Genomic Analysis Defines Increased Circulating, Leukemia-Induced Macrophages That Promote Immune Suppression in Mouse Models of FGFR1-Driven Leukemogenesis
Abstract
1. Background
2. Methods
2.1. In Vivo Studies
2.2. CyTOF Analysis of Peripheral Blood Samples
2.3. Flow Cytometry Analysis
2.4. T-Cell Proliferation Assay
2.5. Electron Microscopy
2.6. RNA-Seq and Quantitative Real-Time PCR
2.7. Single-Cell RNA Genomic Analysis
2.8. Statistical Methods
3. Results
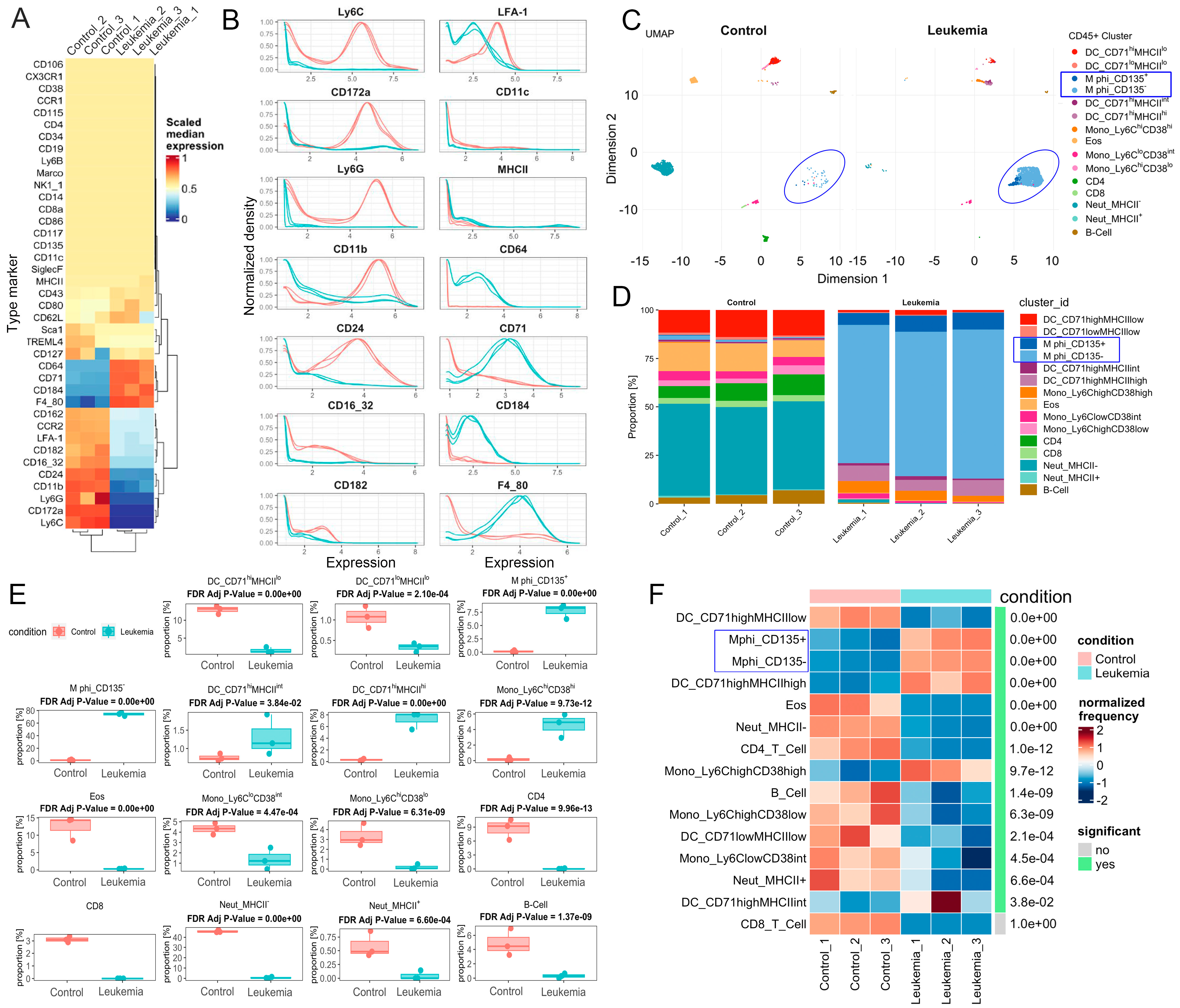
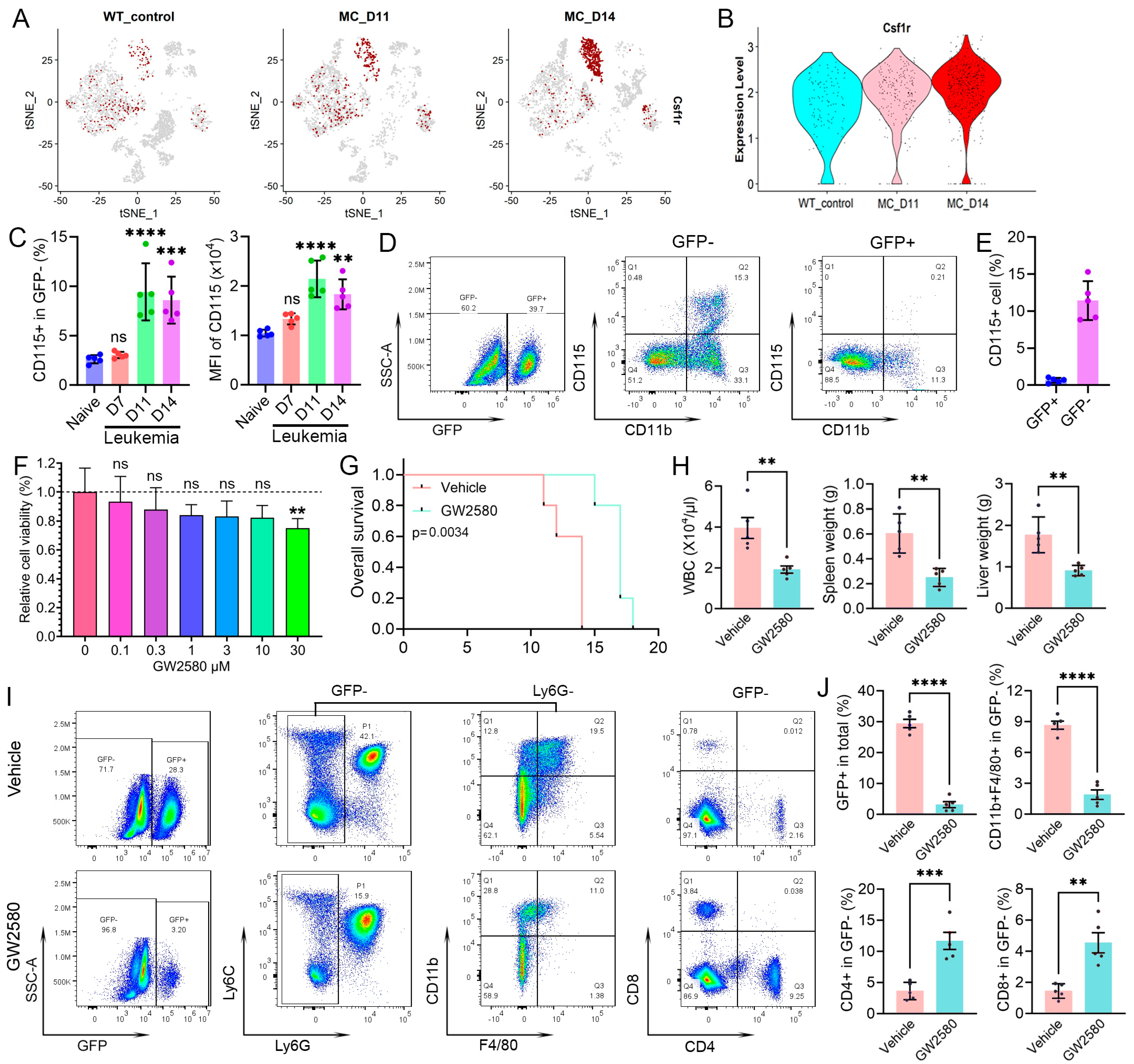
3.1. CyTOF Analysis Identifies Leukemia-Induced Macrophage Populations
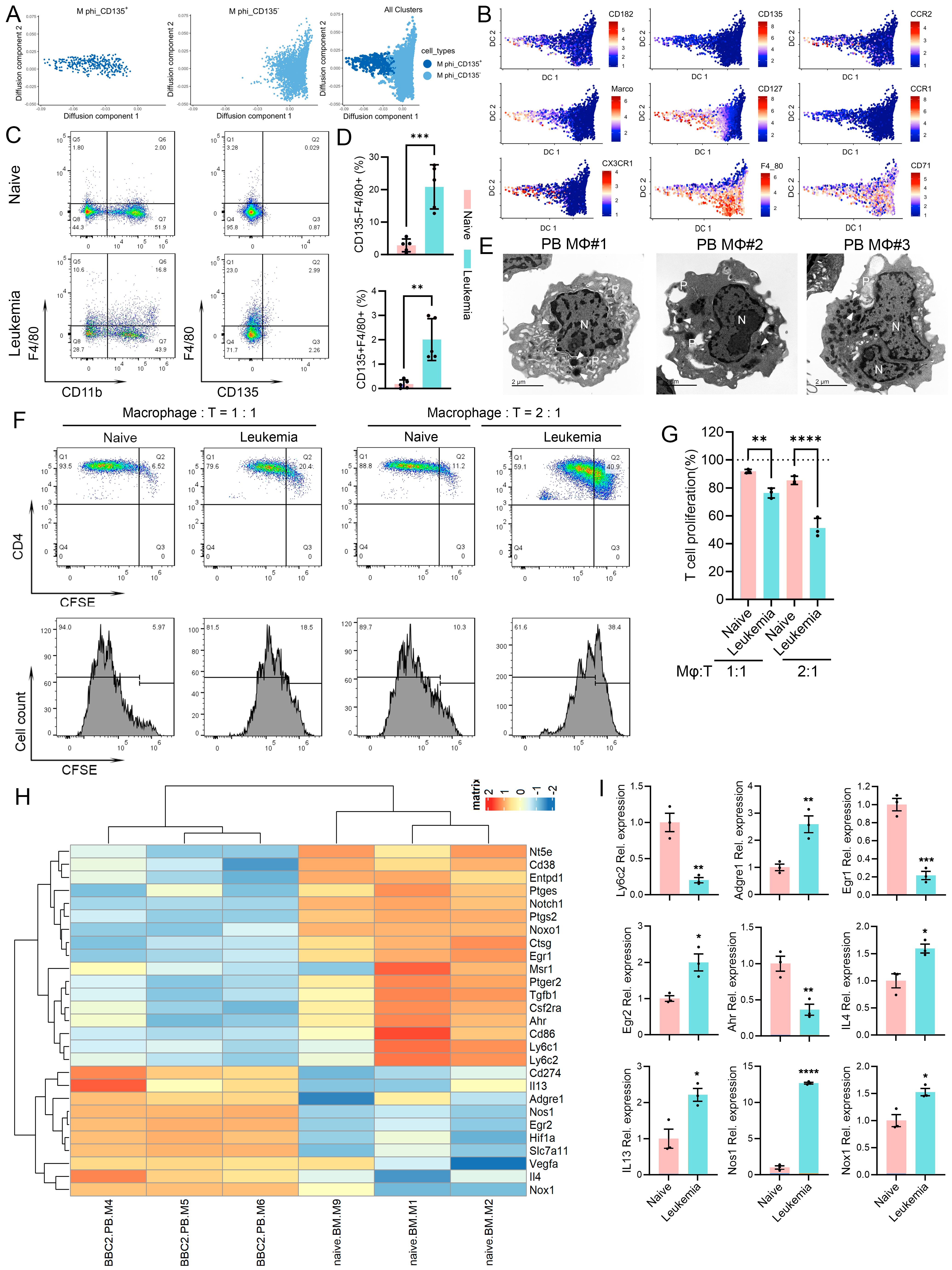
3.2. Leukemia-Induced Immunosuppressive Macrophages in Peripheral Blood
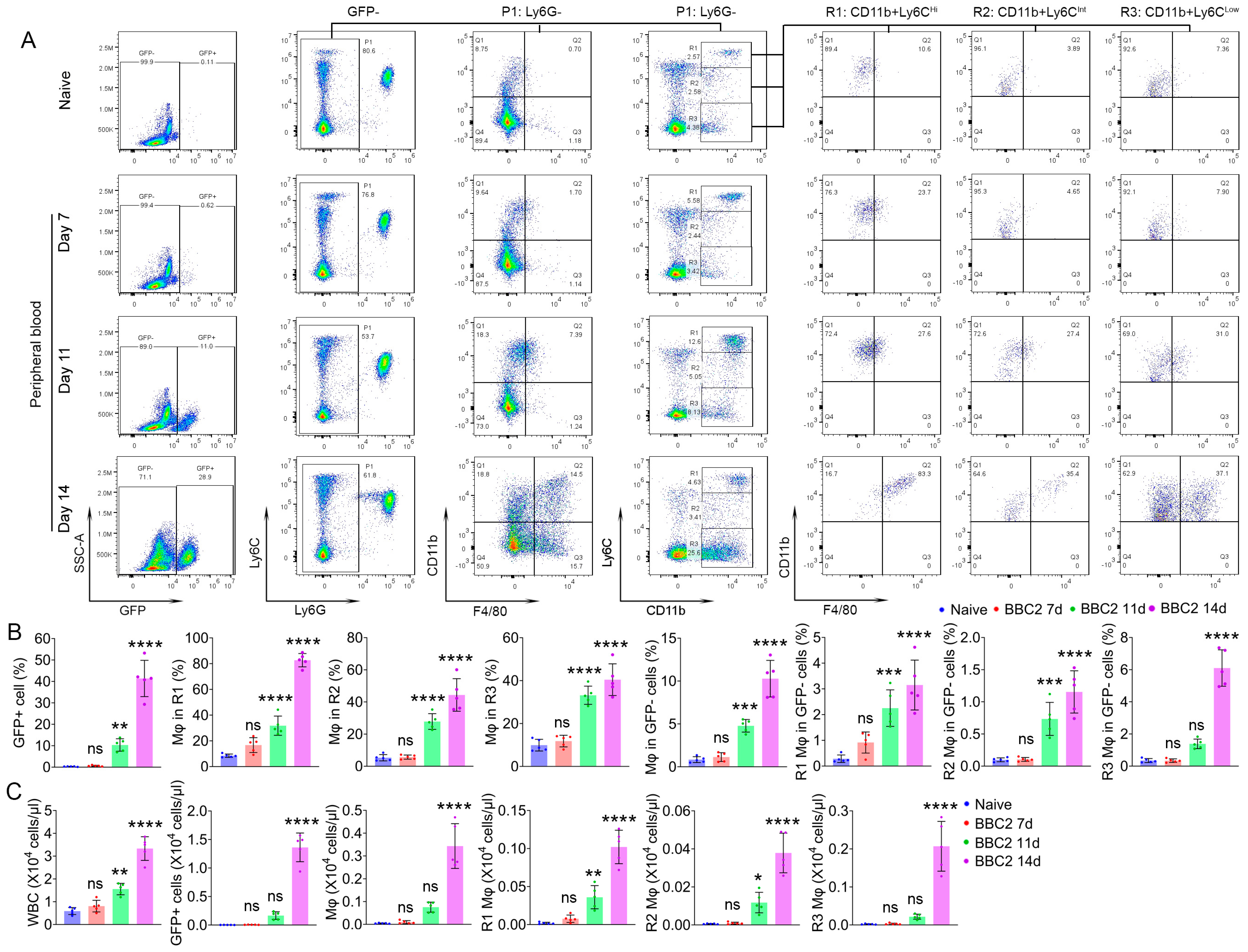
3.3. M-MDSC Partially Contribute to Leukemia-Induced Macrophages in Peripheral Blood
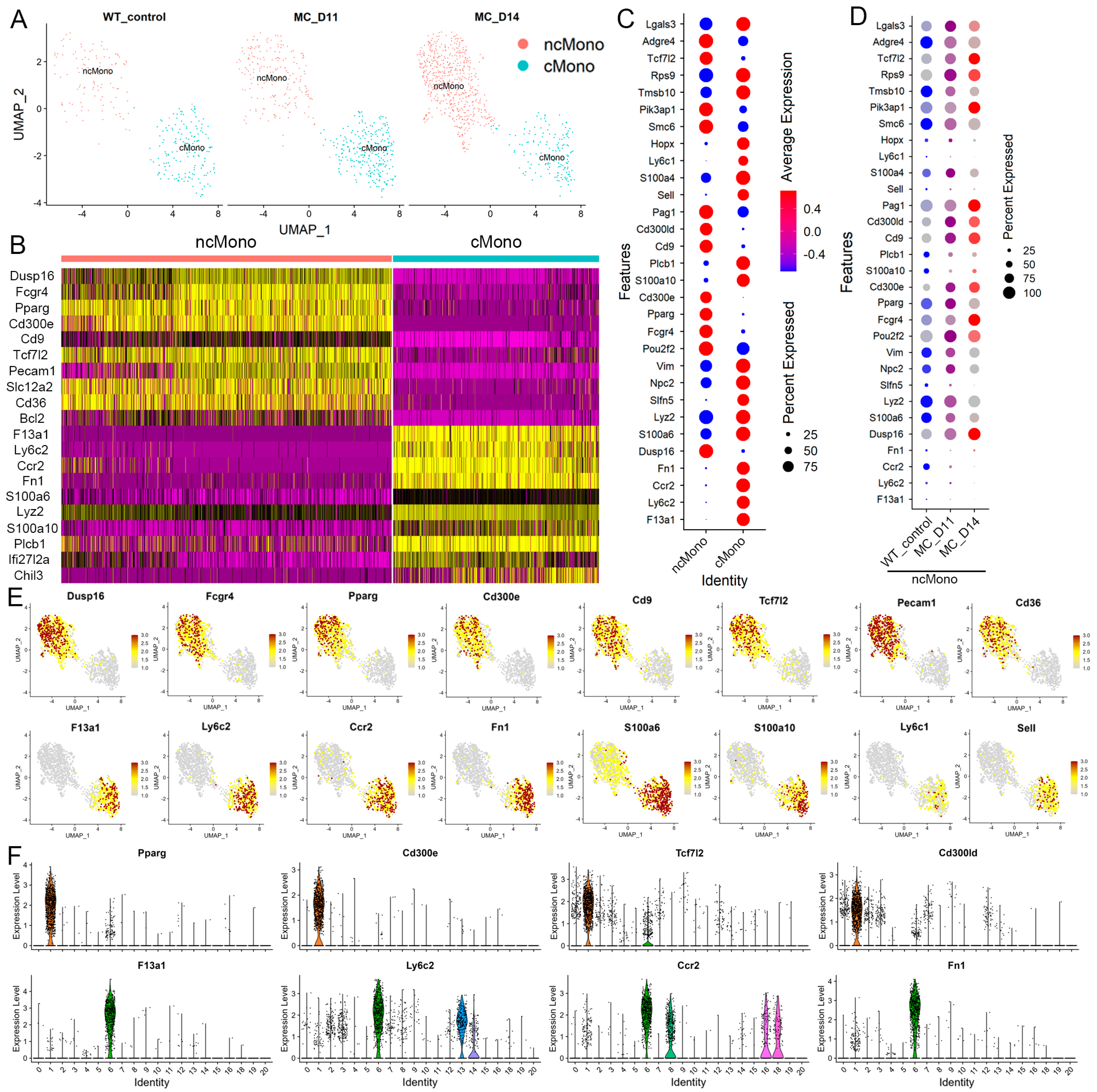
3.4. Molecular Characterization of Leukemia-Induced Monocytes
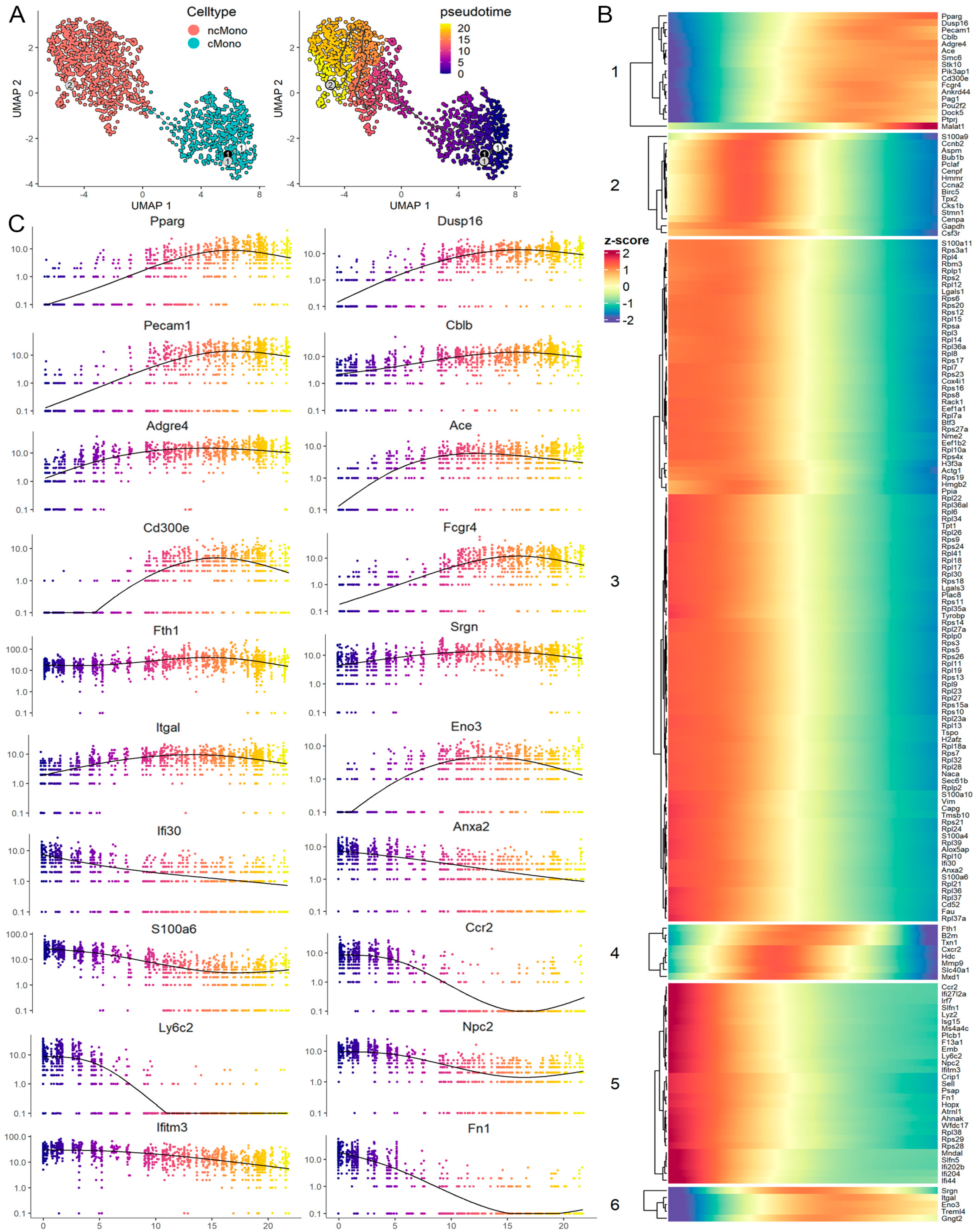
3.5. Leukemia-Induced Macrophage-like ncMonos Undergo Global Transcriptional Reprogramming
3.6. Trajectory Analysis Reveals a Transition from cMono to Macrophage-like ncMono
3.7. Targeting Leukemia-Induced Macrophages Can Impair Immunosuppression and Improve Survival
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jackson, C.C.; Medeiros, L.J.; Miranda, R.N. 8p11 myeloproliferative syndrome: A review. Hum. Pathol. 2010, 41, 461–476. [Google Scholar] [CrossRef]
- Li, T.; Zhang, G.; Zhang, X.; Liu, Q. The 8p11 myeloproliferative syndrome: Genotypic and phenotypic classification and targeted therapy. Front. Oncol. 2022, 12, 1015792. [Google Scholar] [CrossRef]
