Conditional ATXN2L-Null in Adult Frontal Cortex CamK2a+ Neurons Does Not Cause Cell Death but Restricts Spontaneous Mobility and Affects the Alternative Splicing Pathway
Abstract
Highlights
- Constitutive knock-out of ATXN2L across LSMAD and PAM2 is embryonically lethal, confirming its essential role in development.
- Conditional deletion of ATXN2L across LSMAD and PAM2 postnatally in cortical neurons reduces spontaneous movement and alters alternative splicing pathways.
- ATXN2L is indispensable for embryonic survival and neuronal function, highlighting its non-redundant role, in contrast to its paralog ATXN2.
- The LSMAD and PAM2 domains of ATXN2L likely impact nuclear splicing, despite the protein’s perinuclear localization.
Abstract
1. Introduction
2. Materials and Methods
2.1. Mouse Breeding and Genotyping
2.2. Assessment of Embryonic Lethality in Constitutive Atxn2l-KO Mice
2.3. Immunohistochemistry
2.4. Imaging
2.5. Locomotor Phenotyping
2.6. Tamoxifen Preparation and Treatment
2.7. Global Proteomics
2.8. Proteome Interrogation for Pathway Enrichments
2.9. Statistics and Graphical Presentation
3. Results
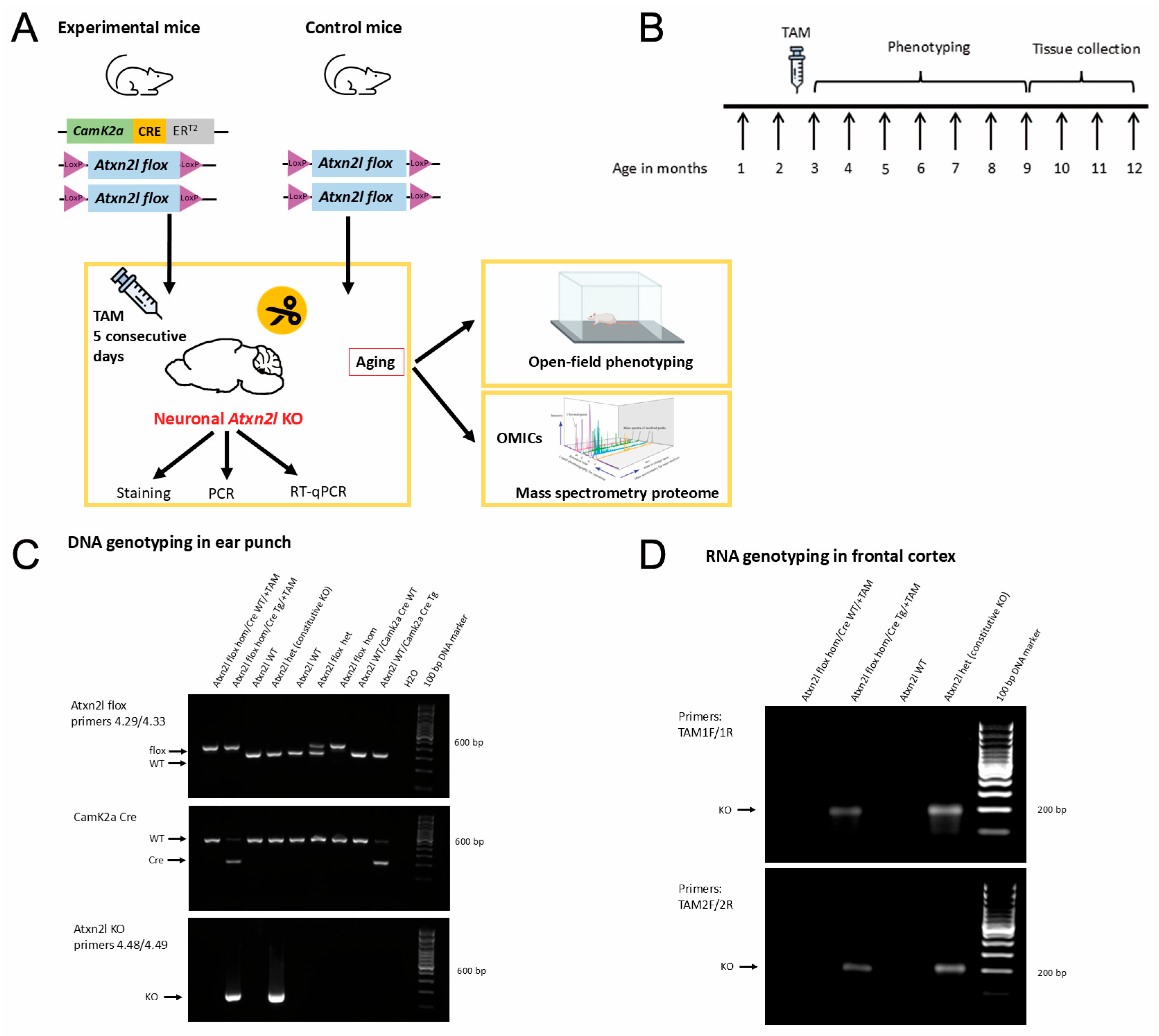
3.1. Generation of Conditional KO Mice via Floxing Exons 10–17 in the Atxn2l Gene
3.2. Crossbreeding with Constitutive Cre-Deleters Confirms Embryonic Lethality of Homozygous ATXN2L-Null Mice
3.3. Crossbreeding with CamK2a-Dependent Cre/ERT2 Mice and Subsequent Tamoxifen Injection Generate Atxn2l-cKO in Adult Frontal Cortex Tissue
3.4. Atxn2l-cKO in Frontal Cortex Tissue Showed Mosaic Expression in CamK2a-Positive Neurons
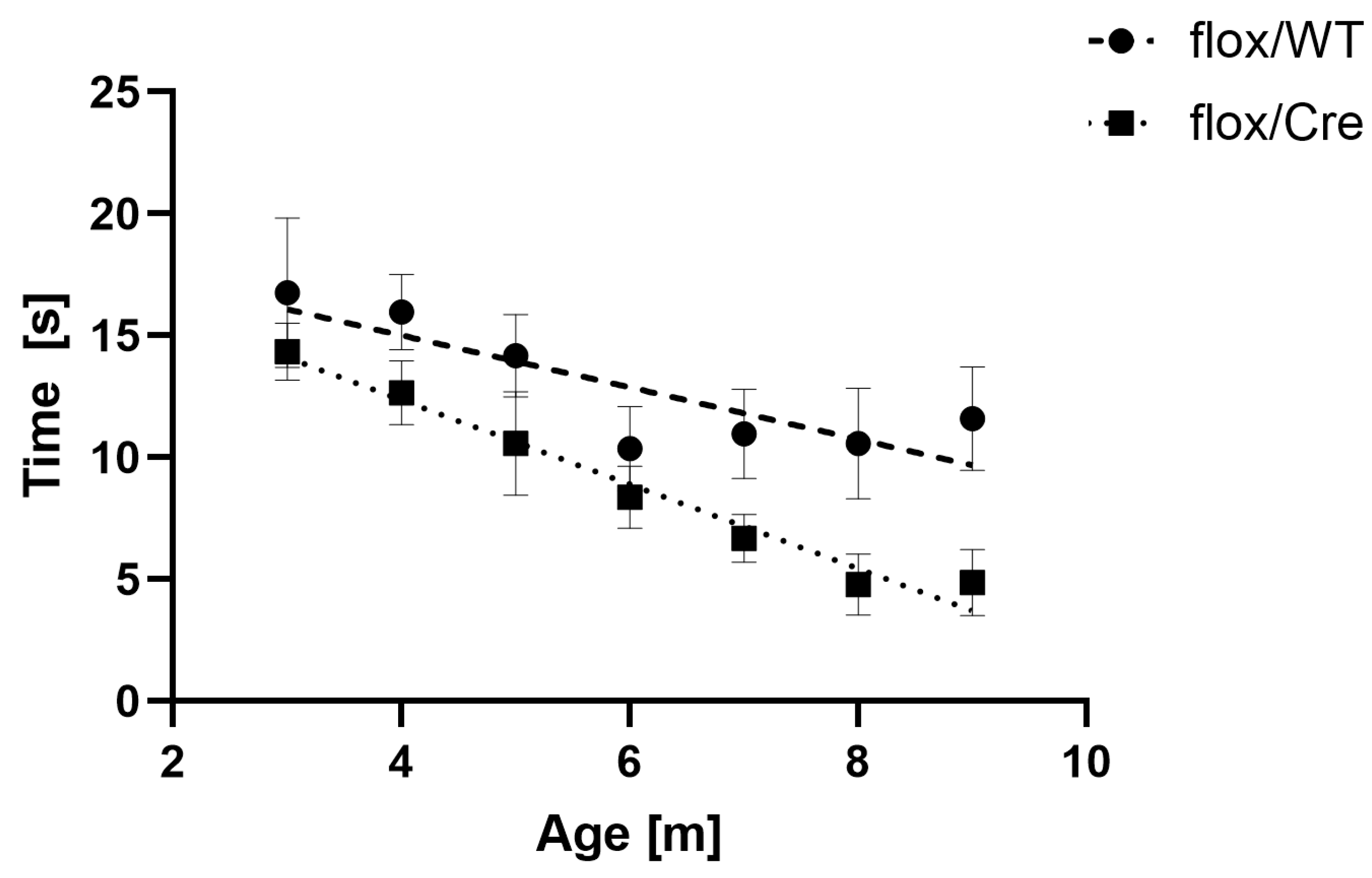
3.5. CamK2a-Dependent Atxn2l-cKO Triggers Deficits in Spontaneous Horizontal Locomotion
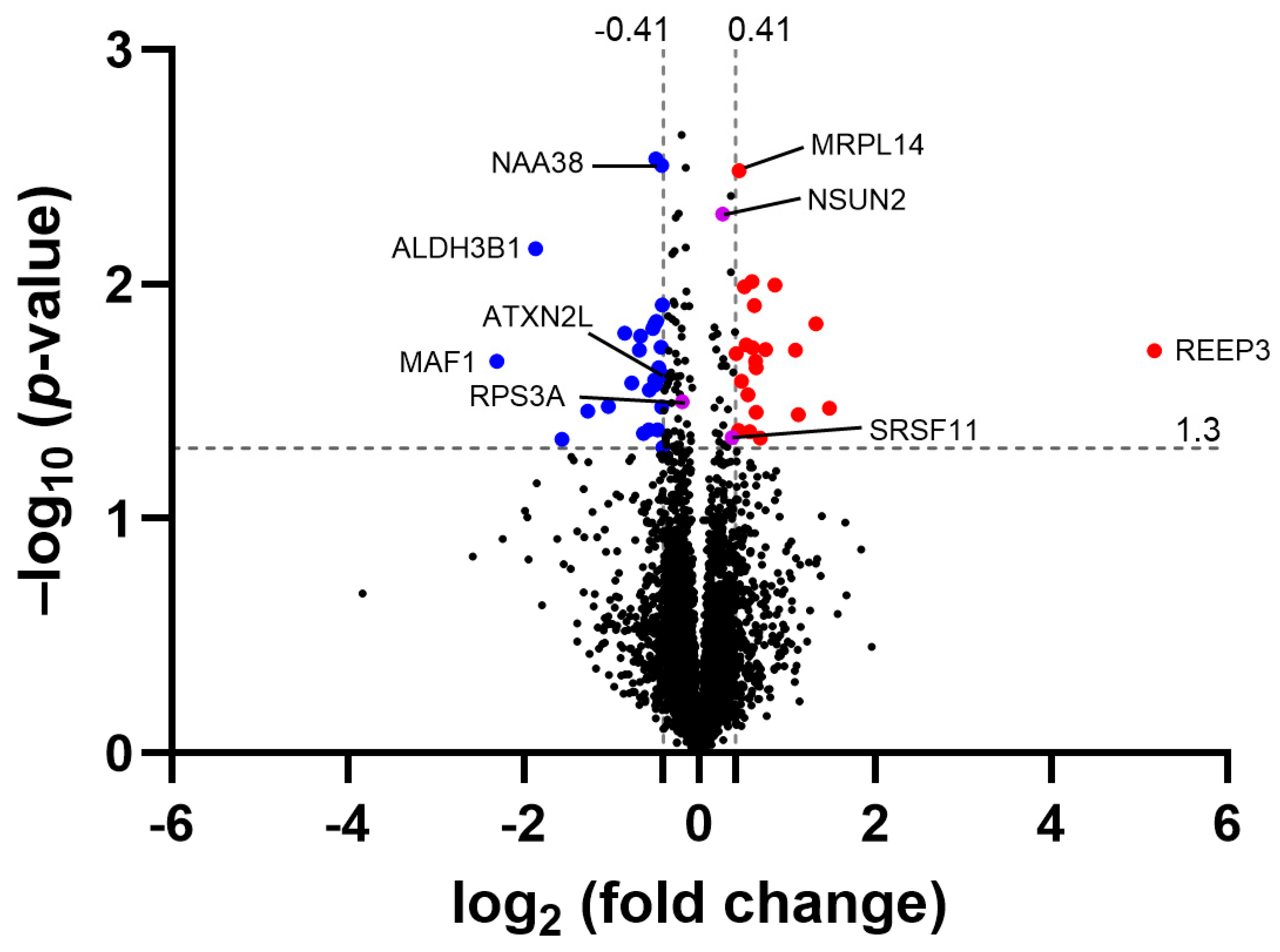
3.6. CamK2a-Dependent Atxn2l-cKO Mouse Frontal Cortex Proteomics Shows ATXN2L Protein Reduction to 75% and Dysregulation of the Alternative Splicing Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AGO3 | Argonaute RISC Catalytic Component 3 |
| ALDH3B1 | Medium-Chain Fatty Aldehyde Dehydrogenase |
| ALS | Amyotrophic Lateral Sclerosis |
| ALYREF | Aly/REF Export Factor |
| ATG | Autophagy-Related Gene |
| ATX | Ataxin |
| ATXN2 | Ataxin-2 |
| ATXN2L | Ataxin-2-Like |
| AU-rich | Adenosin-Uridine Rich |
| BCA | Bicinchoninic Acid |
| bp | Base Pair |
| CAG | Cytosine–Adenine–Guanine Trinucleotide Repeat |
| CamK2a | Calcium/Calmodulin-Dependent Protein Kinase II Alpha |
| cKO | Conditional Knock-Out |
| Cre | Cyclization Recombination Enzyme |
| DEAD | Asp-Glu-Ala-Asp (DEAD-box helicase family) |
| DIA | Data-Independent Acquisition |
| DNA | Deoxyribo-Nucleic Acid |
| DTA | Diphtheria Toxin Fragment A |
| DxxD | Aspartate–Any Amino Acid–Any Amino Acid–Aspartate |
| EDC4 | Enhancer of mRNA Decapping 4 |
| EGF | Epidermal Growth Factor |
| ER | Endoplasmic Reticulum |
| ERT2 | Tamoxifen-Inducible Estrogen Receptor Domain, Improved Version |
| ES | Embryonic Stem Cell |
| FDR | False Discovery Rate |
| FEBS | Federation of European Biochemical Societies |
| FIJI | FIJI Is Just ImageJ |
| Floxed | Sequence Flanked by Two loxP Sites |
| FMRP | Fragile X Mental Retardation Protein |
| FMR1 | Fragile X Messenger Ribonucleoprotein 1 |
| FXR2 | Fragile X Mental Retardation, Autosomal Homolog 2 |
| GO | Gene Ontology |
| G3BP2 | GTPase Activating Protein (SH3 Domain) Binding Protein 2 |
| hATXN2 | Human Ataxin-2 |
| het | Heterozygous |
| hom | Homozygous |
| HSC | Hematopoietic Stem Cell |
| kb | Kilo-Base |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KO | Knock-Out |
| LARP7 | La Ribonucleoprotein 7, Transcription Regulator, Binds U6 snRNA |
| LC | Liquid Chromatography |
| LED | Light Emitting Diode |
| LoxP | Locus of X-Over (Crossing-Over) in Bacteriophage P1 |
| Lsm | Like-Sm Domain |
| LsmAD | Like-Sm-Associated Domain |
| LSM3 | LSM3 Homolog, U6 Small Nuclear RNA-Associated |
| MAF1 | MAF1 Homolog, Negative Regulator of RNA Polymerase III |
| MPL | Myelo-Proliferative Leukaemia Protein |
| mRNA | Messenger Ribo-Nucleic Acid |
| MRPL14 | Large Ribosomal Subunit Protein UL14m |
| MS | Mass Spectrometry |
| mTOR | Mechanistic Target of Rapamycin |
| mTORC1 | Mechanistic Target of Rapamycin-Complex-Associated Protein 1 |
| m5C | 5-methyl-Cytosine |
| NAA38 | N-Alpha-Acetyltransferase 38, NatC Auxiliary Subunit |
| NGS | Normal Goat Serum |
