Reversible Upregulation of the Senescence-Associated Beta-Galactosidase Marker Induced by Cell Detachment in Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
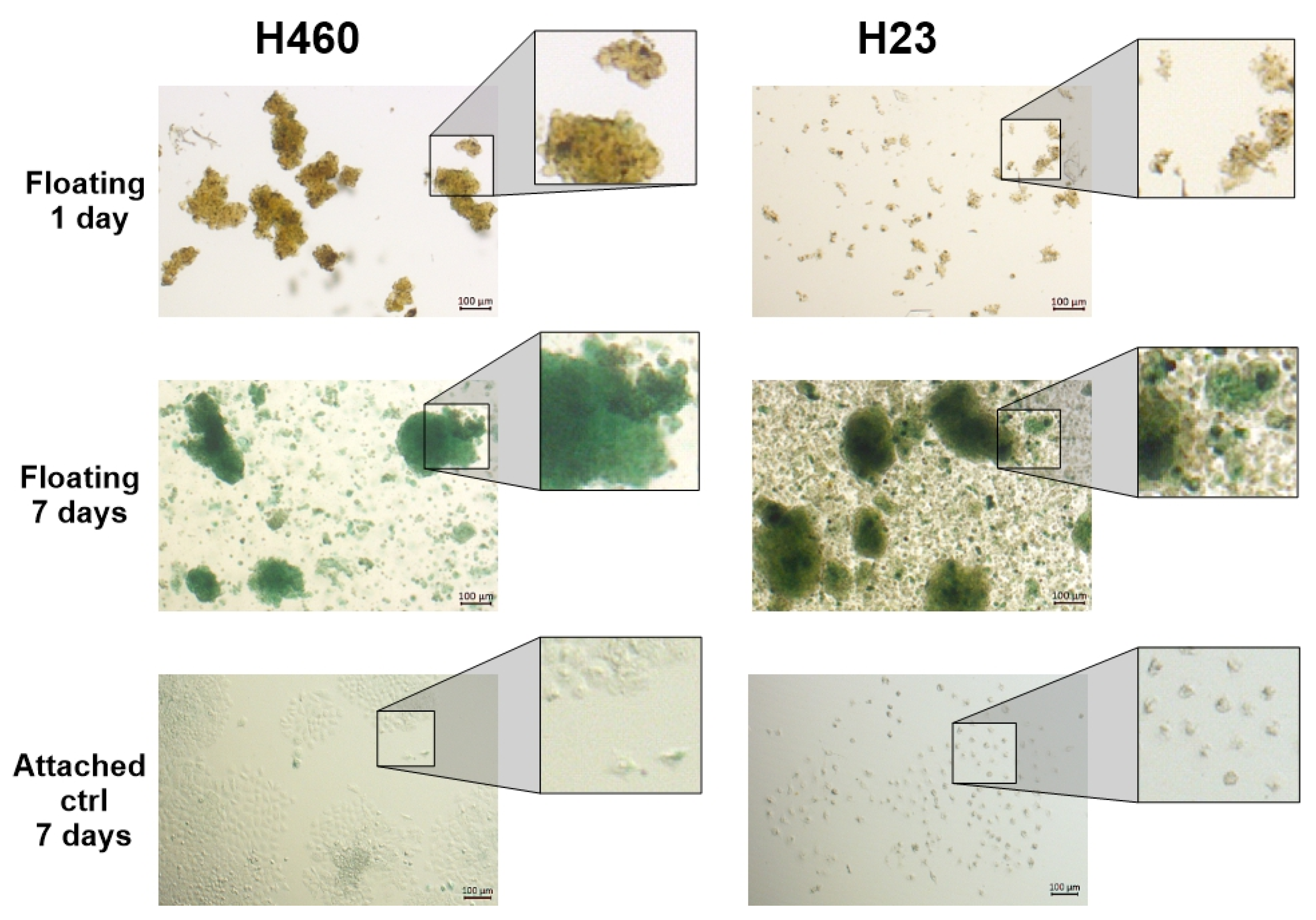
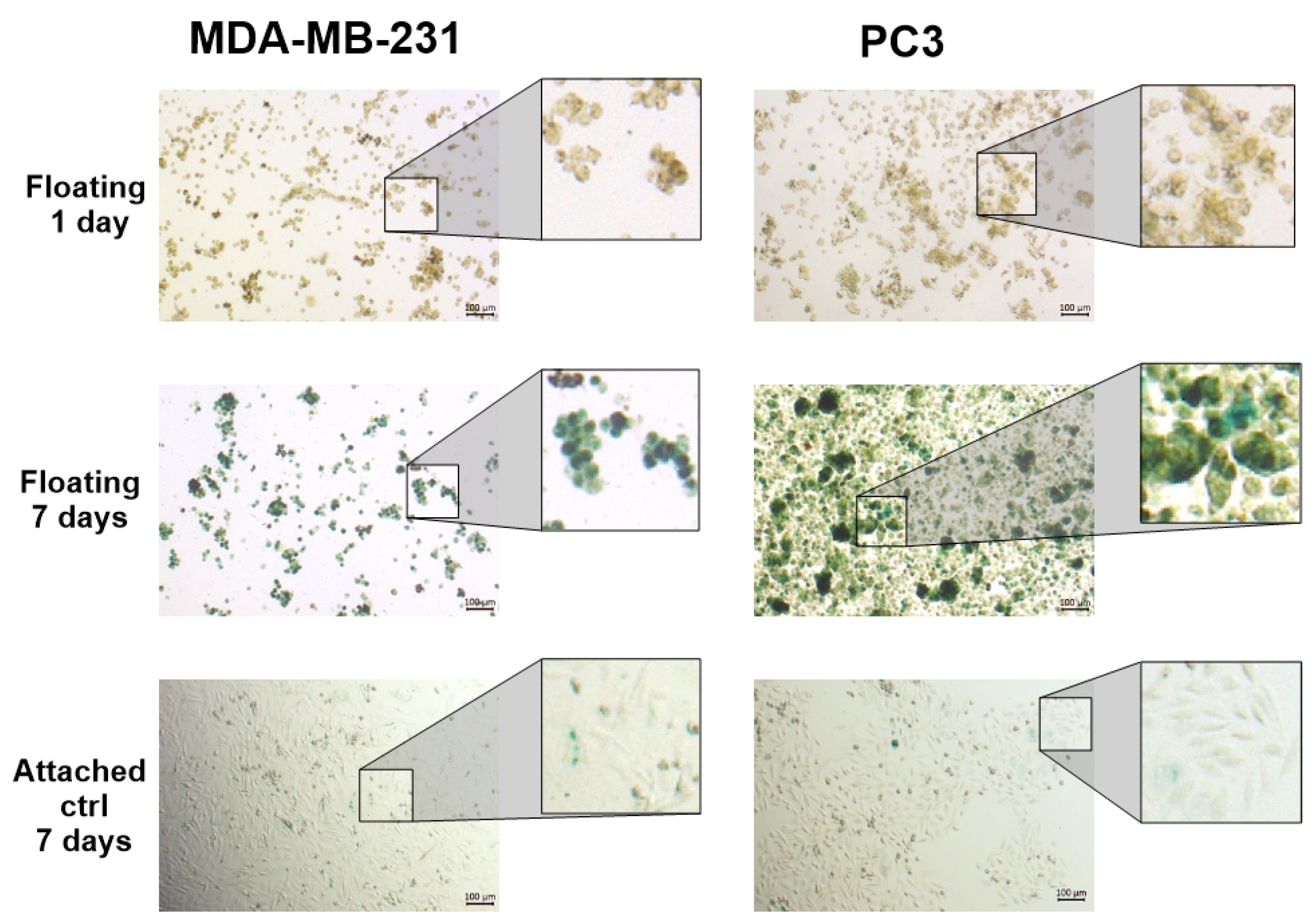
3.1. Floating Spheroids Growing Under AICs Display High Levels of SA-βG Activity
3.2. SA-βG Activity Increases in Floating Cells Independently of Their Ability to Form Complex 3D Structures
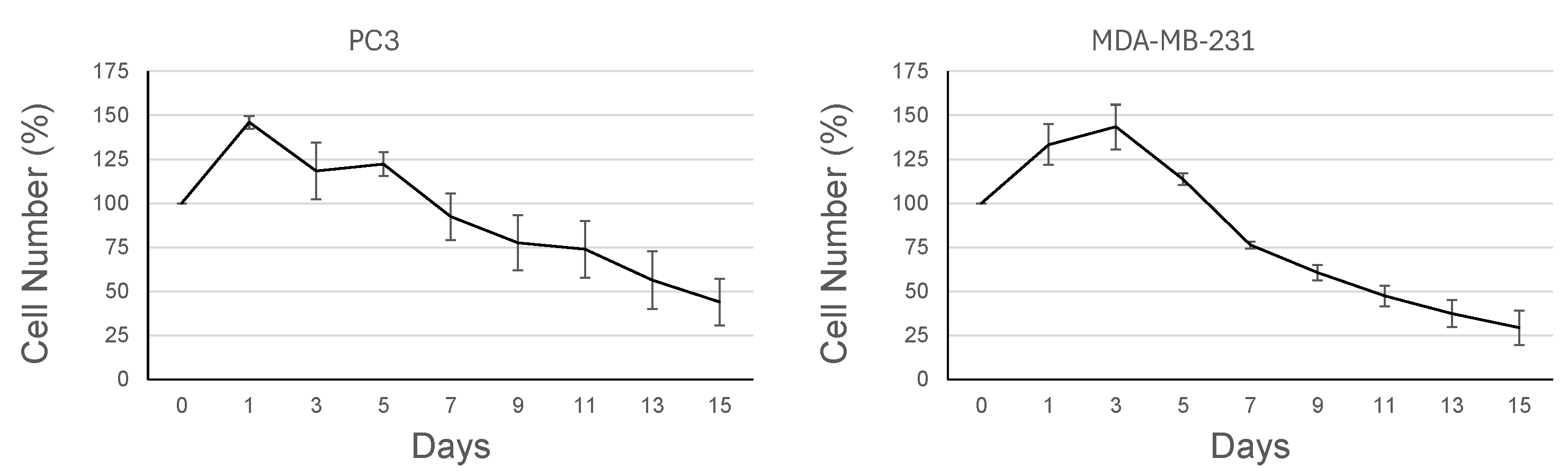
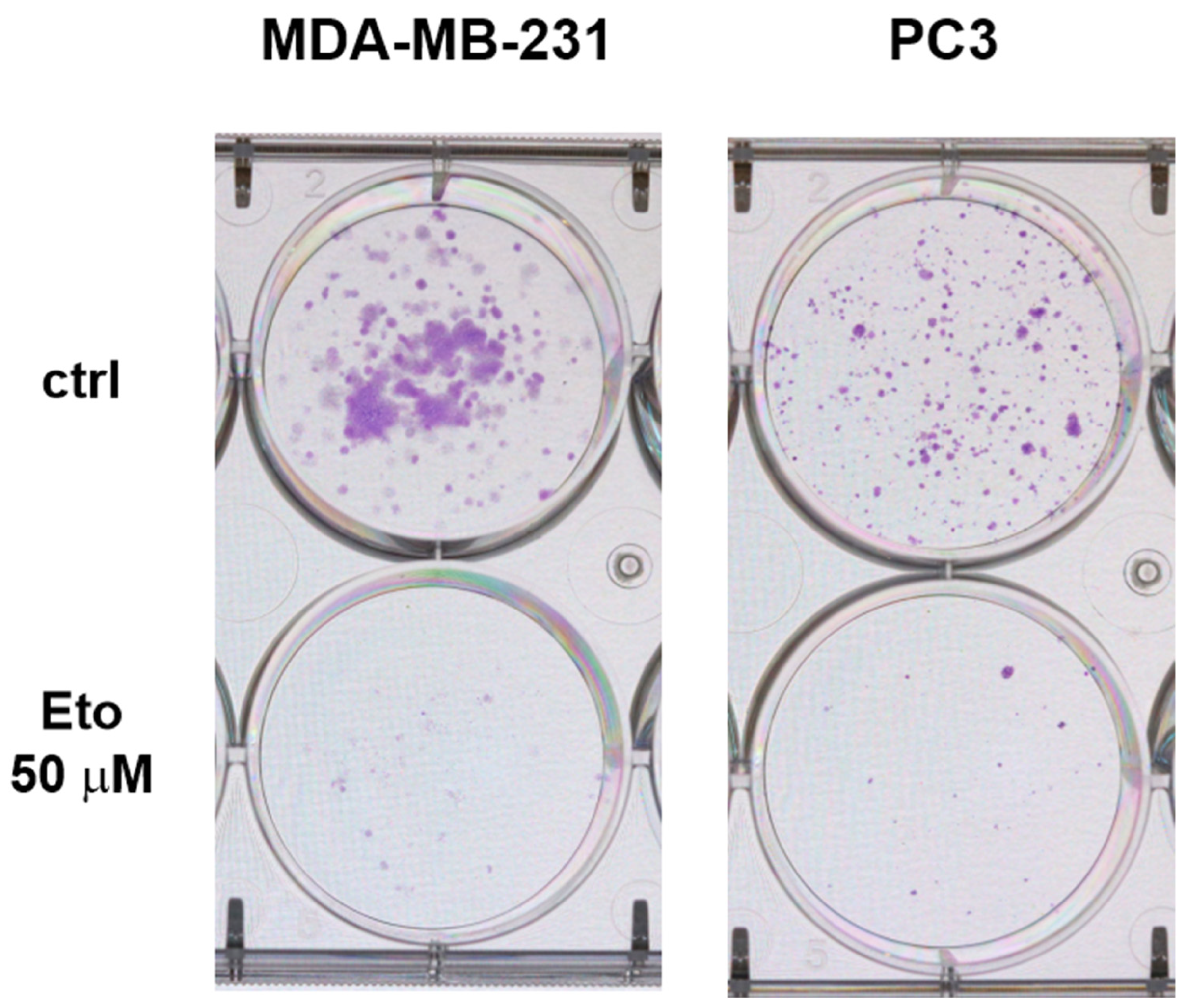
3.3. Proliferative Activity Is Reduced Under AICs
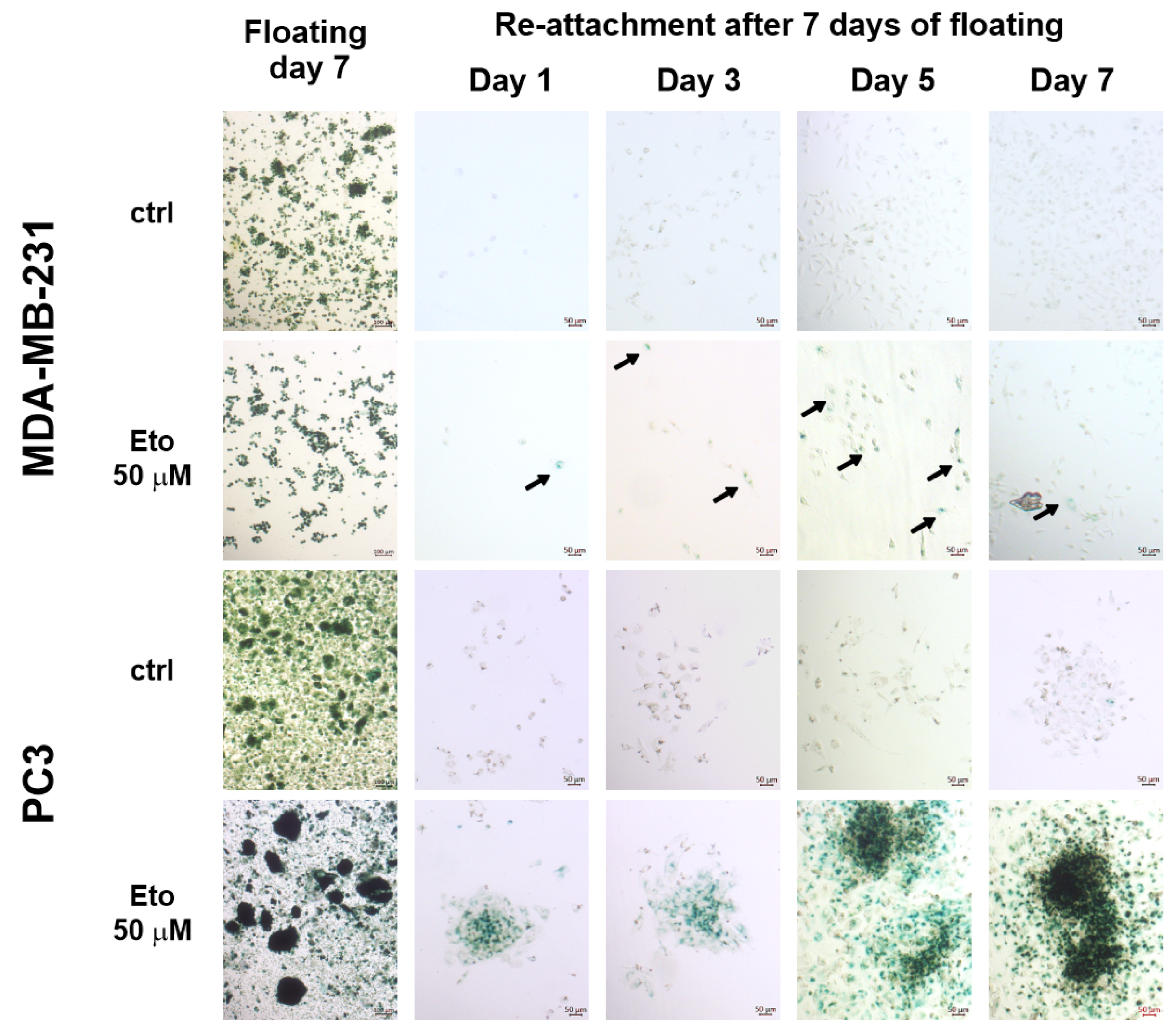
3.4. Cells Grown Under AICs Escape the Q/S State in a Plastic Rather than Stochastic Manner
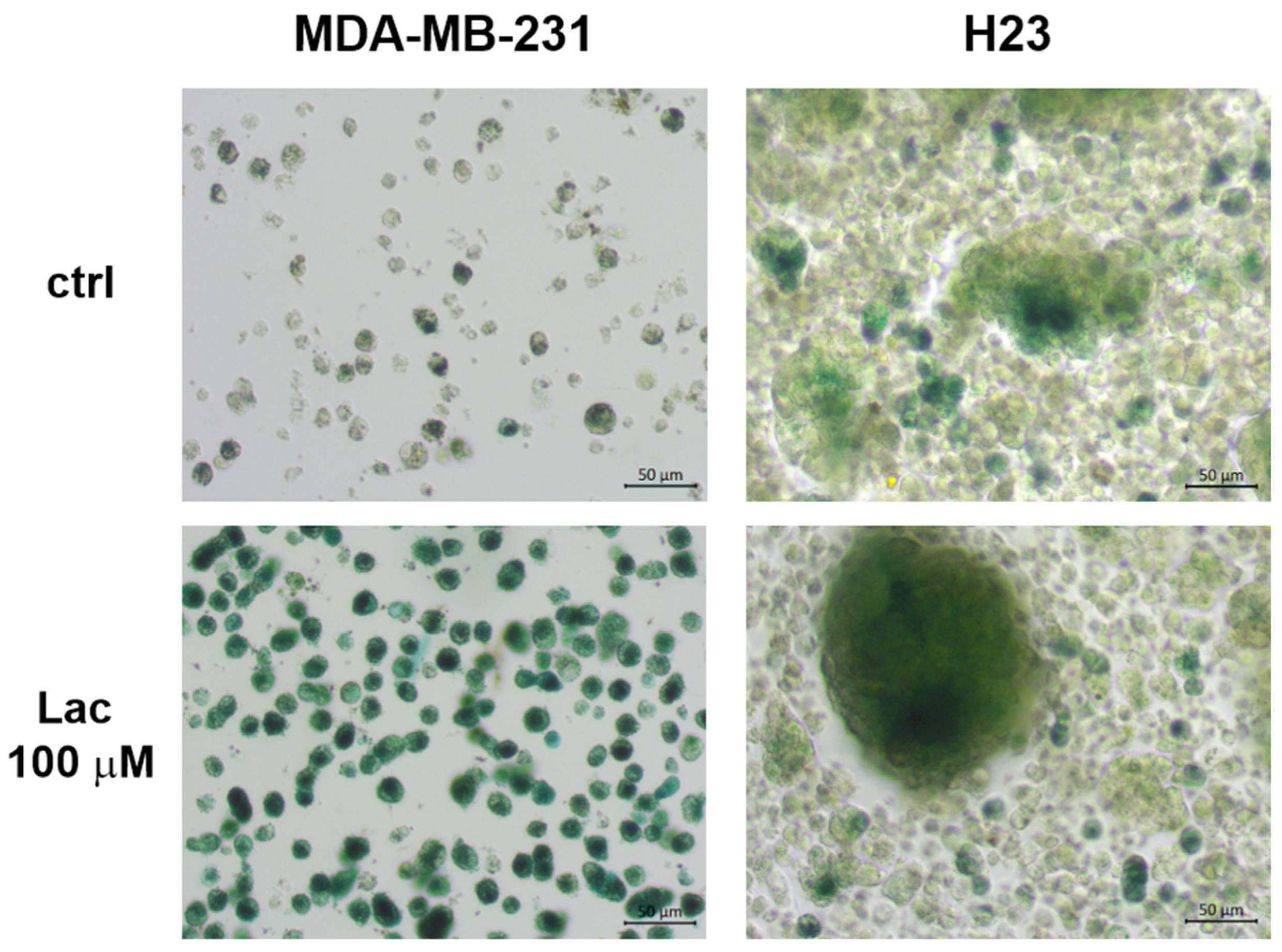
3.5. Lactose Increases the Intensity of β-Gal Expression in Cells Grown Under AICs
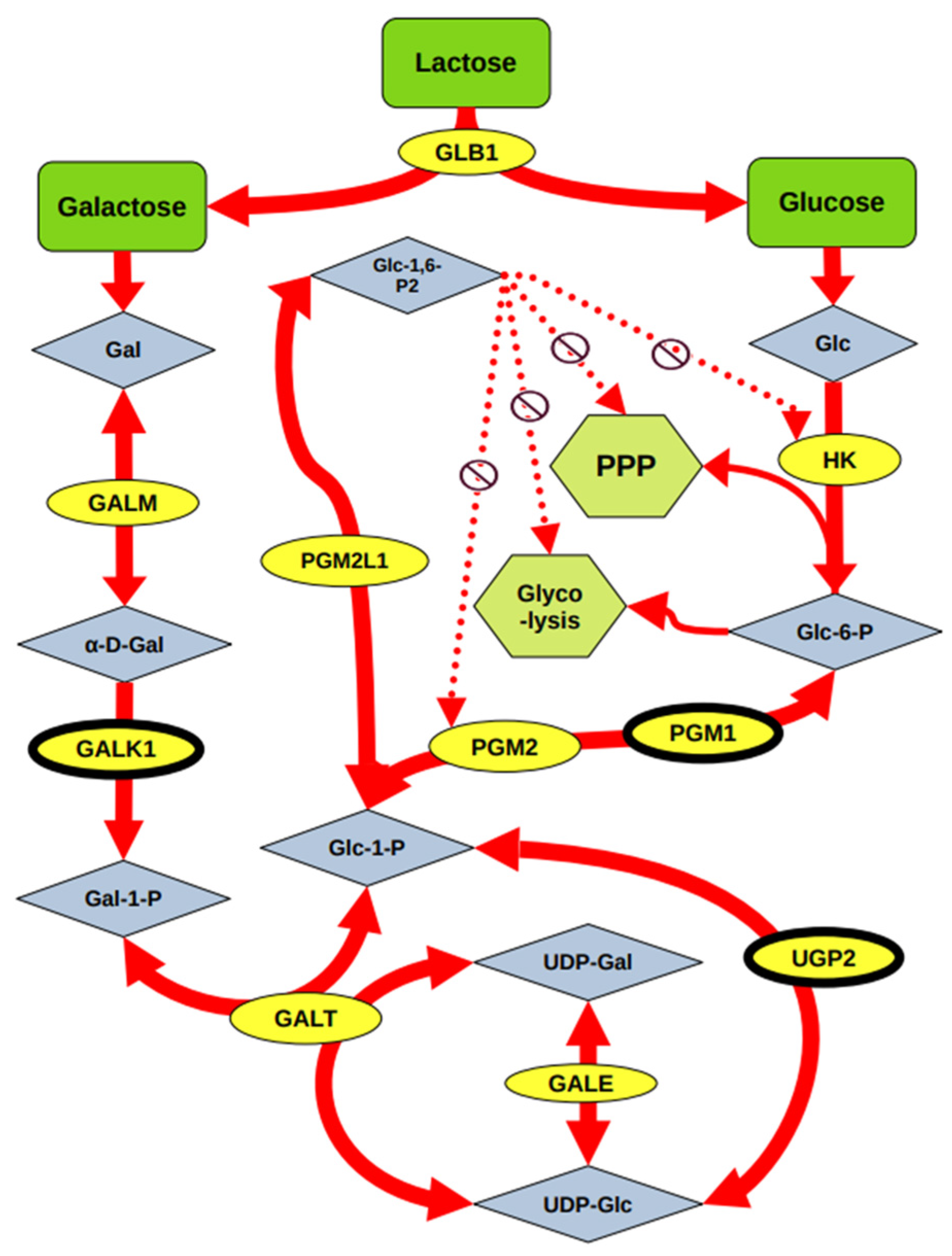
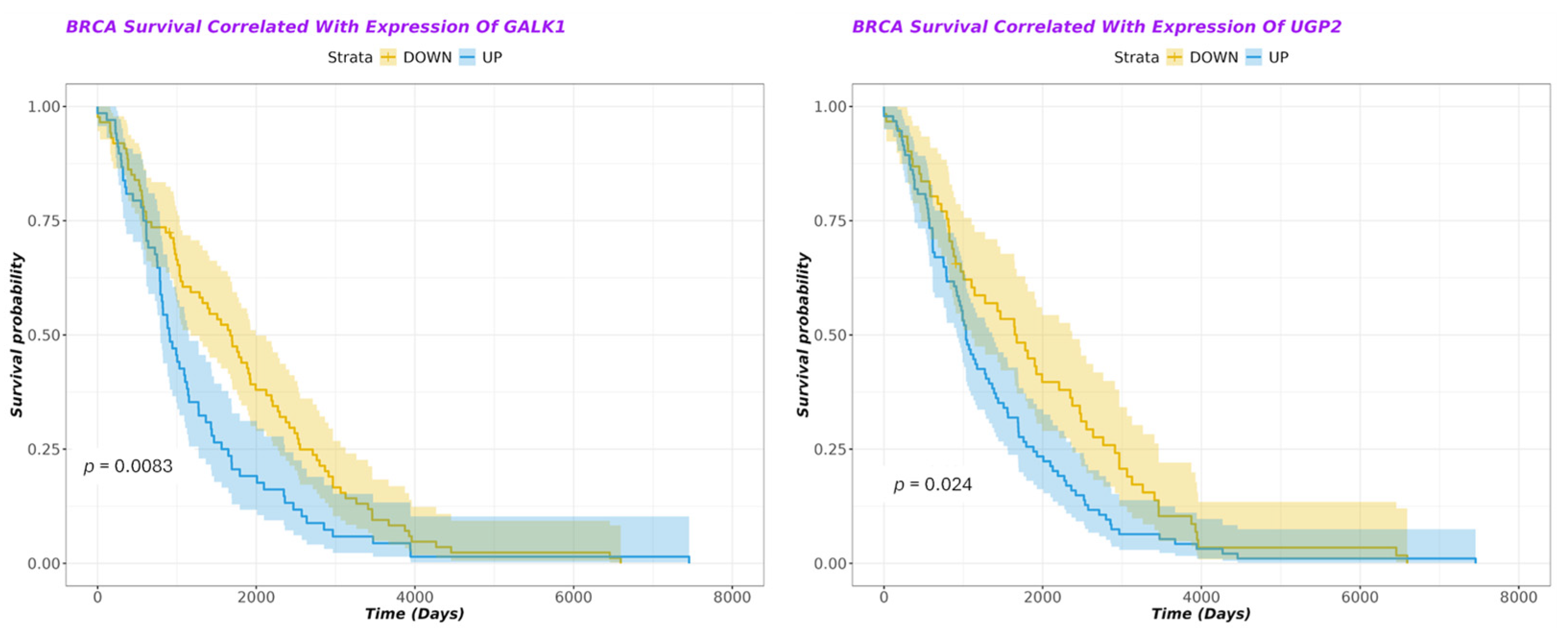
3.6. Dysregulation of Protein Expression Involved in Lactose Metabolism Is Associated with Poor Prognosis in Breast Cancer
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AICs | Anchorage-independent culture conditions |
| ADCs | Anchorage-dependent culture conditions |
| ULAPs | Ultra-low attachment plates |
| Q/S | Quiescent/senescent |
References
- Andryszkiewicz, W.; Gąsiorowska, J.; Kübler, M.; Kublińska, K.; Pałkiewicz, A.; Wiatkowski, A.; Szwedowicz, U.; Choromańska, A. Glucose Metabolism and Tumor Microenvironment: Mechanistic Insights and Therapeutic Implications. Int. J. Mol. Sci. 2025, 26, 1879. [Google Scholar] [CrossRef]
- Jan, A.; Sofi, S.; Jan, N.; Mir, M.A. An update on cancer stem cell survival pathways involved in chemoresistance in triple-negative breast cancer. Future Oncol. 2025, 21, 715–735. [Google Scholar] [CrossRef] [PubMed]