- Ren, M.; Li, X.; Cowell, J.K. Genetic fingerprinting of the development and progression of T-cell lymphoma in a murine model of atypical myeloproliferative disorder initiated by the ZNF198-fibroblast growth factor receptor-1 chimeric tyrosine kinase. Blood 2009, 114, 1576–1584. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Tidwell, J.A.; Sharma, S.; Cowell, J.K. Acute progression of BCR-FGFR1 induced murine B-lympho/myeloproliferative disorder suggests involvement of lineages at the pro-B cell stage. PLoS ONE 2012, 7, e38265. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Qin, H.; Kitamura, E.; Cowell, J.K. Dysregulated signaling pathways in the development of CNTRL-FGFR1-induced myeloid and lymphoid malignancies associated with FGFR1 in human and mouse models. Blood 2013, 122, 1007–1016. [Google Scholar] [CrossRef]
- Hu, T.; Wu, Q.; Chong, Y.; Qin, H.; Poole, C.J.; van Riggelen, J.; Ren, M.; Cowell, J.K. FGFR1 fusion kinase regulation of MYC expression drives development of stem cell leukemia/lymphoma syndrome. Leukemia 2018, 32, 2363–2373. [Google Scholar] [CrossRef]
- Hu, T.; Chong, Y.; Cai, B.; Liu, Y.; Lu, S.; Cowell, J.K. DNA methyltransferase 1-mediated CpG methylation of the miR-150-5p promoter contributes to fibroblast growth factor receptor 1-driven leukemogenesis. J. Biol. Chem. 2019, 294, 18122–18130. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Chong, Y.; Lu, S.; McGuiness, M.; Williaams, D.A.; Cowell, J.K. Rac1/2 activation promotes FGFR1 driven leukemogenesis in stem cell leukemia/lymphoma syndrome. Haematologica 2020, 105, e68–e71. [Google Scholar] [CrossRef]
- Iyer, A.; Hamers, A.A.J.; Pillai, A.B. CyTOF for the Masses. Front. Immunol. 2022, 13, 815828. [Google Scholar] [CrossRef]
- Lv, M.; Wang, K.; Huang, X.J. Myeloid-derived suppressor cells in hematological malignancies: Friends or foes. J. Hematol. Oncol. 2019, 12, 105. [Google Scholar] [CrossRef]
