tapak, L.; Rastgoo Haghi, A.; Ahghari, P.; Moradi, S.; Afshar, S. MiR-15b and let-7a as Non-invasive Diagnostic Biomarkers of Alzheimer’s Disease Using an Artificial Neural Network. Avicenna J. Med. Biochem. 2023, 11, 138–145. [Google Scholar] [CrossRef]
- Choi, H.-J.; Jeong, Y.J.; Kim, J.; Hoe, H.-S. EGFR is a potential dual molecular target for cancer and Alzheimer’s disease. Front. Pharmacol. 2023, 14, 1238639. [Google Scholar] [CrossRef] [PubMed]
- Moustafa, N.A.; El-Sayed, M.A.; Abdallah, S.H.; Hazem, N.M.; Aidaros, M.A.; Abdelmoety, D.A. Effect of Letrozole on hippocampal Let-7 microRNAs and their correlation with working memory and phosphorylated Tau protein in an Alzheimer’s disease-like rat model. Egypt. J. Neurol. Psychiatry Neurosurg. 2022, 58, 70. [Google Scholar] [CrossRef]
- Ueta, M.; Nishigaki, H.; Komai, S.; Mizushima, K.; Tamagawa-Mineoka, R.; Naito, Y.; Katoh, N.; Sotozono, C.; Kinoshita, S. Positive regulation of innate immune response by miRNA-let-7a-5p. Front. Genet. 2023, 13, 1025539. [Google Scholar] [CrossRef]
- Wang, W.; Xu, D.; Huang, Y.; Tao, X.; Fan, Y.; Li, Z.; Ding, X. Identification of the role of autophagy-related TNFSF10/ hsa-let-7a-5p axis in vitiligo development and potential herbs exploring based on a bioinformatics analysis. Heliyon 2023, 9, e23220. [Google Scholar] [CrossRef]
- Kafshdooz, T.; Farajnia, S.; Sharifi, R.; Najmi, S. Hsa-let-7g-5p, a circulating microRNA, as a biomarker for Alzheimer’s disease. Inform. Med. Unlocked 2023, 38, 101203. [Google Scholar] [CrossRef]
- Poursaei, E.; Abolghasemi, M.; Bornehdeli, S.; Shanehbandi, D.; Asadi, M.; Sadeghzadeh, M.; Rahmanpour, D.; Sadeh, R.N. Evaluation of hsa-let-7d-5p, hsa-let-7g-5p and hsa-miR-15b-5p plasma levels in patients with Alzheimer’s disease. Psychiatr. Genet. 2022, 32, 25–29. [Google Scholar] [CrossRef]
- Satoh, J.-I.; Kino, Y.; Niida, S. MicroRNA-Seq Data Analysis Pipeline to Identify Blood Biomarkers for Alzheimer’s Disease from Public Data. Biomark. Insights 2015, 10, BMI.S25132. [Google Scholar] [CrossRef]
- Gleason, C.E.; Zuelsdorff, M.; Gooding, D.C.; Kind, A.J.H.; Johnson, A.L.; James, T.T.; Lambrou, N.H.; Wyman, M.F.; Ketchum, F.B.; Gee, A.; et al. Alzheimer’s disease biomarkers in Black and non-Hispanic White cohorts: A contextualized review of the evidence. Alzheimer’s Dement. 2022, 18, 1545–1564. [Google Scholar] [CrossRef]
- Garrett, S.L.; McDaniel, D.; Obideen, M.; Trammell, A.R.; Shaw, L.M.; Goldstein, F.C.; Hajjar, I. Racial Disparity in Cerebrospinal Fluid Amyloid and Tau Biomarkers and Associated Cutoffs for Mild Cognitive Impairment. JAMA Netw. Open 2019, 2, e1917363. [Google Scholar] [CrossRef]
- Arakelyan, A.; Fitzgerald, W.; Vagida, M.; Vasilieva, E.; Margolis, L.; Grivel, J.-C. Addition of thrombin reduces the recovery of extracellular vesicles from blood plasma. J. Circ. Biomark. 2016, 5, 184945441666364. [Google Scholar] [CrossRef]