- Kilmister, E.J.; Tan, S.T. Cancer Stem Cells and the Renin-Angiotensin System in the Tumor Microenvironment of Melanoma: Implications on Current Therapies. Int. J. Mol. Sci. 2025, 26, 1389. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Hassani, M.; Moghaddam, M.B.; Fazilat, A.; Ojarudi, M.; Valilo, M. Contribution of tumor microenvironment (TME) to tumor apoptosis, angiogenesis, metastasis, and drug resistance. Med. Oncol. 2025, 42, 108. [Google Scholar] [CrossRef]
- Atat, O.E.; Farzaneh, Z.; Pourhamzeh, M.; Taki, F.; Abi-Habib, R.; Vosough, M.; El-Sibai, M. 3D modeling in cancer studies. Hum. Cell 2022, 35, 23–36. [Google Scholar] [CrossRef] [PubMed]
- Dzobo, K. Taking a Full Snapshot of Cancer Biology: Deciphering the Tumor Microenvironment for Effective Cancer Therapy in the Oncology Clinic. Omics 2020, 24, 175–179. [Google Scholar] [CrossRef]
- Ponomarev, A.S.; Gilazieva, Z.E.; Solovyova, V.V.; Rizvanov, A.A. Molecular Mechanisms of Tumor Cell Stemness Modulation during Formation of Spheroids. Biochemistry 2023, 88, 979–994. [Google Scholar] [CrossRef]
- Shehzad, A.; Ravinayagam, V.; AlRumaih, H.; Aljafary, M.; Almohazey, D.; Almofty, S.; Al-Rashid, N.A.; Al-Suhaimi, E.A. Application of Three-dimensional (3D) Tumor Cell Culture Systems and Mechanism of Drug Resistance. Curr. Pharm. Des. 2019, 25, 3599–3607. [Google Scholar] [CrossRef]
- Yang, M.; Liu, P.; Huang, P. Cancer stem cells, metabolism, and therapeutic significance. Tumour Biol. 2016, 37, 5735–5742. [Google Scholar] [CrossRef]
- Kaushik, V.; Yakisich, J.S.; Way, L.F.; Azad, N.; Iyer, A.K.V. Chemoresistance of cancer floating cells is independent of their ability to form 3D structures: Implications for anticancer drug screening. J. Cell Physiol. 2019, 234, 4445–4453. [Google Scholar] [CrossRef]