- Bizymi, N.; Bjelica, S.; Kittang, A.O.; Mojsilovic, S.; Velegraki, M.; Pontikoglou, C.; Roussel, M.; Ersvær, E.; Santibañez, J.F.; Lipoldová, M.; et al. Myeloid-Derived Suppressor Cells in Hematologic Diseases: Promising Biomarkers and Treatment Targets. Hemasphere 2019, 3, e168. [Google Scholar] [CrossRef]
- Wolf, A.A.; Yáñez, A.; Barman, P.K.; Goodridge, H.S. The Ontogeny of Monocyte Subsets. Front. Immunol. 2019, 10, 1642. [Google Scholar] [CrossRef]
- Cai, B.; Liu, Y.; Chong, Y.; Zhang, H.; Matsunaga, A.; Fang, X.; Pacholczyk, R.; Zhou, G.; Cowell, J.K.; Hu, T. IRAK1-regulated IFN-γ signaling induces MDSC to facilitate immune evasion in FGFR1-driven hematological malignancies. Mol. Cancer 2021, 20, 165. [Google Scholar] [CrossRef]
- Zhu, Y.P.; Padgett, L.; Dinh, H.Q.; Marcovecchio, P.; Blatchley, A.; Wu, R.; Ehinger, E.; Kim, C.; Mikulski, Z.; Seumois, G.; et al. Identification of an Early Unipotent Neutrophil Progenitor with Pro-tumoral Activity in Mouse and Human Bone Marrow. Cell Rep. 2018, 24, 2329–2341. [Google Scholar] [CrossRef]
- Finck, R.; Simonds, E.F.; Jager, A.; Krishnaswamy, S.; Sachs, K.; Fantl, W.; Pe’er, D.; Nolan, G.P.; Bendall, S.C. Normalization of mass cytometry data with bead standards. Cytom. A 2013, 83, 483–494. [Google Scholar] [CrossRef]
- Van Gassen, S.; Callebaut, B.; Van Helden, M.J.; Lambrecht, B.N.; Demeester, P.; Dhaene, T.; Saeys, Y. SOM: Using self-organizing maps for visualization and interpretation of cytometry data. Cytom. A 2015, 87, 636–645. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Saul, N.; Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 2018, 3, 861. [Google Scholar] [CrossRef]
