ATHB1 Interacts with Hormone-Related Gene Regulatory Networks Involved in Biotic and Abiotic Stress Responses in Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Growth Conditions
2.3. MeJA Treatment
2.4. Gene Expression Analysis
2.5. Microarray Analysis
3. Results
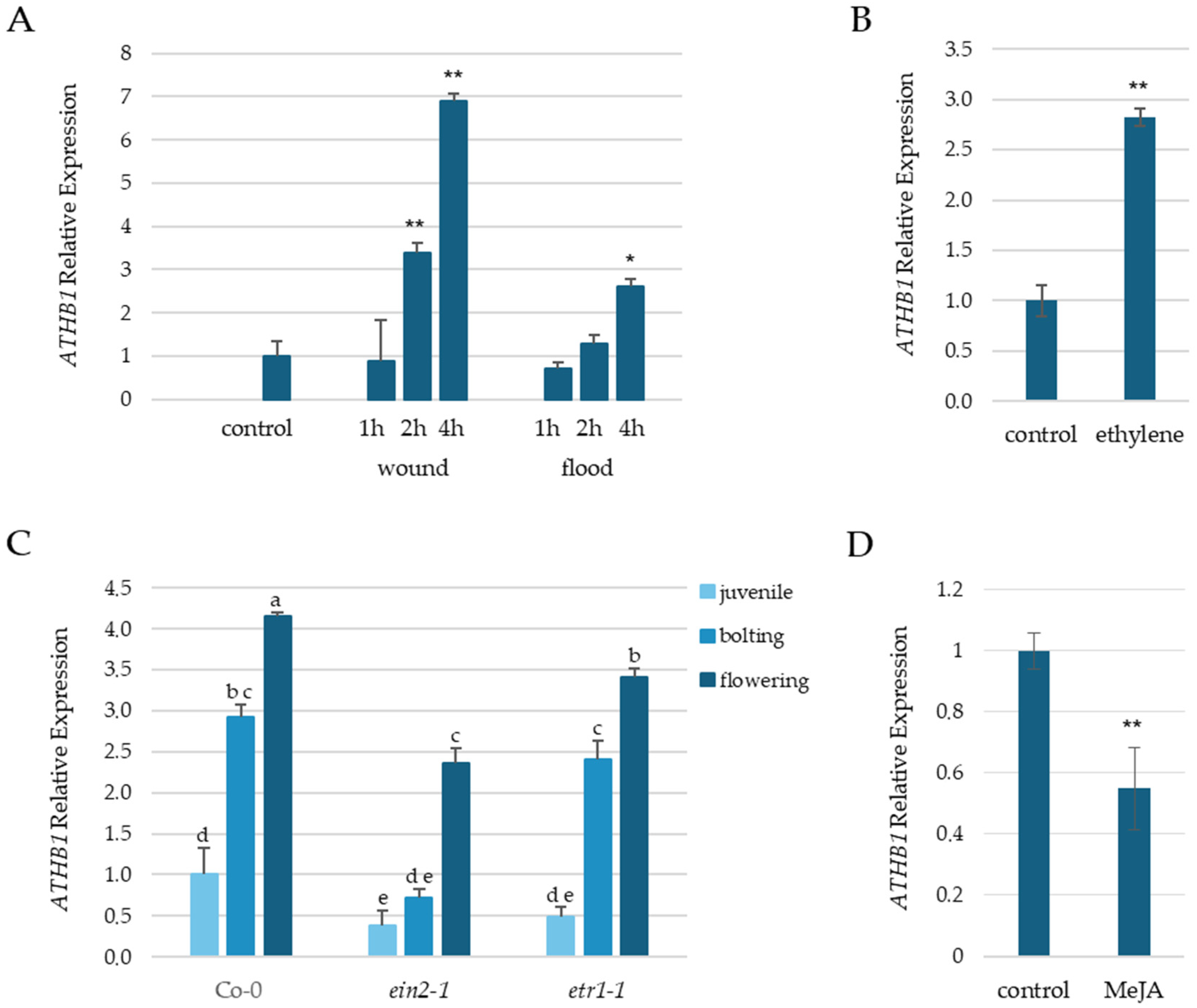
3.1. ATHB1 Expression Is Stress Inducible and Regulated by Ethylene
3.2. ATHB1 Enhances Sensitivity to MeJA Treatment
3.3. ATHB1 Affects the Expression of Genes Responsive to MeJA Treatment
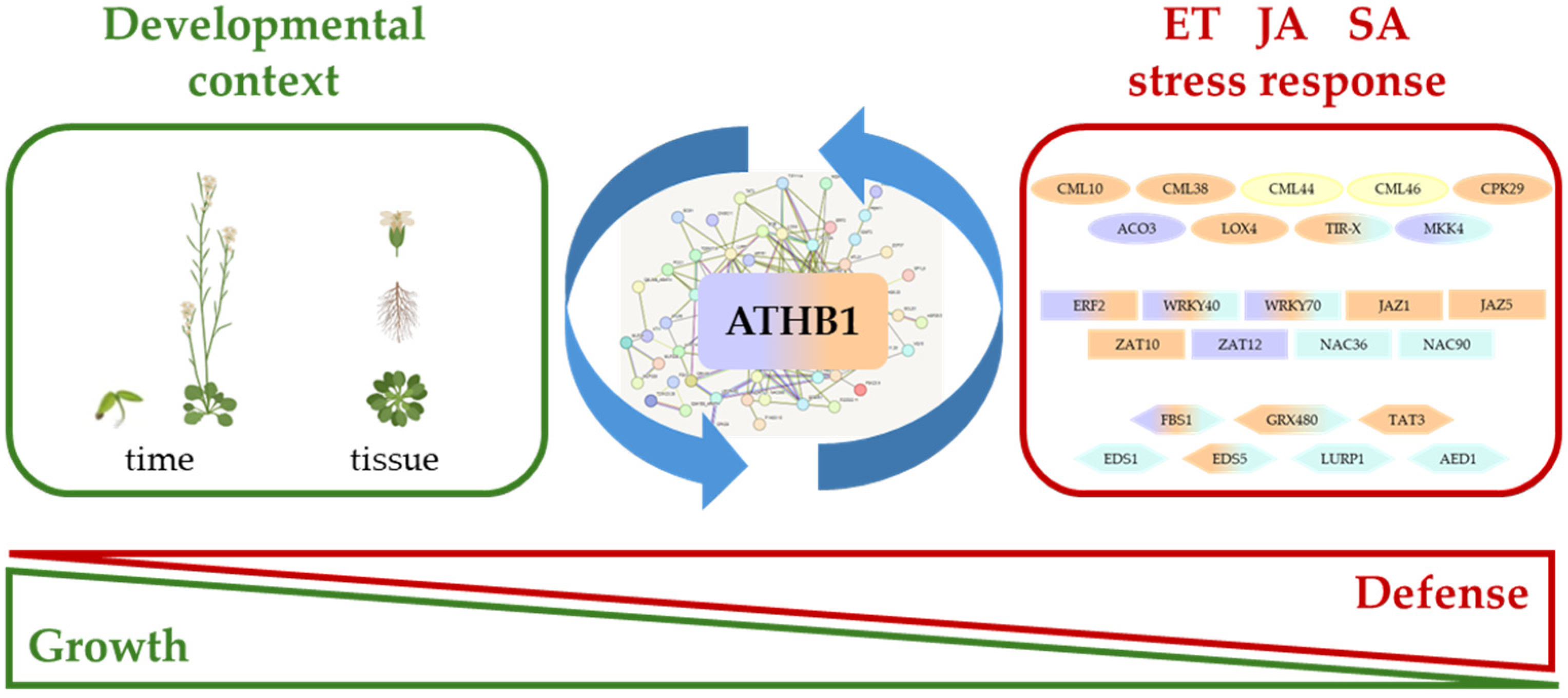
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.-K. Abiotic stress responses in plants. Nat. Rev. Genet. 2022, 23, 104–119. [Google Scholar] [CrossRef]
- Aerts, N.; Pereira Mendes, M.; Van Wees, S.C.M. Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 2021, 105, 489–504. [Google Scholar] [CrossRef] [PubMed]
- Waadt, R.; Seller, C.A.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Khan, S.-A.; Li, M.-Z.; Wang, S.-M.; Yin, H.-J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef]
- Yin, L.; Zander, M.; Carol Huang, S.; Xie, M.; Paola Saldierna Guzmán, J.; Hann, E.; Shanbhag, B.K.; Ng, S.; Jain, S.; Janssen, B.J.; et al. Transcription Factor Dynamics in Cross-Regulation of Plant Hormone Signaling Pathways. bioRxiv 2023. [Google Scholar] [CrossRef]
- Henriksson, E.; Olsson, A.S.B.; Johannesson, H.; Johansson, H.; Hanson, J.; Engström, P.; SödErman, E. Homeodomain Leucine Zipper Class I Genes in Arabidopsis. Expression Patterns and Phylogenetic Relationships. Plant Physiol. 2005, 139, 509–518. [Google Scholar] [CrossRef]
- Dezar, C.A.; Gago, G.M.; González, D.H.; Chan, R.L. Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res. 2005, 14, 429–440. [Google Scholar] [CrossRef]
- Deng, X.; Phillips, J.; Bräutigam, A.; Engström, P.; Johannesson, H.; Ouwerkerk, P.B.F.; Ruberti, I.; Salinas, J.; Vera, P.; Iannacone, R.; et al. A Homeodomain Leucine Zipper Gene from Craterostigma plantagineum Regulates Abscisic Acid Responsive Gene Expression and Physiological Responses. Plant Mol. Biol. 2006, 61, 469–489. [Google Scholar] [CrossRef]