- Lindell, E.; Zhong, L.; Zhang, X. Quiescent Cancer Cells-A Potential Therapeutic Target to Overcome Tumor Resistance and Relapse. Int. J. Mol. Sci. 2023, 24, 3762. [Google Scholar] [CrossRef]
- Chakrabarty, A.; Chakraborty, S.; Bhattacharya, R.; Chowdhury, G. Senescence-Induced Chemoresistance in Triple Negative Breast Cancer and Evolution-Based Treatment Strategies. Front. Oncol. 2021, 11, 674354. [Google Scholar] [CrossRef]
- Dodig, S.; Čepelak, I.; Pavić, I. Hallmarks of senescence and aging. Biochem. Med. 2019, 29, 030501. [Google Scholar] [CrossRef] [PubMed]
- Beauséjour, C.M.; Krtolica, A.; Galimi, F.; Narita, M.; Lowe, S.W.; Yaswen, P.; Campisi, J. Reversal of human cellular senescence: Roles of the p53 and p16 pathways. EMBO J. 2003, 22, 4212–4222. [Google Scholar] [CrossRef]
- Milanovic, M.; Fan, D.N.Y.; Belenki, D.; Däbritz, J.H.M.; Zhao, Z.; Yu, Y.; Dörr, J.R.; Dimitrova, L.; Lenze, D.; Monteiro Barbosa, I.A.; et al. Senescence-associated reprogramming promotes cancer stemness. Nature 2018, 553, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Hwang, E.S. Status of mTOR activity may phenotypically differentiate senescence and quiescence. Mol. Cells 2012, 33, 597–604. [Google Scholar] [CrossRef]
- Kim, H.; Sung, J.Y.; Park, E.K.; Kho, S.; Koo, K.H.; Park, S.Y.; Goh, S.H.; Jeon, Y.K.; Oh, S.; Park, B.K.; et al. Regulation of anoikis resistance by NADPH oxidase 4 and epidermal growth factor receptor. Br. J. Cancer 2017, 116, 370–381. [Google Scholar] [CrossRef]