- Gomes, D.E.; Witwer, K.W. L1CAM-associated extracellular vesicles: A systematic review of nomenclature, sources, separation, and characterization. J. Extracell. Biol. 2022, 1, e35. [Google Scholar] [CrossRef]
- Badhwar, A.; Hirschberg, Y.; Valle-Tamayo, N.; Iulita, M.F.; Udeh-Momoh, C.T.; Matton, A.; Tarawneh, R.M.; Rissman, R.A.; Ledreux, A.; Winston, C.N.; et al. Assessment of brain-derived extracellular vesicle enrichment for blood biomarker analysis in age-related neurodegenerative diseases: An international overview. Alzheimer’s Dement. 2024, 20, 4411–4422. [Google Scholar] [CrossRef]






| CI (AD or MCI) | NC | |||
|---|---|---|---|---|
| Population Group (n) | MA (n = 24) | NHW (n = 24) | MA (n = 23) | NHW (n = 25) |
| Metabolic comorbidities, * count 0/1/2/3 comorbidities | 13/0/8/3 | 8/3/8/5 | 8/3/6/5 | 14/4/1/6 |
| Gender, count F/count M | 14/10 | 8/16 | 15/7 | 10/14 |
| Age, Mean (SD) | 70.6 (8.2) | 71.2 (5.1) | 68.7 (5.4) | 72.8 (7.8) |
| APOE e4 alleles, count 0 alleles/1 allele/2 alleles | 9/13/2 | 10/8/6 | 17/5/0 | 13/11/1 |
| miRNA | QIAGEN | DESeq | ||||
|---|---|---|---|---|---|---|
| Fold Change | FDR p-Value | p-Value | Fold Change | FDR p-Value | p-Value | |
| MA–Visit 1 | ||||||
| hsa-miR-122-5p | −2.6589 | 0.0951 | 0.006906 | −2.6095 | 1.37 × 10−6 | 1.48793 × 10−8 |
| MA–Visit 2 | ||||||
| hsa-miR-122-5p | −3.4067 | 0.00366 | 0.0000809 | −1.0303 | 0.084205 | 0.009356078 |
| hsa-let-7a-5p | 1.7354 | 0.03074 | 0.001768 | 0.8062 | 0.005604 | 0.000124533 |
| NHW–Visit 1 | ||||||
| hsa-miR-122-5p | −3.5135 | 0.00019 | 0.000002973 | −1.6541 | 0.002667 | 9.30459 × 10−5 |
| NHW–Visit 2 | ||||||
| hsa-miR-122-5p | −2.7712 | 0.00771 | 0.0001398 | −1.6402 | 0.005497 | 0.000279485 |
| hsa-let-7a-5p | 1.6885 | 0.0973 | 0.002646 | 1.1073 | 0.002107 | 5.95071 × 10−5 |
| miRNA | QIAGEN | DESeq | DESeq Controlled for CV | |||
|---|---|---|---|---|---|---|
| Fold Change | p-Value | Fold Change | p-Value | Fold Change | p-Value | |
| Visit 1 | ||||||
| hsa-miR-26a-5p | 2.2595 | 0.000378 | 0.8642 | 0.010922669 | 0.8726 | 0.01832371 |
| hsa-let-7f-5p | 1.8955 | 0.000396 | 0.7564 | 0.000507709 | 0.6397 | 0.006657008 |
| hsa-miR-320b | 2.3782 | 0.001096 | 0.9056 | 0.006483572 | 0.7316 | 0.045400752 |
| hsa-miR-320c | 2.2238 | 0.001604 | 0.9066 | 0.011943306 | N/A | N/A |
| hsa-let-7d-5p | 1.7734 | 0.002541 | 0.5524 | 0.011829511 | 0.5260 | 0.03158259 |
| hsa-miR-139-5p | 2.4935 | 0.007471 | 1.1965 | 0.016015759 | N/A | N/A |
| hsa-miR-155-5p | 3.0543 | 0.007599 | 1.7612 | 0.005540475 | 1.7734 | 0.009433616 |
| hsa-miR-320d | 1.9904 | 0.009145 | 0.8377 | 0.031186727 | N/A | N/A |
| Visit 2 | ||||||
| hsa-miR-26a-5p | 3.3986 | 1.34 × 10−6 | 1.6468 | 3.22755 × 10−6 | 1.7479 | 1.77526 × 10−5 |