- Agalou, A.; Purwantomo, S.; Övernäs, E.; Johannesson, H.; Zhu, X.; Estiati, A.; de Kam, R.J.; Engström, P.; Slamet-Loedin, I.H.; Zhu, Z.; et al. A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 2008, 66, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Li, H.; Teng, R.; Wang, Y.; Wang, W.; Zhuang, J. Genomic and transcriptomic analyses of HD-Zip family transcription factors and their responses to abiotic stress in tea plant (Camellia sinensis). Genomics 2019, 111, 1142–1151. [Google Scholar] [CrossRef]
- Romani, F.; Ribone, P.A.; Capella, M.; Miguel, V.N.; Chan, R.L. A matter of quantity: Common features in the drought response of transgenic plants overexpressing HD-Zip I transcription factors. Plant Sci. 2016, 251, 139–154. [Google Scholar] [CrossRef]
- Perotti, M.F.; Ribone, P.A.; Chan, R.L. Plant transcription factors from the homeodomain-leucine zipper family I. Role in development and stress responses. IUBMB Life 2017, 69, 280–289. [Google Scholar] [CrossRef]
- Ruberti, I.; Sessa, G.; Lucchetti, S.; Morelli, G. A novel class of plant proteins containing a homeodomain with a closely linked leucine zipper motif. EMBO J. 1991, 10, 1787–1791. [Google Scholar] [CrossRef]
- Aoyama, T.; Dong, C.-H.; Wu, Y.; Carabelli, M.; Sessa, G.; Ruberti, I.; Morelli, G.; Chua, N.-H. Ectopic Expression of the Arabidopsis Transcriptional Activator Athb-I Alters Leaf Cell Fate in Tobacco. Plant Cell 1995, 7, 1773–1785. [Google Scholar] [PubMed]
- Miguel, V.N.; Manavella, P.A.; Chan, R.L.; Capella, M. The AtHB1 Transcription Factor Controls the miR164-CUC2 Regulatory Node to Modulate Leaf Development. Plant Cell Physiol. 2020, 61, 659–670. [Google Scholar] [CrossRef]
- Capella, M.; Ribone, P.A.; Arce, A.L.; Chan, R.L. Arabidopsis thaliana HomeoBox 1 (AtHB1), a Homedomain-Leucine Zipper I (HD-Zip I) transcription factor, is regulated by PHYTOCHROME-INTERACTING FACTOR 1 to promote hypocotyl elongation. New Phytol. 2015, 207, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Zhong, G.Y.; Burns, J.K. Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol. Biol. 2003, 53, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Hong, Y.; Yin, M.; Li, C.; Zhang, K.; Grierson, D. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 2008, 55, 301–310. [Google Scholar] [CrossRef]