- Dimri, G.P.; Lee, X.; Basile, G.; Acosta, M.; Scott, G.; Roskelley, C.; Medrano, E.E.; Linskens, M.; Rubelj, I.; Pereira-Smith, O.; et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 1995, 92, 9363–9367. [Google Scholar] [CrossRef]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. Curr. Protoc. Immunol. 2015, 111, A3.B.1–A3.B.3. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, M.A.; Ijare, O.B.; Baskin, D.S.; Baskin, A.M.; Baskin, B.N.; Pichumani, K. The Leloir Cycle in Glioblastoma: Galactose Scavenging and Metabolic Remodeling. Cancers 2021, 13, 1815. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef] [PubMed]
- Mounir, M.; Lucchetta, M.; Silva, T.C.; Olsen, C.; Bontempi, G.; Chen, X.; Noushmehr, H.; Colaprico, A.; Papaleo, E. New functionalities in the TCGAbiolinks package for the study and integration of cancer data from GDC and GTEx. PLoS Comput. Biol. 2019, 15, e1006701. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G.K. Robust Hyperparameter Estimation Protects Against Hypervariable Genes And Improves Power To Detect Differential Expression. Ann. Appl. Stat. 2016, 10, 946–963. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: New York, NY, USA, 2000; p. 350. [Google Scholar]
- Lee, H.G.; Kim, J.H.; Sun, W.; Chi, S.G.; Choi, W.; Lee, K.J. Senescent tumor cells building three-dimensional tumor clusters. Sci. Rep. 2018, 8, 10503. [Google Scholar] [CrossRef]
- Rovere, M.; Reverberi, D.; Arnaldi, P.; Palamà, M.E.F.; Gentili, C. Spheroid size influences cellular senescence and angiogenic potential of mesenchymal stromal cell-derived soluble factors and extracellular vesicles. Front. Bioeng. Biotechnol. 2023, 11, 1297644. [Google Scholar] [CrossRef]
- Xu, S.; Ma, L.; Wu, T.; Tian, Y.; Wu, L. Assessment of cellular senescence potential of PM2.5 using 3D human lung fibroblast spheroids in vitro model. Toxicol. Res. 2024, 13, tfae037. [Google Scholar] [CrossRef] [PubMed]