| NSUN2 | NOP2/Sun RNA Methyltransferase 2 |
| NUFIP2 | Nuclear Fragile X Mental Retardation Protein Interacting Protein 2 |
| oligo(U) | Several (Uridine Bases) |
| OR | Odds Ratio |
| PABP | Poly(A)-Binding Protein |
| PAM2 | Poly(A)-Binding protein association Motif type 2 |
| PASEF | Parallel Accumulation Serial Fragmentation |
| Pat1 | Yeast Factor for Protection of mRNA 3′-UTRs from Trimming |
| P-bodies | Processing Bodies in the Cytosol, Where RNA Is Degraded |
| Pbp1 | Poly(A)-Binding Protein 1 in Yeast |
| PBS | Phosphate-Buffered Saline |
| PCR | Polymerase Chain Reaction |
| Poly(A) | many (Adenine bases) |
| polyQ | Many Glutamine Amino Acids |
| PRM | Proline-Rich Motif |
| REEP3 | Receptor Expression-Enhancing Protein 3 |
| RNA | Ribo-Nucleic Acid |
| RNP | Ribo-Nucleoprotein |
| RoxP | loxP-Analogous Site, Recognized by Phage Integrase Dre |
| RPS3 | Small Ribosomal Subunit Protein US3 |
| RRM | RNA Recognition Motif |
| rRNA | Ribosomal Ribo-Nucleic Acid |
| RT | Reverse Transcription |
| SCA2 | Spinocerebellar Ataxia type 2 |
| SD | Standard Deviation |
| SEM | Standard Error of the Mean |
| SG | Stress Granule |
| siRNA | Small Interfering RNA |
| SN | Substantia Nigra |
| SNP | Single Nucleotide Polymorphism |
| snRNA | Small Nuclear Ribo-Nucleic Acid |
| snRNP | Small Nuclear Ribo-Nucleic-Acid Binding Protein |
| SR | Serine/Arginine-Rich Protein, Family of Splicing Factors |
| SRSF11 | Serine and Arginine Rich Splicing Factor 11 |
| SYNE2 | Spectrin Repeat Containing Nuclear Envelope Protein 2 |
| TAM | Tamoxifen |
| TDP-43 | TAR DNA-Binding Protein-43 |
| Tg | Transgenic |
| tRNA | Transfer Ribo-Nucleic Acid |
| UPR | Unfolded Protein Response |
| UTR | Untranslated Region of mRNA |
| U2AF65 | U2 Small Nuclear RNA Auxiliary Factor 2 |
| WB | Western Blot |
| WT | Wild-Type |
| YBX1 | Y-Box-Binding Protein 1, CCAAT-Binding Transcription Factor I |
| ZFE | Zentrale Forschungseinrichtung (Central Animal Facility) |
References
- Achsel, T.; Stark, H.; Luhrmann, R. The Sm domain is an ancient RNA-binding motif with oligo(U) specificity. Proc. Natl. Acad. Sci. USA 2001, 98, 3685–3689. [Google Scholar] [CrossRef]
- Tharun, S.; He, W.; Mayes, A.E.; Lennertz, P.; Beggs, J.D.; Parker, R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 2000, 404, 515–518. [Google Scholar] [CrossRef]
- Mura, C.; Randolph, P.S.; Patterson, J.; Cozen, A.E. Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function. RNA Biol. 2013, 10, 636–651. [Google Scholar] [CrossRef]
- Swisher, K.D.; Parker, R. Localization to, and effects of Pbp1, Pbp4, Lsm12, Dhh1, and Pab1 on stress granules in Saccharomyces cerevisiae. PLoS ONE 2010, 5, e10006. [Google Scholar] [CrossRef] [PubMed]
- Seidel, G.; Meierhofer, D.; Sen, N.E.; Guenther, A.; Krobitsch, S.; Auburger, G. Quantitative Global Proteomics of Yeast PBP1 Deletion Mutants and Their Stress Responses Identifies Glucose Metabolism, Mitochondrial, and Stress Granule Changes. J. Proteome Res. 2017, 16, 504–515. [Google Scholar] [CrossRef]
- Yang, Y.S.; Kato, M.; Wu, X.; Litsios, A.; Sutter, B.M.; Wang, Y.; Hsu, C.H.; Wood, N.E.; Lemoff, A.; Mirzaei, H.; et al. Yeast Ataxin-2 Forms an Intracellular Condensate Required for the Inhibition of TORC1 Signaling during Respiratory Growth. Cell 2019, 177, 697–710.E17. [Google Scholar] [CrossRef]
- Kato, M.; Yang, Y.S.; Sutter, B.M.; Wang, Y.; McKnight, S.L.; Tu, B.P. Redox State Controls Phase Separation of the Yeast Ataxin-2 Protein via Reversible Oxidation of Its Methionine-Rich Low-Complexity Domain. Cell 2019, 177, 711–721.E8. [Google Scholar] [CrossRef]
- Kiehl, T.R.; Shibata, H.; Pulst, S.M. The ortholog of human ataxin-2 is essential for early embryonic patterning in C. elegans. J. Mol. Neurosci. 2000, 15, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Ciosk, R.; DePalma, M.; Priess, J.R. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development 2004, 131, 4831–4841. [Google Scholar] [CrossRef]
- Bar, D.Z.; Charar, C.; Dorfman, J.; Yadid, T.; Tafforeau, L.; Lafontaine, D.L.; Gruenbaum, Y. Cell size and fat content of dietary-restricted Caenorhabditis elegans are regulated by ATX-2, an mTOR repressor. Proc. Natl. Acad. Sci. USA 2016, 113, E4620–E4629. [Google Scholar] [CrossRef] [PubMed]
- Stubenvoll, M.D.; Medley, J.C.; Irwin, M.; Song, M.H. ATX-2, the C. elegans Ortholog of Human Ataxin-2, Regulates Centrosome Size and Microtubule Dynamics. PLoS Genet. 2016, 12, e1006370. [Google Scholar] [CrossRef]