- Sundaresan, V.; Springer, P.; Volpe, T.; Haward, S.; Jones, J.D.; Dean, C.; Ma, H.; Martienssen, R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 1995, 9, 1797–1810. [Google Scholar] [CrossRef]
- Ribone, P.A.; Capella, M.; Arce, A.L.; Chan, R.L. A uORF Represses the Transcription Factor AtHB1 in Aerial Tissues to Avoid a Deleterious Phenotype. Plant Physiol. 2017, 175, 1238–1253. [Google Scholar] [CrossRef] [PubMed]
- Baima, S.; Nobili, F.; Sessa, G.; Lucchetti, S.; Ruberti, I.; Morelli, G. The expression of the Athb-8 homeobox gene is restricted to provascular cells in Arabidopsis thaliana. Development 1995, 121, 4171–4182. [Google Scholar] [CrossRef]
- Baima, S.; Possenti, M.; Matteucci, A.; Wisman, E.; Altamura, M.M.; Ruberti, I.; Morelli, G. The arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol. 2001, 126, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Willems, E.; Leyns, L.; Vandesompele, J. Standardization of real-time PCR gene expression data from independent biological replicates. Anal. Biochem. 2008, 379, 127–129. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 1–25. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 9 September 2025).
- Santana-Garcia, W.; Castro-Mondragon, J.A.; Padilla-Gálvez, M.; Nguyen, N.T.T.; Elizondo-Salas, A.; Ksouri, N.; Gerbes, F.; Thieffry, D.; Vincens, P.; Contreras-Moreira, B.; et al. RSAT 2022: Regulatory sequence analysis tools. Nucleic Acids Res. 2022, 50, W670–W676. [Google Scholar] [CrossRef]
- Graham, L.E.; Schippers, J.H.M.; Dijkwel, P.P.; Wagstaff, C. Ethylene and Senescence Processes. In Annual Plant Reviews Volume 44: The Plant Hormone Ethylene; Wiley: Hoboken, NJ, USA, 2012; pp. 305–341. [Google Scholar]
- Patterson, S.E.; Bleecker, A.B. Ethylene-Dependent and-Independent Processes Associated with Floral Organ Abscission in Arabidopsis. Plant Physiol. 2004, 134, 194–203. [Google Scholar] [CrossRef]
- Roberts, J.A.; Whitelaw, C.A.; Gonzalez-Carranza, Z.H.; McManus, M.T. Cell Separation Processes in Plants–Models, Mechanisms and Manipulation. Ann. Bot. 2000, 86, 223–235. [Google Scholar] [CrossRef]
- Zhu, Z. Molecular basis for jasmonate and ethylene signal interactions in Arabidopsis. J. Exp. Bot. 2014, 65, 5743–5748. [Google Scholar] [CrossRef]