- Terzi, M.Y.; Izmirli, M.; Gogebakan, B. The cell fate: Senescence or quiescence. Mol. Biol. Rep. 2016, 43, 1213–1220. [Google Scholar] [CrossRef] [PubMed]
- Foster, D.A.; Yellen, P.; Xu, L.; Saqcena, M. Regulation of G1 Cell Cycle Progression: Distinguishing the Restriction Point from a Nutrient-Sensing Cell Growth Checkpoint(s). Genes. Cancer 2010, 1, 1124–1131. [Google Scholar] [CrossRef]
- Delwar, Z.; Avramidis, D.; Ake, S.; Cruz, M.; Paulsson, K.; Yakisich, J.S. Low Concentration of Salinomycin Prevents Regrowth and Partially Depletes Human Glioma Cells Surviving High Concentrations of Alkylating Agents. Clin. Cancer Drugs 2014, 1, 72–77. [Google Scholar] [CrossRef]
- Delwar, Z.M.; Avramidis, D.; Siden, A.; Cruz, M.; Yakisich, J.S. Depletion of drug-surviving glioma cells by a second phase treatment with low concentration of salinomycin. Drugs Ther. Stud. 2011, 1, e7. [Google Scholar] [CrossRef]
- Bang, M.; Kim, D.G.; Gonzales, E.L.; Kwon, K.J.; Shin, C.Y. Etoposide Induces Mitochondrial Dysfunction and Cellular Senescence in Primary Cultured Rat Astrocytes. Biomol. Ther. 2019, 27, 530–539. [Google Scholar] [CrossRef] [PubMed]
- Pioger, A.; Loison, I.; Metatla, I.; Spruyt, N.; Abbadie, C.; Dehennaut, V. Comparative analysis of senescence induction by different chemotherapeutic agents in HCT116 colon cancer cells. Biochem. Biophys. Res. Commun. 2025, 752, 151482. [Google Scholar] [CrossRef]
- Yakisich, J.S.; Azad, N.; Kaushik, V.; Iyer, A.K.V. Cancer Cell Plasticity: Rapid Reversal of Chemosensitivity and Expression of Stemness Markers in Lung and Breast Cancer Tumorspheres. J. Cell Physiol. 2017, 232, 2280–2286. [Google Scholar] [CrossRef]