- Del Castillo, U.; Gnazzo, M.M.; Sorensen Turpin, C.G.; Nguyen, K.C.Q.; Semaya, E.; Lam, Y.; de Cruz, M.A.; Bembenek, J.N.; Hall, D.H.; Riggs, B.; et al. Conserved role for Ataxin-2 in mediating endoplasmic reticulum dynamics. Traffic 2019, 20, 436–447. [Google Scholar] [CrossRef]
- Beath, E.A.; Bailey, C.; Mahantesh Magadam, M.; Qiu, S.; McNally, K.L.; McNally, F.J. Katanin, kinesin-13, and ataxin-2 inhibit premature interaction between maternal and paternal genomes in C. elegans zygotes. Elife 2024, 13, RP97812. [Google Scholar] [CrossRef]
- Satterfield, T.F.; Jackson, S.M.; Pallanck, L.J. A Drosophila homolog of the polyglutamine disease gene SCA2 is a dosage-sensitive regulator of actin filament formation. Genetics 2002, 162, 1687–1702. [Google Scholar] [CrossRef] [PubMed]
- Satterfield, T.F.; Pallanck, L.J. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum. Mol. Genet. 2006, 15, 2523–2532. [Google Scholar] [CrossRef]
- Al-Ramahi, I.; Perez, A.M.; Lim, J.; Zhang, M.; Sorensen, R.; de Haro, M.; Branco, J.; Pulst, S.M.; Zoghbi, H.Y.; Botas, J. dAtaxin-2 mediates expanded Ataxin-1-induced neurodegeneration in a Drosophila model of SCA1. PLoS Genet. 2007, 3, e234. [Google Scholar] [CrossRef]
- McCann, C.; Holohan, E.E.; Das, S.; Dervan, A.; Larkin, A.; Lee, J.A.; Rodrigues, V.; Parker, R.; Ramaswami, M. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc. Natl. Acad. Sci. USA 2011, 108, E655–E662. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.; Allada, R. ATAXIN-2 activates PERIOD translation to sustain circadian rhythms in Drosophila. Science 2013, 340, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ling, J.; Yuan, C.; Dubruille, R.; Emery, P. A role for Drosophila ATX2 in activation of PER translation and circadian behavior. Science 2013, 340, 879–882. [Google Scholar] [CrossRef]
- Sudhakaran, I.P.; Hillebrand, J.; Dervan, A.; Das, S.; Holohan, E.E.; Hulsmeier, J.; Sarov, M.; Parker, R.; VijayRaghavan, K.; Ramaswami, M. FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. Proc. Natl. Acad. Sci. USA 2014, 111, E99–E108. [Google Scholar] [CrossRef]
- Vianna, M.C.; Poleto, D.C.; Gomes, P.F.; Valente, V.; Paco-Larson, M.L. Drosophila ataxin-2 gene encodes two differentially expressed isoforms and its function in larval fat body is crucial for development of peripheral tissues. FEBS Open Bio 2016, 6, 1040–1053. [Google Scholar] [CrossRef]
- Lee, J.; Yoo, E.; Lee, H.; Park, K.; Hur, J.H.; Lim, C. LSM12 and ME31B/DDX6 Define Distinct Modes of Posttranscriptional Regulation by ATAXIN-2 Protein Complex in Drosophila Circadian Pacemaker Neurons. Mol. Cell 2017, 66, 129–140.e7. [Google Scholar] [CrossRef] [PubMed]
- Bakthavachalu, B.; Huelsmeier, J.; Sudhakaran, I.P.; Hillebrand, J.; Singh, A.; Petrauskas, A.; Thiagarajan, D.; Sankaranarayanan, M.; Mizoue, L.; Anderson, E.N.; et al. RNP-Granule Assembly via Ataxin-2 Disordered Domains Is Required for Long-Term Memory and Neurodegeneration. Neuron 2018, 98, 754–766. [Google Scholar] [CrossRef]
- Cha, I.J.; Lee, D.; Park, S.S.; Chung, C.G.; Kim, S.Y.; Jo, M.G.; Kim, S.Y.; Lee, B.H.; Lee, Y.S.; Lee, S.B. Ataxin-2 Dysregulation Triggers a Compensatory Fragile X Mental Retardation Protein Decrease in Drosophila C4da Neurons. Mol. Cells 2020, 43, 870–879. [Google Scholar] [CrossRef]
- Singh, A.; Hulsmeier, J.; Kandi, A.R.; Pothapragada, S.S.; Hillebrand, J.; Petrauskas, A.; Agrawal, K.; Rt, K.; Thiagarajan, D.; Jayaprakashappa, D.; et al. Antagonistic roles for Ataxin-2 structured and disordered domains in RNP condensation. Elife 2021, 10, e60326. [Google Scholar] [CrossRef]
- Del Castillo, U.; Norkett, R.; Lu, W.; Serpinskaya, A.; Gelfand, V.I. Ataxin-2 is essential for cytoskeletal dynamics and neurodevelopment in Drosophila. iScience 2022, 25, 103536. [Google Scholar] [CrossRef]
- Corgiat, E.B.; List, S.M.; Rounds, J.C.; Yu, D.; Chen, P.; Corbett, A.H.; Moberg, K.H. The Nab2 RNA-binding protein patterns dendritic and axonal projections through a planar cell polarity-sensitive mechanism. G3 2022, 12, jkac100. [Google Scholar] [CrossRef] [PubMed]