| hsa-miR-98-5p | 2.6111 | 0.000258 | 1.1289 | 0.005393586 | 0.9795 | 0.038377946 |
| hsa-miR-16-5p | −1.9802 | 0.000356 | −0.8942 | 9.9069 × 10−5 | −0.7785 | 0.002250029 |
| hsa-miR-744-5p | 2.7088 | 0.000648 | 1.5357 | 0.002876615 | 1.5806 | 0.009644383 |
| hsa-miR-432-5p | 3.3424 | 0.000753 | 1.3822 | 0.023778746 | N/A | N/A |
| hsa-miR-25-3p | −2.0924 | 0.001285 | −0.9827 | 0.003175884 | −0.8134 | 0.033630161 |
| hsa-miR-29c-3p | −2.1563 | 0.001582 | −0.7637 | 0.046790757 | N/A | N/A |
| hsa-let-7f-5p | 1.8117 | 0.002175 | 0.8449 | 0.000388528 | 0.8964 | 0.001432407 |
| hsa-let-7e-5p | 1.9017 | 0.002485 | 0.9063 | 0.001089766 | 0.9029 | 0.005154222 |
| hsa-miR-652-3p | 2.9118 | 0.003041 | 1.7948 | 0.008419156 | N/A | N/A |
| hsa-miR-451a | −2.1719 | 0.003222 | −0.8719 | 0.040413382 | N/A | N/A |
| hsa-miR-181a-5p | 2.9416 | 0.00499 | 2.0489 | 0.00157171 | 1.8985 | 0.010124267 |
| hsa-miR-382-5p | 2.5017 | 0.006856 | 1.2004 | 0.040084269 | N/A | N/A |
| hsa-miR-32-5p | −2.7186 | 0.006956 | −2.0868 | 0.013834039 | −3.0281 | 0.003596737 |
| hsa-miR-144-3p | −2.0347 | 0.011515 | −1.1413 | 0.024266645 | −1.2024 | 0.039307561 |
| hsa-miR-26b-5p | 1.8652 | 0.011878 | 0.8465 | 0.014921276 | 0.8327 | 0.040978652 |
| hsa-miR-139-5p | 1.9814 | 0.014604 | 1.1088 | 0.010990028 | 0.9906 | 0.048569444 |
| hsa-miR-628-3p | 2.2481 | 0.015016 | 1.5135 | 0.030302303 | 1.8781 | 0.026313543 |
| miRNA | QIAGEN | DESeq | DESeq Controlled for CV | |||
|---|---|---|---|---|---|---|
| Fold Change | p-Value | Fold Change | p-Value | Fold Change | p-Value | |
| Visit 1 | ||||||
| hsa-miR-184 | 22.8945 | 1.31 × 10−12 | 2.8623 | 2.55508 × 10−5 | 2.7454 | 0.001225839 |
| hsa-miR-486-5p | −2.5550 | 8.21 × 10−6 | −1.0808 | 3.81796 × 10−5 | −1.1442 | 1.17529 × 10−5 |
| hsa-miR-133a-3p | 4.8088 | 0.000136 | 3.1374 | 0.000782628 | N/A | N/A |
| hsa-miR-1260b | 3.1867 | 0.000232 | 1.7908 | 0.000977265 | 1.1588 | 0.035758812 |
| hsa-miR-125b-5p | 4.0213 | 0.000302 | 2.3179 | 0.005661509 | 2.5050 | 0.003892795 |
| hsa-miR-1260a | 2.7921 | 0.000428 | 1.7995 | 0.000943134 | 1.1201 | 0.042932868 |
| hsa-miR-451a | −2.9846 | 0.000709 | −1.5134 | 0.003312469 | −1.3089 | 0.013690742 |
| hsa-miR-150-5p | −2.3846 | 0.000724 | −1.3585 | 0.010751915 | −1.4706 | 0.009600561 |
| hsa-miR-30a-3p | −2.5455 | 0.001874 | −1.2236 | 0.04022529 | −1.2556 | 0.03173225 |
| hsa-miR-4732-5p | −2.8367 | 0.001951 | −1.2917 | 0.021940416 | −1.3245 | 0.022124385 |
| hsa-miR-629-5p | −1.9755 | 0.003393 | -0.9841 | 0.006541369 | −0.9703 | 0.008959777 |
| hsa-miR-100-5p | 3.0964 | 0.004191 | 2.4792 | 0.000331924 | N/A | N/A |
| hsa-let-7i-5p | −1.5609 | 0.007761 | −0.3969 | 0.025137485 | −0.3864 | 0.019854645 |
| Visit 2 | ||||||
| hsa-miR-31-5p | 15.4400 | 2.63 × 10−6 | 3.4495 | 0.027186298 | N/A | N/A |