- Yang, N.C.; Hu, M.L. The limitations and validities of senescence associated-beta-galactosidase activity as an aging marker for human foreskin fibroblast Hs68 cells. Exp. Gerontol. 2005, 40, 813–819. [Google Scholar] [CrossRef]
- Yakisich, J.S.; Venkatadri, R.; Azad, N.; Iyer, A.K.V. Chemoresistance of Lung and Breast Cancer Cells Growing Under Prolonged Periods of Serum Starvation. J. Cell Physiol. 2017, 232, 2033–2043. [Google Scholar] [CrossRef]
- Scialo, T.E.; Pace, C.M.; Abrams, D.I. The Dairy and Cancer Controversy: Milking the Evidence. Curr. Oncol. Rep. 2024, 26, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Arthur, P.G.; Kent, J.C.; Potter, J.M.; Hartmann, P.E. Lactose in blood in nonpregnant, pregnant, and lactating women. J. Pediatr. Gastroenterol. Nutr. 1991, 13, 254–259. [Google Scholar] [CrossRef]
- Xing, S.; Zhang, L.; Lin, H.; Mao, Z.; Bao, K.; Hao, P.; Pei, Z.; Li, J.; Hu, Z. Lactose induced redox-dependent senescence and activated Nrf2 pathway. Int. J. Clin. Exp. Pathol. 2019, 12, 2034–2045. [Google Scholar]
- Wolfe, A.L.; Zhou, Q.; Toska, E.; Galeas, J.; Ku, A.A.; Koche, R.P.; Bandyopadhyay, S.; Scaltriti, M.; Lebrilla, C.B.; McCormick, F.; et al. UDP-glucose pyrophosphorylase 2, a regulator of glycogen synthesis and glycosylation, is critical for pancreatic cancer growth. Proc. Natl. Acad. Sci. USA 2021, 118, e2103592118. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Han, B.; Luo, H.; Ling, H.; Hu, X. Integrated Multi-Omics Profiling of Young Breast Cancer Patients Reveals a Correlation between Galactose Metabolism Pathway and Poor Disease-Free Survival. Cancers 2023, 15, 4637. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Zhang, K.; Liang, Z.; Zhang, S.; Dai, Y.; Cong, Y.; Qiao, G. Integrated transcriptome analysis identifies APPL1/RPS6KB2/GALK1 as immune-related metastasis factors in breast cancer. Open Med. 2023, 18, 20230732. [Google Scholar] [CrossRef]