- Pérez-Llorca, M.; Pollmann, S.; Müller, M. Ethylene and Jasmonates Signaling Network Mediating Secondary Metabolites under Abiotic Stress. Int. J. Mol. Sci. 2023, 24, 5990. [Google Scholar] [CrossRef]
- Farhangi-Abriz, S.; Ghassemi-Golezani, K. Jasmonates: Mechanisms and functions in abiotic stress tolerance of plants. Biocatal. Agric. Biotechnol. 2019, 20, 101210. [Google Scholar] [CrossRef]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of Jasmonic Acid in Plant Regulation and Response to Abiotic Stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef]
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.S.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2021, 40, 1513–1541. [Google Scholar] [CrossRef]
- Hu, S.; Yu, K.; Yan, J.; Shan, X.; Xie, D. Jasmonate perception: Ligand–receptor interaction, regulation, and evolution. Mol. Plant 2023, 16, 23–42. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Brader, G.; Palva, E.T. The WRKY70 Transcription Factor: A Node of Convergence for Jasmonate-Mediated and Salicylate-Mediated Signals in Plant Defense. Plant Cell 2004, 16, 319–331. [Google Scholar] [CrossRef]
- Brotman, Y.; Landau, U.; Cuadros-Inostroza, Á.; Takayuki, T.; Fernie, A.R.; Chet, I.; Viterbo, A.; Willmitzer, L. Trichoderma-Plant Root Colonization: Escaping Early Plant Defense Responses and Activation of the Antioxidant Machinery for Saline Stress Tolerance. PLoS Pathog. 2013, 9, e1003221. [Google Scholar] [CrossRef]
- Kroes, A.; van Loon, J.J.A.; Dicke, M. Density-Dependent Interference of Aphids with Caterpillar-Induced Defenses in Arabidopsis: Involvement of Phytohormones and Transcription Factors. Plant Cell Physiol. 2015, 56, 98–106. [Google Scholar] [CrossRef]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef]
- Xie, M.; Sun, J.; Gong, D.; Kong, Y. The Roles of Arabidopsis C1-2i Subclass of C2H2-type Zinc-Finger Transcription Factors. Genes 2019, 10, 653. [Google Scholar] [CrossRef]
- McGrath, K.C.; Dombrecht, B.; Manners, J.M.; Schenk, P.M.; Edgar, C.I.; Maclean, D.J.; Scheible, W.-R.; Udvardi, M.K.; Kazan, K. Repressor- and Activator-Type Ethylene Response Factors Functioning in Jasmonate Signaling and Disease Resistance Identified via a Genome-Wide Screen of Arabidopsis Transcription Factor Gene Expression. Plant Physiol. 2005, 139, 949–959. [Google Scholar] [CrossRef]