- Petrauskas, A.; Fortunati, D.L.; Kandi, A.R.; Pothapragada, S.S.; Agrawal, K.; Singh, A.; Huelsmeier, J.; Hillebrand, J.; Brown, G.; Chaturvedi, D.; et al. Structured and disordered regions of Ataxin-2 contribute differently to the specificity and efficiency of mRNP granule formation. PLoS Genet. 2024, 20, e1011251. [Google Scholar] [CrossRef] [PubMed]
- Auburger, G.; Sen, N.E.; Meierhofer, D.; Basak, A.N.; Gitler, A.D. Efficient Prevention of Neurodegenerative Diseases by Depletion of Starvation Response Factor Ataxin-2. Trends Neurosci. 2017, 40, 507–516. [Google Scholar] [CrossRef]
- Jimenez-Lopez, D.; Guzman, P. Insights into the evolution and domain structure of Ataxin-2 proteins across eukaryotes. BMC Res. Notes 2014, 7, 453. [Google Scholar] [CrossRef]
- Pulst, S.M.; Nechiporuk, A.; Nechiporuk, T.; Gispert, S.; Chen, X.N.; Lopes-Cendes, I.; Pearlman, S.; Starkman, S.; Orozco-Diaz, G.; Lunkes, A.; et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat. Genet. 1996, 14, 269–276. [Google Scholar] [CrossRef]
- Sanpei, K.; Takano, H.; Igarashi, S.; Sato, T.; Oyake, M.; Sasaki, H.; Wakisaka, A.; Tashiro, K.; Ishida, Y.; Ikeuchi, T.; et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat. Genet. 1996, 14, 277–284. [Google Scholar] [CrossRef]
- Imbert, G.; Saudou, F.; Yvert, G.; Devys, D.; Trottier, Y.; Garnier, J.M.; Weber, C.; Mandel, J.L.; Cancel, G.; Abbas, N.; et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat. Genet. 1996, 14, 285–291. [Google Scholar] [CrossRef]
- Velazquez-Perez, L.; Seifried, C.; Santos-Falcon, N.; Abele, M.; Ziemann, U.; Almaguer, L.E.; Martinez-Gongora, E.; Sanchez-Cruz, G.; Canales, N.; Perez-Gonzalez, R.; et al. Saccade velocity is controlled by polyglutamine size in spinocerebellar ataxia 2. Ann. Neurol. 2004, 56, 444–447. [Google Scholar] [CrossRef]
- Rub, U.; Gierga, K.; Brunt, E.R.; de Vos, R.A.; Bauer, M.; Schols, L.; Burk, K.; Auburger, G.; Bohl, J.; Schultz, C.; et al. Spinocerebellar ataxias types 2 and 3: Degeneration of the pre-cerebellar nuclei isolates the three phylogenetically defined regions of the cerebellum. J. Neural Transm. 2005, 112, 1523–1545. [Google Scholar] [CrossRef] [PubMed]
- Gierga, K.; Burk, K.; Bauer, M.; Orozco Diaz, G.; Auburger, G.; Schultz, C.; Vuksic, M.; Schols, L.; de Vos, R.A.; Braak, H.; et al. Involvement of the cranial nerves and their nuclei in spinocerebellar ataxia type 2 (SCA2). Acta Neuropathol. 2005, 109, 617–631. [Google Scholar] [CrossRef] [PubMed]
- Ralser, M.; Nonhoff, U.; Albrecht, M.; Lengauer, T.; Wanker, E.E.; Lehrach, H.; Krobitsch, S. Ataxin-2 and huntingtin interact with endophilin-A complexes to function in plastin-associated pathways. Hum. Mol. Genet. 2005, 14, 2893–2909. [Google Scholar] [CrossRef]
- Tuin, I.; Voss, U.; Kang, J.S.; Kessler, K.; Rub, U.; Nolte, D.; Lochmuller, H.; Tinschert, S.; Claus, D.; Krakow, K.; et al. Stages of sleep pathology in spinocerebellar ataxia type 2 (SCA2). Neurology 2006, 67, 1966–1972. [Google Scholar] [CrossRef] [PubMed]
- Lastres-Becker, I.; Brodesser, S.; Lutjohann, D.; Azizov, M.; Buchmann, J.; Hintermann, E.; Sandhoff, K.; Schurmann, A.; Nowock, J.; Auburger, G. Insulin receptor and lipid metabolism pathology in ataxin-2 knock-out mice. Hum. Mol. Genet. 2008, 17, 1465–1481. [Google Scholar] [CrossRef]
- Lastres-Becker, I.; Rub, U.; Auburger, G. Spinocerebellar ataxia 2 (SCA2). Cerebellum 2008, 7, 115–124. [Google Scholar] [CrossRef]
- Nonis, D.; Schmidt, M.H.H.; van de Loo, S.; Eich, F.; Dikic, I.; Nowock, J.; Auburger, G. Ataxin-2 associates with the endocytosis complex and affects EGF receptor trafficking. Cell. Signal 2008, 20, 1725–1739. [Google Scholar] [CrossRef] [PubMed]
- Rub, U.; Del Turco, D.; Del Tredici, K.; de Vos, R.A.; Brunt, E.R.; Reifenberger, G.; Seifried, C.; Schultz, C.; Auburger, G.; Braak, H. Thalamic involvement in a spinocerebellar ataxia type 2 (SCA2) and a spinocerebellar ataxia type 3 (SCA3) patient, and its clinical relevance. Brain 2003, 126, 2257–2272. [Google Scholar] [CrossRef]
- van de Loo, S.; Eich, F.; Nonis, D.; Auburger, G.; Nowock, J. Ataxin-2 associates with rough endoplasmic reticulum. Exp. Neurol. 2009, 215, 110–118. [Google Scholar] [CrossRef]
- Almaguer-Mederos, L.E.; Falcon, N.S.; Almira, Y.R.; Zaldivar, Y.G.; Almarales, D.C.; Gongora, E.M.; Herrera, M.P.; Batallan, K.E.; Arminan, R.R.; Manresa, M.V.; et al. Estimation of the age at onset in spinocerebellar ataxia type 2 Cuban patients by survival analysis. Clin. Genet. 2010, 78, 169–174. [Google Scholar] [CrossRef]