- Hu, Q.; Shen, S.; Li, J.; Liu, L.; Liu, X.; Zhang, Y.; Zhou, Y.; Zhu, W.; Yu, Y.; Cui, G. Low UGP2 Expression Is Associated with Tumour Progression and Predicts Poor Prognosis in Hepatocellular Carcinoma. Dis. Markers 2020, 2020, 3231273. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Guo, Y.; Ning, W.; Wang, X.; Li, S.; Chen, Y.; Ma, L.; Qu, Y.; Song, Y.; Zhang, H. Comprehensive Analyses of Glucose Metabolism in Glioma Reveal the Glioma-Promoting Effect of GALM. Front. Cell Dev. Biol. 2021, 9, 717182. [Google Scholar] [CrossRef]
| Gene Symbol | Pathway | Expression | LogFC | Effect on Survival | p-Value (Survival) |
|---|---|---|---|---|---|
| ACOT8 | Dicarboxylate metabolism | DOWN | −2.3 | Deleterious | 0.00012 |
| GSTO2 | Ascorbate metabolism | DOWN | −1.9 | Deleterious | 0.00099 |
| IDH1 | TCA pathway | DOWN | −2.3 | Deleterious | 0.0024 |
| KYAT3 | Dicarboxylate metabolism | UP | 0.95 | Improved | 0.0042 |
| ALDOC | Glycolytic processes | UP | 0.77 | Improved | 0.0056 |
| HK3 | Glycolytic processes | UP | 0.56 | Deleterious | 0.0059 |
| DLST | TCA pathway | DOWN | −0.42 | Deleterious | 0.0063 |
| ADHFE1 | Dicarboxylate metabolism | UP | 0.7 | Deleterious | 0.0067 |
| GALK1 | Galactose metabolism | UP | 0.6 | Deleterious | 0.0083 |
| QPRT | Dicarboxylate metabolism | UP | 0.8 | Deleterious | 0.0087 |
| PFKL | Glycolytic processes | UP | 0.41 | Deleterious | 0.017 |
| SLC37A4 | Gluconeogenesis pathway | UP | 0.5 | Improved | 0.017 |
| IDH3B | TCA pathway | UP | 1.1 | Deleterious | 0.019 |
| UGP2 | Galactose metabolism | DOWN | −1.03 | Improved | 0.024 |