- Setty, M.; Tadmor, M.D.; Reich-Zeliger, S.; Angel, O.; Salame, T.M.; Kathail, P.; Choi, K.; Bendall, S.; Friedman, N.; Pe’er, D. Wishbone identifies bifurcating developmental trajectories from single-cell data. Nat. Biotechnol. 2016, 34, 637–645. [Google Scholar] [CrossRef]
- Hu, T.; Cheng, B.; Matsunaga, A.; Zhang, T.; Lu, X.; Fang, H.; Mori, S.F.; Fang, X.; Wang, G.; Xu, H.; et al. Single-cell analysis defines highly specific leukemia-induced neutrophils and links MMP8 expression to recruitment of tumor associated neutrophils during FGFR1 driven leukemogenesis. Exp. Hematol. Oncol. 2024, 13, 49. [Google Scholar] [CrossRef] [PubMed]
- Kotsakis, A.; Harasymczuk, M.; Schilling, B.; Georgoulias, V.; Argiris, A.; Whiteside, T.L. Myeloid-derived suppressor cell measurements in fresh and cryopreserved blood samples. J. Immunol. Methods 2012, 381, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Patel, S.; Tcyganov, E.; Gabrilovich, D.I. The nature of myeloid-derived suppressor cells in the tumor microenvironment. Trends Immunol. 2016, 37, 208–220. [Google Scholar] [CrossRef]
- Veglia, F.; Perego, M.; Gabrilovich, D. Myeloid-derived suppressor cells coming of age. Nat. Immunol. 2018, 19, 108–119. [Google Scholar] [CrossRef]
- Spector, A.A. Arachidonic acid cytochrome P450 epoxygenase pathway. J. Lipid Res. 2009, 50, S52–S56. [Google Scholar] [CrossRef]
- Song, M.Y.; Lee, D.Y.; Chun, K.S.; Kim, E.H. The Role of NRF2/KEAP1 Signaling Pathway in Cancer Metabolism. Int. J. Mol. Sci. 2021, 22, 4376. [Google Scholar] [CrossRef]
- Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.A.; Zhang, Y.; Fullerton, J.N.; Boelen, L.; Rongvaux, A.; Maini, A.A.; Bigley, V.; Flavell, R.A.; Gilroy, D.W.; Asquith, B.; et al. The fate and lifespan of human monocyte subsets in steady state and systemic inflammation. J. Exp. Med. 2017, 214, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Tak, T.; Drylewicz, J.; Conemans, L.; de Boer, L.; Borghams, J.A.M.; Tesselaar, K. Circulatory and maturation kinetics of human monocyte subsets in vivo. Blood 2017, 130, 1474–1477. [Google Scholar] [CrossRef] [PubMed]