- Auburger, G.W. Spinocerebellar ataxia type 2. Handb. Clin. Neurol. 2012, 103, 423–436. [Google Scholar] [CrossRef]
- Rub, U.; Schols, L.; Paulson, H.; Auburger, G.; Kermer, P.; Jen, J.C.; Seidel, K.; Korf, H.W.; Deller, T. Clinical features, neurogenetics and neuropathology of the polyglutamine spinocerebellar ataxias type 1, 2, 3, 6 and 7. Prog. Neurobiol. 2013, 104, 38–66. [Google Scholar] [CrossRef]
- Schols, L.; Reimold, M.; Seidel, K.; Globas, C.; Brockmann, K.; Hauser, T.K.; Auburger, G.; Burk, K.; den Dunnen, W.; Reischl, G.; et al. No parkinsonism in SCA2 and SCA3 despite severe neurodegeneration of the dopaminergic substantia nigra. Brain 2015, 138, 3316–3326. [Google Scholar] [CrossRef] [PubMed]
- Meierhofer, D.; Halbach, M.; Sen, N.E.; Gispert, S.; Auburger, G. Ataxin-2 (Atxn2)-Knock-Out Mice Show Branched Chain Amino Acids and Fatty Acids Pathway Alterations. Mol. Cell. Proteom. 2016, 15, 1728–1739. [Google Scholar] [CrossRef]
- Halbach, M.V.; Gispert, S.; Stehning, T.; Damrath, E.; Walter, M.; Auburger, G. Atxn2 Knockout and CAG42-Knock-in Cerebellum Shows Similarly Dysregulated Expression in Calcium Homeostasis Pathway. Cerebellum 2017, 16, 68–81. [Google Scholar] [CrossRef]
- Lastres-Becker, I.; Nonis, D.; Eich, F.; Klinkenberg, M.; Gorospe, M.; Kotter, P.; Klein, F.A.; Kedersha, N.; Auburger, G. Mammalian ataxin-2 modulates translation control at the pre-initiation complex via PI3K/mTOR and is induced by starvation. Biochim. Biophys. Acta 2016, 1862, 1558–1569. [Google Scholar] [CrossRef] [PubMed]
- Seidel, K.; Siswanto, S.; Fredrich, M.; Bouzrou, M.; den Dunnen, W.F.A.; Ozerden, I.; Korf, H.W.; Melegh, B.; de Vries, J.J.; Brunt, E.R.; et al. On the distribution of intranuclear and cytoplasmic aggregates in the brainstem of patients with spinocerebellar ataxia type 2 and 3. Brain Pathol. 2017, 27, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Sen, N.E.; Canet-Pons, J.; Halbach, M.V.; Arsovic, A.; Pilatus, U.; Chae, W.H.; Kaya, Z.E.; Seidel, K.; Rollmann, E.; Mittelbronn, M.; et al. Generation of an Atxn2-CAG100 knock-in mouse reveals N-acetylaspartate production deficit due to early Nat8l dysregulation. Neurobiol. Dis. 2019, 132, 104559. [Google Scholar] [CrossRef]
- Xu, F.; Kula-Eversole, E.; Iwanaszko, M.; Lim, C.; Allada, R. Ataxin2 functions via CrebA to mediate Huntingtin toxicity in circadian clock neurons. PLoS Genet. 2019, 15, e1008356. [Google Scholar] [CrossRef]
- Elden, A.C.; Kim, H.J.; Hart, M.P.; Chen-Plotkin, A.S.; Johnson, B.S.; Fang, X.; Armakola, M.; Geser, F.; Greene, R.; Lu, M.M.; et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010, 466, 1069–1075. [Google Scholar] [CrossRef]
- Becker, L.A.; Huang, B.; Bieri, G.; Ma, R.; Knowles, D.A.; Jafar-Nejad, P.; Messing, J.; Kim, H.J.; Soriano, A.; Auburger, G.; et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 2017, 544, 367–371. [Google Scholar] [CrossRef]
- Gispert, S.; Kurz, A.; Waibel, S.; Bauer, P.; Liepelt, I.; Geisen, C.; Gitler, A.D.; Becker, T.; Weber, M.; Berg, D.; et al. The modulation of Amyotrophic Lateral Sclerosis risk by ataxin-2 intermediate polyglutamine expansions is a specific effect. Neurobiol. Dis. 2012, 45, 356–361. [Google Scholar] [CrossRef]
- Lahut, S.; Omur, O.; Uyan, O.; Agim, Z.S.; Ozoguz, A.; Parman, Y.; Deymeer, F.; Oflazer, P.; Koc, F.; Ozcelik, H.; et al. ATXN2 and its neighbouring gene SH2B3 are associated with increased ALS risk in the Turkish population. PLoS ONE 2012, 7, e42956. [Google Scholar] [CrossRef]
- Lee, T.; Li, Y.R.; Ingre, C.; Weber, M.; Grehl, T.; Gredal, O.; de Carvalho, M.; Meyer, T.; Tysnes, O.B.; Auburger, G.; et al. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum. Mol. Genet. 2011, 20, 1697–1700. [Google Scholar] [CrossRef] [PubMed]
- Ralser, M.; Albrecht, M.; Nonhoff, U.; Lengauer, T.; Lehrach, H.; Krobitsch, S. An integrative approach to gain insights into the cellular function of human ataxin-2. J. Mol. Biol. 2005, 346, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Nonhoff, U.; Ralser, M.; Welzel, F.; Piccini, I.; Balzereit, D.; Yaspo, M.L.; Lehrach, H.; Krobitsch, S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol. Biol. Cell 2007, 18, 1385–1396. [Google Scholar] [CrossRef] [PubMed]