| CSKMT | TCA cycle pathway | UP | 0.35 | Improved | 0.027 |
| AGXT | Glyoxylate metabolism | DOWN | −0.43 | Improved | 0.029 |
| ASL | Dicarboxylate metabolism | UP | 0.81 | Improved | 0.029 |
| PGK1 | Glycolytic processes | UP | 0.35 | Deleterious | 0.03 |
| MTHFD1L | Dicarboxylate metabolism | DOWN | −1.2 | Improved | 0.03 |
| PGAM1 | Glycolytic processes | UP | 0.39 | Deleterious | 0.032 |
| UGT2B15 | Pentose glucuronate interconversions | UP | 0.15 | Improved | 0.034 |
| DHTKD1 | Glycolytic processes | DOWN | −0.16 | Deleterious | 0.037 |
| GAA | Sucrose metabolism | UP | 0.28 | Improved | 0.049 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Semenova, N.; Yakisich, J.S.; Ascue, R.; Iyer, A.K.V.; Azad, N. Reversible Upregulation of the Senescence-Associated Beta-Galactosidase Marker Induced by Cell Detachment in Cancer Cells. Cells 2025, 14, 1667. https://doi.org/10.3390/cells14211667
Semenova N, Yakisich JS, Ascue R, Iyer AKV, Azad N. Reversible Upregulation of the Senescence-Associated Beta-Galactosidase Marker Induced by Cell Detachment in Cancer Cells. Cells. 2025; 14(21):1667. https://doi.org/10.3390/cells14211667
Chicago/Turabian StyleSemenova, Nina, Juan Sebastian Yakisich, Robyn Ascue, Anand K. V. Iyer, and Neelam Azad. 2025. "Reversible Upregulation of the Senescence-Associated Beta-Galactosidase Marker Induced by Cell Detachment in Cancer Cells" Cells 14, no. 21: 1667. https://doi.org/10.3390/cells14211667
APA StyleSemenova, N., Yakisich, J. S., Ascue, R., Iyer, A. K. V., & Azad, N. (2025). Reversible Upregulation of the Senescence-Associated Beta-Galactosidase Marker Induced by Cell Detachment in Cancer Cells. Cells, 14(21), 1667. https://doi.org/10.3390/cells14211667