- Sanley, E.R.; Chitu, V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol. 2014, 6, a021857. [Google Scholar] [CrossRef]
- Conway, J.G.; McDonald, B.; Parham, J.; Keith, B.; Rusnak, D.W.; Shaw, E.; Jansen, M.; Lin, P.; Payne, A.; Crosby, R.M.; et al. Inhibition of colony-stimulating-factor-1 signaling in vivo with the orally bioavailable cFMS kinase inhibitor GW2580. Proc. Natl. Acad. Sci. USA 2005, 102, 16078–16083. [Google Scholar] [CrossRef]
- Wynn, T.A.; Chawla, A.; Pollard, J.W. Macrophage biology in development, homeostasis and disease. Nature 2013, 496, 445–455. [Google Scholar] [CrossRef]
- Komohara, Y.; Niino, D.; Ohnishi, K.; Ohshima, K.; Takeya, M. Role of tumor-associated macrophages in hematological malignancies. Pathol. Int. 2015, 65, 170–176. [Google Scholar] [CrossRef]
- Xie, Y.; Yang, H.; Yang, C.; He, L.; Zhang, X.; Peng, L.; Zhu, H.; Gao, L. Role and Mechanisms of Tumor-Associated Macrophages in Hematological Malignancies. Front. Oncol. 2022, 12, 933666. [Google Scholar] [CrossRef]
- Petty, A.J.; Yang, Y. Tumor-Associated Macrophages in Hematologic Malignancies: New Insights and Targeted Therapies. Cells 2019, 8, 1526. [Google Scholar] [CrossRef]
- Al-Matary, Y.S.; Botezatu, L.; Opalka, B.; Hönes, J.M.; Lams, R.F.; Thivakaran, A.; Schütte, J.; Köster, R.; Lennartz, K.; Schroeder, T.; et al. Acute myeloid leukemia cells polarize macrophages towards a leukemia supporting state in a Growth factor independence 1 dependent manner. Haematologica 2016, 101, 1216–1227. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; He, G.; Wang, J.; Guo, X.; Zhao, Z.; Gao, J. Hypoxia induces inflammatory microenvironment by priming specific macrophage polarization and modifies LSC behaviour via VEGF-HIF1α signaling. Transl. Pediatr. 2021, 10, 1792–1804. [Google Scholar] [CrossRef]