- Damrath, E.; Heck, M.V.; Gispert, S.; Azizov, M.; Nowock, J.; Seifried, C.; Rub, U.; Walter, M.; Auburger, G. ATXN2-CAG42 sequesters PABPC1 into insolubility and induces FBXW8 in cerebellum of old ataxic knock-in mice. PLoS Genet. 2012, 8, e1002920. [Google Scholar] [CrossRef]
- Kaehler, C.; Isensee, J.; Nonhoff, U.; Terrey, M.; Hucho, T.; Lehrach, H.; Krobitsch, S. Ataxin-2-like is a regulator of stress granules and processing bodies. PLoS ONE 2012, 7, e50134. [Google Scholar] [CrossRef]
- Yokoshi, M.; Li, Q.; Yamamoto, M.; Okada, H.; Suzuki, Y.; Kawahara, Y. Direct binding of Ataxin-2 to distinct elements in 3′ UTRs promotes mRNA stability and protein expression. Mol. Cell 2014, 55, 186–198. [Google Scholar] [CrossRef]
- Fittschen, M.; Lastres-Becker, I.; Halbach, M.V.; Damrath, E.; Gispert, S.; Azizov, M.; Walter, M.; Muller, S.; Auburger, G. Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate. Neurogenetics 2015, 16, 181–192. [Google Scholar] [CrossRef]
- Inagaki, H.; Hosoda, N.; Tsuiji, H.; Hoshino, S.I. Direct evidence that Ataxin-2 is a translational activator mediating cytoplasmic polyadenylation. J. Biol. Chem. 2020, 295, 15810–15825. [Google Scholar] [CrossRef]
- Canet-Pons, J.; Sen, N.E.; Arsovic, A.; Almaguer-Mederos, L.E.; Halbach, M.V.; Key, J.; Doring, C.; Kerksiek, A.; Picchiarelli, G.; Cassel, R.; et al. Atxn2-CAG100-KnockIn mouse spinal cord shows progressive TDP43 pathology associated with cholesterol biosynthesis suppression. Neurobiol. Dis. 2021, 152, 105289. [Google Scholar] [CrossRef] [PubMed]
- Boeynaems, S.; Dorone, Y.; Zhuang, Y.; Shabardina, V.; Huang, G.; Marian, A.; Kim, G.; Sanyal, A.; Sen, N.E.; Griffith, D.; et al. Poly(A)-binding protein is an ataxin-2 chaperone that regulates biomolecular condensates. Mol. Cell 2023, 83, 2020–2034.e6. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Y.; Li, Z.; Xiong, S.; Sun, C.; Li, B.; Wu, S.A.; Lyu, J.; Shi, X.; Yang, L.; Chen, Y.; et al. Circadian clocks are modulated by compartmentalized oscillating translation. Cell 2023, 186, 3245–3260.e23. [Google Scholar] [CrossRef]
- Wang, J.Y.; Liu, Y.J.; Zhang, X.L.; Liu, Y.H.; Jiang, L.L.; Hu, H.Y. PolyQ-expanded ataxin-2 aggregation impairs cellular processing-body homeostasis via sequestering the RNA helicase DDX6. J. Biol. Chem. 2024, 300, 107413. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, Y.; Chen, T.; Hu, H.Y.; Lu, C. The LSmAD Domain of Ataxin-2 Modulates the Structure and RNA Binding of Its Preceding LSm Domain. Cells 2025, 14, 383. [Google Scholar] [CrossRef] [PubMed]
- Drost, J.; Nonis, D.; Eich, F.; Leske, O.; Damrath, E.; Brunt, E.R.; Lastres-Becker, I.; Heumann, R.; Nowock, J.; Auburger, G. Ataxin-2 modulates the levels of Grb2 and SRC but not ras signaling. J. Mol. Neurosci. 2013, 51, 68–81. [Google Scholar] [CrossRef]
- Meunier, C.; Bordereaux, D.; Porteu, F.; Gisselbrecht, S.; Chretien, S.; Courtois, G. Cloning and characterization of a family of proteins associated with Mpl. J. Biol. Chem. 2002, 277, 9139–9147. [Google Scholar] [CrossRef] [PubMed]
- Coller, J.; Parker, R. General translational repression by activators of mRNA decapping. Cell 2005, 122, 875–886. [Google Scholar] [CrossRef]
- Lobel, J.H.; Gross, J.D. Pdc2/Pat1 increases the range of decay factors and RNA bound by the Lsm1-7 complex. RNA 2020, 26, 1380–1388. [Google Scholar] [CrossRef]
- Hurst, Z.; Liu, W.; Shi, Q.; Herman, P.K. A distinct P-body-like granule is induced in response to the disruption of microtubule integrity in Saccharomyces cerevisiae. Genetics 2022, 222, iyac105. [Google Scholar] [CrossRef]
- Vijjamarri, A.K.; Gupta, N.; Onu, C.; Niu, X.; Zhang, F.; Kumar, R.; Lin, Z.; Greenberg, M.L.; Hinnebusch, A.G. mRNA decapping activators Pat1 and Dhh1 regulate transcript abundance and translation to tune cellular responses to nutrient availability. Nucleic Acids Res. 2023, 51, 9314–9336. [Google Scholar] [CrossRef]
- Bahassou-Benamri, R.; Davin, A.H.; Gaillard, J.C.; Alonso, B.; Odorico, M.; Pible, O.; Armengaud, J.; Godon, C. Subcellular localization and interaction network of the mRNA decay activator Pat1 upon UV stress. Yeast 2013, 30, 353–363. [Google Scholar] [CrossRef]
- Vindry, C.; Marnef, A.; Broomhead, H.; Twyffels, L.; Ozgur, S.; Stoecklin, G.; Llorian, M.; Smith, C.W.; Mata, J.; Weil, D.; et al. Dual RNA Processing Roles of Pat1b via Cytoplasmic Lsm1-7 and Nuclear Lsm2-8 Complexes. Cell Rep. 2017, 20, 1187–1200. [Google Scholar] [CrossRef] [PubMed]