- Yang, T.-H.; Lenglet-Hilfiker, A.; Stolz, S.; Glauser, G.; Farmer, E.E. Jasmonate Precursor Biosynthetic Enzymes LOX3 and LOX4 Control Wound-Response Growth Restriction. Plant Physiol. 2020, 184, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.S.; Islam, F.; Chen, H.; Chang, M.; Wang, D.; Liu, F.; Fu, Z.Q.; Chen, J. Transcriptional Coactivators: Driving Force of Plant Immunity. Front. Plant Sci. 2022, 13, 823937. [Google Scholar] [CrossRef]
- Mine, A.; Nobori, T.; Salazar-Rondon, M.C.; Winkelmüller, T.M.; Anver, S.; Becker, D.; Tsuda, K. An incoherent feed-forward loop mediates robustness and tunability in a plant immune network. EMBO Rep. 2017, 18, 464–476. [Google Scholar] [CrossRef]
- Uhrig, J.F.; Huang, L.-J.; Barghahn, S.; Willmer, M.; Thurow, C.; Gatz, C. CC-type glutaredoxins recruit the transcriptional co-repressor TOPLESS to TGA-dependent target promoters in Arabidopsis thaliana. Biochim. Biophys. Acta-Gene Regul. Mech. 2017, 1860, 218–226. [Google Scholar] [CrossRef]
- Kato, H.; Saito, T.; Ito, H.; Komeda, Y.; Kato, A. Overexpression of the TIR-X gene results in a dwarf phenotype and activation of defense-related gene expression in Arabidopsis thaliana. J. Plant Physiol. 2014, 171, 382–388. [Google Scholar] [CrossRef]
- Yan, Y.; Stolz, S.; Chételat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A Downstream Mediator in the Growth Repression Limb of the Jasmonate Pathway. Plant Cell 2007, 19, 2470–2483. [Google Scholar] [CrossRef]
- Breitenbach, H.H.; Wenig, M.; Wittek, F.; Jordá, L.; Maldonado-Alconada, A.M.; Sarioglu, H.; Colby, T.; Knappe, C.; Bichlmeier, M.; Pabst, E.; et al. Contrasting Roles of the Apoplastic Aspartyl Protease Apoplastic, Enhanced Disease Susceptibility1-Dependent1 and Legume Lectin-Like Protein1 in Arabidopsis Systemic Acquired Resistance. Plant Physiol. 2014, 165, 791–809. [Google Scholar] [CrossRef]
- Knoth, C.; Eulgem, T. The oomycete response gene LURP1 is required for defense against Hyaloperonospora parasitica in Arabidopsis thaliana. Plant J. 2008, 55, 53–64. [Google Scholar] [CrossRef]