- Kloosterman, D.J.; Akkari, L. Macrophages at the interface of the co-evolving cancer ecosystem. Cell 2023, 186, 1627–1651. [Google Scholar] [CrossRef]
- Hume, D.A.; Millard, S.M.; Pettit, A.R. Macrophage heterogeneity in the single-cell era: Facts and artifacts. Blood 2023, 142, 1339–1347. [Google Scholar] [CrossRef]
- Cassetta, L.; Pollard, J.W. A timeline of tumour-associated macrophage biology. Nat. Rev. Cancer 2023, 23, 238–257. [Google Scholar] [CrossRef]
- Thomas, G.; Tacke, R.; Hedrick, C.C.; Hanna, R.N. Nonclassical patrolling monocyte function in the vasculature. Arter. Thromb. Vasc. Biol. 2015, 35, 1306–1316. [Google Scholar] [CrossRef]
- Tacke, F.; Alvarez, D.; Kaplan, T.J.; Jakubzick, C.; Spanbroek, R.; Llodra, J.; Garin, A.; Liu, J.; Mack, M.; Van Rooijen, N.; et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J. Clin. Investig. 2007, 117, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.D.; Hanna, R.N.; Vasudevan, N.T.; Hamers, A.A.; Romanoski, C.E.; McArdle, S.; Ross, K.D.; Blatchley, A.; Yoakum, D.; Hamilton, B.A.; et al. Deleting an Nr4a1 Super-Enhancer Subdomain Ablates Ly6C(low) Monocytes while Preserving Macrophage Gene Function. Immunity 2016, 45, 975–987. [Google Scholar] [CrossRef]
- Hanna, R.N.; Carlin, L.M.; Hubbeling, H.G.; Nackiewicz, D.; Green, A.M.; Punt, J.A.; Geissmann, F.; Hedrick, C.C. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat. Immunol. 2011, 12, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Bronte, V.; Brandau, S.; Chen, S.H.; Colombo, M.P.; Frey, A.B.; Greten, T.F.; Mandruzzato, S.; Murray, P.J.; Ochoa, A.; Ostrand-Rosenberg, S.; et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016, 7, 12150. [Google Scholar] [CrossRef]