- Vindry, C.; Weil, D.; Standart, N. Pat1 RNA-binding proteins: Multitasking shuttling proteins. Wiley Interdiscip. Rev. RNA 2019, 10, e1557. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.J.; Nesler, K.R.; Rosen, S.F.; Kato, Y.; Nakamura, A.; Ramaswami, M.; Barbee, S.A. The conserved P body component HPat/Pat1 negatively regulates synaptic terminal growth at the larval Drosophila neuromuscular junction. J. Cell Sci. 2012, 125, 6105–6116. [Google Scholar] [CrossRef]
- Kaehler, C.; Guenther, A.; Uhlich, A.; Krobitsch, S. PRMT1-mediated arginine methylation controls ATXN2L localization. Exp. Cell Res. 2015, 334, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Key, J.; Almaguer-Mederos, L.E.; Kandi, A.R.; Sen, N.E.; Gispert, S.; Kopf, G.; Meierhofer, D.; Auburger, G. ATXN2L primarily interacts with NUFIP2, the absence of ATXN2L results in NUFIP2 depletion, and the ATXN2-polyQ expansion triggers NUFIP2 accumulation. Neurobiol. Dis. 2025, 209, 106903. [Google Scholar] [CrossRef]
- Key, J.; Harter, P.N.; Sen, N.E.; Gradhand, E.; Auburger, G.; Gispert, S. Mid-Gestation lethality of Atxn2l-Ablated Mice. Int. J. Mol. Sci. 2020, 21, 5124. [Google Scholar] [CrossRef] [PubMed]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Martens, L.; Hermjakob, H.; Jones, P.; Adamski, M.; Taylor, C.; States, D.; Gevaert, K.; Vandekerckhove, J.; Apweiler, R. PRIDE: The proteomics identifications database. Proteomics 2005, 5, 3537–3545. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Li, X.; Wang, H.Y.; Steven Zheng, X.F. Beyond regulation of pol III: Role of MAF1 in growth, metabolism, aging and cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 338–343. [Google Scholar] [CrossRef]
- Schlaitz, A.L.; Thompson, J.; Wong, C.C.; Yates, J.R., 3rd; Heald, R. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev. Cell 2013, 26, 315–323. [Google Scholar] [CrossRef]
- Burke, B. PREEParing for mitosis. Dev. Cell 2013, 26, 221–222. [Google Scholar] [CrossRef]
- Urlaub, H.; Raker, V.A.; Kostka, S.; Luhrmann, R. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 2001, 20, 187–196. [Google Scholar] [CrossRef]
- Decker, C.J.; Parker, R. P-bodies and stress granules: Possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 2012, 4, a012286. [Google Scholar] [CrossRef]
- Irimia, M.; Rukov, J.L.; Penny, D.; Roy, S.W. Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing. BMC Evol. Biol. 2007, 7, 188. [Google Scholar] [CrossRef]
- Cardoso, B.R.; Roberts, B.R.; Bush, A.I.; Hare, D.J. Selenium, selenoproteins and neurodegenerative diseases. Metallomics 2015, 7, 1213–1228. [Google Scholar] [CrossRef]
- Ding, W.; Wang, S.; Gu, J.; Yu, L. Selenium and human nervous system. Chin. Chem. Lett. 2023, 34, 108043. [Google Scholar] [CrossRef]
- Hayashi, S.; McMahon, A.P. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 2002, 244, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Eckardt, D.; Theis, M.; Doring, B.; Speidel, D.; Willecke, K.; Ott, T. Spontaneous ectopic recombination in cell-type-specific Cre mice removes loxP-flanked marker cassettes in vivo. Genesis 2004, 38, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, S.; Keck, T.; Roelandse, M.; Hartman, R.; Jeromin, A.; Perry, S.; Hofer, S.B.; Mrsic-Flogel, T.; Levelt, C.N. Cre-dependent expression of multiple transgenes in isolated neurons of the adult forebrain. PLoS ONE 2008, 3, e3059. [Google Scholar] [CrossRef] [PubMed]
- Senserrich, J.; Batsivari, A.; Rybtsov, S.; Gordon-Keylock, S.; Souilhol, C.; Buchholz, F.; Hills, D.; Zhao, S.; Medvinsky, A. Analysis of Runx1 Using Induced Gene Ablation Reveals Its Essential Role in Pre-liver HSC Development and Limitations of an In Vivo Approach. Stem Cell Rep. 2018, 11, 784–794. [Google Scholar] [CrossRef]
- Crawley, J.N. Behavioral phenotyping strategies for mutant mice. Neuron 2008, 57, 809–818. [Google Scholar] [CrossRef]
- Kwon, D.Y.; Xu, B.; Hu, P.; Zhao, Y.T.; Beagan, J.A.; Nofziger, J.H.; Cui, Y.; Phillips-Cremins, J.E.; Blendy, J.A.; Wu, H.; et al. Neuronal Yin Yang1 in the prefrontal cortex regulates transcriptional and behavioral responses to chronic stress in mice. Nat. Commun. 2022, 13, 55. [Google Scholar] [CrossRef]
- Mastwal, S.; Li, X.; Stowell, R.; Manion, M.; Zhang, W.; Kim, N.S.; Yoon, K.J.; Song, H.; Ming, G.L.; Wang, K.H. Adolescent neurostimulation of dopamine circuit reverses genetic deficits in frontal cortex function. Elife 2023, 12, RP87414. [Google Scholar] [CrossRef]