| hsa-miR-100-5p | 7.2821 | 4.86 × 10−6 | 3.4026 | 8.0048 × 10−6 | 2.4187 | 0.002164492 |
| hsa-miR-4674 | 5.6239 | 7.35 × 10−6 | 3.0092 | 0.000164365 | 2.2139 | 0.008434245 |
| hsa-miR-133a-3p | 6.0388 | 4.13 × 10−5 | 3.4721 | 1.53595 × 10−5 | 2.3877 | 0.00527116 |
| hsa-miR-125b-5p | 5.7115 | 8.03 × 10−5 | 2.1817 | 0.006523807 | N/A | N/A |
| hsa-miR-101-3p | 3.9138 | 0.000603 | 2.6056 | 0.010452733 | 2.7051 | 0.015040877 |
| hsa-miR-7-5p | 2.5373 | 0.000813 | 1.4101 | 0.003260672 | 1.4041 | 0.003602864 |
| QIAGEN RNA-Seq Portal | DESeq2 | ||||
|---|---|---|---|---|---|
| miRNA | Fold Change | p-Value | miRNA | Fold Change | p-Value |
| MA with CI | |||||
| Overrepresented | Overrepresented | ||||
| hsa-miR-499a-5p | 21.1916 | 0.00003385 | hsa-miR-324-5p | 1.8707 | 0.00164 |
| hsa-miR-26a-5p | 0.9759 | 0.00236 | |||
| hsa-miR-1307-3p | 1.2662 | 0.01342 | |||
| hsa-miR-98-5p | 0.9709 | 0.01389 | |||
| hsa-miR-326 | 2.2103 | 0.01759 | |||
| hsa-miR-744-5p | 1.1047 | 0.02441 | |||
| hsa-miR-12136 | 2.8788 | 0.03226 | |||
| hsa-miR-26b-5p | 0.6761 | 0.03857 | |||
| hsa-miR-186-5p | 1.8926 | 0.04413 | |||
| hsa-miR-142-3p | 0.9846 | 0.0454 | |||
| Underrepresented | |||||
| hsa-miR-184 | −2.2055 | 0.00095 | |||
| hsa-miR-1268b | −2.7423 | 0.00795 | |||
| hsa-miR-133b | −3.1213 | 0.03241 | |||
| hsa-miR-4787-5p | −2.4008 | 0.04179 | |||
| NHW with CI | |||||
| Underrepresented | Overrepresented | ||||
| hsa-miR-1-3p | −11.80037074 | 7.598 × 10−9 | hsa-miR-877-5p | 1.419898676 | 0.01123 |
| hsa-miR-7-5p | −4.325563298 | 0.0001131 | hsa-miR-1908-5p | 1.52342791 | 0.02548 |
| hsa-miR-184 | −5.240625245 | 0.0002988 | hsa-miR-483-5p | 1.452485598 | 0.02763 |
| hsa-miR-3182 | −5.759964762 | 0.001183 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subasinghe, K.; Hall, C.; Rowe, M.; Zhou, Z.; Barber, R.; Phillips, N. Neuronal Enriched Extracellular Vesicle miR-122-5p as a Potential Biomarker for Alzheimer’s Disease. Cells 2025, 14, 1784. https://doi.org/10.3390/cells14221784
Subasinghe K, Hall C, Rowe M, Zhou Z, Barber R, Phillips N. Neuronal Enriched Extracellular Vesicle miR-122-5p as a Potential Biomarker for Alzheimer’s Disease. Cells. 2025; 14(22):1784. https://doi.org/10.3390/cells14221784
Chicago/Turabian StyleSubasinghe, Kumudu, Courtney Hall, Megan Rowe, Zhengyang Zhou, Robert Barber, and Nicole Phillips. 2025. "Neuronal Enriched Extracellular Vesicle miR-122-5p as a Potential Biomarker for Alzheimer’s Disease" Cells 14, no. 22: 1784. https://doi.org/10.3390/cells14221784
APA StyleSubasinghe, K., Hall, C., Rowe, M., Zhou, Z., Barber, R., & Phillips, N. (2025). Neuronal Enriched Extracellular Vesicle miR-122-5p as a Potential Biomarker for Alzheimer’s Disease. Cells, 14(22), 1784. https://doi.org/10.3390/cells14221784