- Mills, C.D.; Kincaid, K.; Alt, J.M.; Heilman, M.J.; Hill, A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000, 164, 6166–6173. [Google Scholar] [CrossRef] [PubMed]
- Cendrowicz, E.; Sas, Z.; Bremer, E.; Rygeil, T.P. The Role of Macrophages in Cancer Development and Therapy. Cancers 2021, 13, 1946. [Google Scholar] [CrossRef]
- Daniel, B.; Czimmerer, Z.; Halasz, L.; Boto, P.; Kolostyak, Z.; Poliska, S.; Berger, W.K.; Tzerpos, P.; Nagy, G.; Horvath, A.; et al. The transcription factor EGR2 is the molecular linchpin connecting STAT6 activation to the late, stable epigenomic program of alternative macrophage polarization. Genes Dev. 2020, 34, 1474–1492. [Google Scholar] [CrossRef]
- Quintana, F.J.; Yeste, A.; Mascanfroni, I.D. Role and therapeutic value of dendritic cells in central nervous system autoimmunity. Cell Death Differ. 2015, 22, 215–224. [Google Scholar] [CrossRef]
- Goudot, C.; Coillard, A.; Villani, A.C.; Gueguen, P.; Cros, A.; Sarkizova, S.; Tang-Huau, T.L.; Bohec, M.; Baulande, S.; Hacohen, N.; et al. Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity. 2017, 47, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Riemschneider, S.; Kohlschmidt, J.; Fueldner, C.; Esser, C.; Hauschild, S.; Lehmann, J. Aryl hydrocarbon receptor activation by benzo(a)pyrene inhibits proliferation of myeloid precursor cells and alters the differentiation state as well as the functional phenotype of murine bone marrow-derived macrophages. Toxicol. Lett. 2018, 296, 106–113. [Google Scholar] [CrossRef]
- Toobian, D.; Ghosh, P.; Katkar, G.D. Parsing the Role of PPARs in Macrophage Processes. Front. Immunol. 2021, 12, 783780. [Google Scholar] [CrossRef]