- Bish, R.; Cuevas-Polo, N.; Cheng, Z.; Hambardzumyan, D.; Munschauer, M.; Landthaler, M.; Vogel, C. Comprehensive Protein Interactome Analysis of a Key RNA Helicase: Detection of Novel Stress Granule Proteins. Biomolecules 2015, 5, 1441–1466. [Google Scholar] [CrossRef]
- Shah, S.; Molinaro, G.; Liu, B.; Wang, R.; Huber, K.M.; Richter, J.D. FMRP Control of Ribosome Translocation Promotes Chromatin Modifications and Alternative Splicing of Neuronal Genes Linked to Autism. Cell Rep. 2020, 30, 4459–4472.e6. [Google Scholar] [CrossRef]
- Brown, A.L.; Wilkins, O.G.; Keuss, M.J.; Kargbo-Hill, S.E.; Zanovello, M.; Lee, W.C.; Bampton, A.; Lee, F.C.Y.; Masino, L.; Qi, Y.A.; et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature 2022, 603, 131–137. [Google Scholar] [CrossRef]
- Takayama, K.I.; Suzuki, T.; Sato, K.; Saito, Y.; Inoue, S. Cooperative nuclear action of RNA-binding proteins PSF and G3BP2 to sustain neuronal cell viability is decreased in aging and dementia. Aging Cell 2024, 23, e14316. [Google Scholar] [CrossRef] [PubMed]
- Goebbels, S.; Bormuth, I.; Bode, U.; Hermanson, O.; Schwab, M.H.; Nave, K.A. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis 2006, 44, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.S.; Xu, J.W.; Lai, W.Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef]
- Chen, X.; Li, A.; Sun, B.F.; Yang, Y.; Han, Y.N.; Yuan, X.; Chen, R.X.; Wei, W.S.; Liu, Y.; Gao, C.C.; et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019, 21, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; McMahon, C.; Blobel, G. Primary structure of a human arginine-rich nuclear protein that colocalizes with spliceosome components. Proc. Natl. Acad. Sci. USA 1991, 88, 8189–8193. [Google Scholar] [CrossRef]
- Twyffels, L.; Gueydan, C.; Kruys, V. Shuttling SR proteins: More than splicing factors. FEBS J. 2011, 278, 3246–3255. [Google Scholar] [CrossRef]
- Zhang, W.J.; Wu, J.Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol. Cell Biol. 1996, 16, 5400–5408. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, C.F.; Kramer, A.; Berget, S.M. A role for SRp54 during intron bridging of small introns with pyrimidine tracts upstream of the branch point. Mol. Cell Biol. 1998, 18, 5425–5434. [Google Scholar] [CrossRef] [PubMed]
- Gehring, N.H.; Roignant, J.Y. Anything but Ordinary—Emerging Splicing Mechanisms in Eukaryotic Gene Regulation. Trends Genet. 2021, 37, 355–372. [Google Scholar] [CrossRef]


| Observed/Expected Number of Live Born Mice with Indicated Genotype | ||||
|---|---|---|---|---|
| +/+ | +/− | −/− | Number of Offspring | |
| Live born female | 33/27 | 39/54 | 0/27 | 72/108 |
| Live born male | 20/27 | 26/54 | 0/27 | 46/108 |
| Live born total | 53/54 | 65/108 | 0/54 | 118/216 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Key, J.; Almaguer-Mederos, L.-E.; Kandi, A.R.; Fellenz, M.; Gispert, S.; Köpf, G.; Meierhofer, D.; Deller, T.; Auburger, G. Conditional ATXN2L-Null in Adult Frontal Cortex CamK2a+ Neurons Does Not Cause Cell Death but Restricts Spontaneous Mobility and Affects the Alternative Splicing Pathway. Cells 2025, 14, 1532. https://doi.org/10.3390/cells14191532
Key J, Almaguer-Mederos L-E, Kandi AR, Fellenz M, Gispert S, Köpf G, Meierhofer D, Deller T, Auburger G. Conditional ATXN2L-Null in Adult Frontal Cortex CamK2a+ Neurons Does Not Cause Cell Death but Restricts Spontaneous Mobility and Affects the Alternative Splicing Pathway. Cells. 2025; 14(19):1532. https://doi.org/10.3390/cells14191532
Chicago/Turabian StyleKey, Jana, Luis-Enrique Almaguer-Mederos, Arvind Reddy Kandi, Meike Fellenz, Suzana Gispert, Gabriele Köpf, David Meierhofer, Thomas Deller, and Georg Auburger. 2025. "Conditional ATXN2L-Null in Adult Frontal Cortex CamK2a+ Neurons Does Not Cause Cell Death but Restricts Spontaneous Mobility and Affects the Alternative Splicing Pathway" Cells 14, no. 19: 1532. https://doi.org/10.3390/cells14191532
APA StyleKey, J., Almaguer-Mederos, L.-E., Kandi, A. R., Fellenz, M., Gispert, S., Köpf, G., Meierhofer, D., Deller, T., & Auburger, G. (2025). Conditional ATXN2L-Null in Adult Frontal Cortex CamK2a+ Neurons Does Not Cause Cell Death but Restricts Spontaneous Mobility and Affects the Alternative Splicing Pathway. Cells, 14(19), 1532. https://doi.org/10.3390/cells14191532