- Kang, J.; Brajanovski, N.; Chan KTXuan, J.; Pearson, R.B.; Sanji, E. Ribosomal proteins and human diseases: Molecular mechanisms and targeted therapy. Sig. Transduct. Target Ther. 2021, 6, 323. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, T.; Matsunaga, A.; Lu, X.; Fang, H.; Chatterjee, N.; Alimadadi, A.; Mori, S.F.; Fang, X.; Wang, G.; Shi, H.; et al. Genomic Analysis Defines Increased Circulating, Leukemia-Induced Macrophages That Promote Immune Suppression in Mouse Models of FGFR1-Driven Leukemogenesis. Cells 2025, 14, 1533. https://doi.org/10.3390/cells14191533
Zhang T, Matsunaga A, Lu X, Fang H, Chatterjee N, Alimadadi A, Mori SF, Fang X, Wang G, Shi H, et al. Genomic Analysis Defines Increased Circulating, Leukemia-Induced Macrophages That Promote Immune Suppression in Mouse Models of FGFR1-Driven Leukemogenesis. Cells. 2025; 14(19):1533. https://doi.org/10.3390/cells14191533
Chicago/Turabian StyleZhang, Ting, Atsuko Matsunaga, Xiaocui Lu, Hui Fang, Nandini Chatterjee, Ahmad Alimadadi, Stephanie F. Mori, Xuexiu Fang, Gavin Wang, Huidong Shi, and et al. 2025. "Genomic Analysis Defines Increased Circulating, Leukemia-Induced Macrophages That Promote Immune Suppression in Mouse Models of FGFR1-Driven Leukemogenesis" Cells 14, no. 19: 1533. https://doi.org/10.3390/cells14191533
APA StyleZhang, T., Matsunaga, A., Lu, X., Fang, H., Chatterjee, N., Alimadadi, A., Mori, S. F., Fang, X., Wang, G., Shi, H., Zhang, L., Hedrick, C. C., Cheng, B., Hu, T., & Cowell, J. K. (2025). Genomic Analysis Defines Increased Circulating, Leukemia-Induced Macrophages That Promote Immune Suppression in Mouse Models of FGFR1-Driven Leukemogenesis. Cells, 14(19), 1533. https://doi.org/10.3390/cells14191533