- Gonzalez, L.E.; Keller, K.; Chan, K.X.; Gessel, M.M.; Thines, B.C. Transcriptome analysis uncovers Arabidopsis F-BOX STRESS INDUCED 1 as a regulator of jasmonic acid and abscisic acid stress gene expression. BMC Genomics 2017, 18, 533. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, J.; Zhu, Z.; Song, Y.; Wang, X.; Wang, X.; Zhou, X. MKK4/MKK5-MPK1/MPK2 cascade mediates SA-activated leaf senescence via phosphorylation of NPR1 in Arabidopsis. Plant Mol. Biol. 2020, 102, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Lokdarshi, A.; Conner, W.C.; McClintock, C.; Li, T.; Roberts, D.M. Arabidopsis CML38, a Calcium Sensor That Localizes to Ribonucleoprotein Complexes under Hypoxia Stress. Plant Physiol. 2016, 170, 1046–1059. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Ganguly, A.; Baik, S.; Cho, H.-T. Calcium-dependent protein kinase 29 modulates PIN-FORMED polarity and Arabidopsis development via its own phosphorylation code. Plant Cell 2021, 33, 3513–3531. [Google Scholar] [CrossRef] [PubMed]
- Maple, R.; Zhu, P.; Hepworth, J.; Wang, J.-W.; Dean, C. Flowering time: From physiology, through genetics to mechanism. Plant Physiol. 2024, 195, 190–212. [Google Scholar] [CrossRef]
- Houben, M.; Van de Poel, B. 1-Aminocyclopropane-1-Carboxylic Acid Oxidase (ACO): The Enzyme That Makes the Plant Hormone Ethylene. Front. Plant Sci. 2019, 10, 695. [Google Scholar] [CrossRef]
- Sessa, G.; Morelli, G.; Ruberti, I. The Athb-1 and-2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J. 1993, 12, 3507–3517. [Google Scholar] [CrossRef]
- Sessa, G.; Morelli, G.; Ruberti, I. DNA-binding specificity of the homeodomain-leucine zipper domain. J. Mol. Biol. 1997, 274, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Palena, C.M.; Gonzalez, D.H.; Chan, R.L. A monomer-dimer equilibrium modulates the interaction of the sunflower homeodomain leucine-zipper protein Hahb-4 with DNA. Biochem. J. 1999, 341, 81–87. [Google Scholar] [CrossRef]
- Perotti, M.F.; Arce, A.L.; Chan, R.L. The underground life of homeodomain-leucine zipper transcription factors. J. Exp. Bot. 2021, 72, 4005–4021. [Google Scholar] [CrossRef]
- Żyła, N.; Babula-Skowrońska, D. Evolutionary Consequences of Functional and Regulatory Divergence of HD-Zip I Transcription Factors as a Source of Diversity in Protein Interaction Networks in Plants. J. Mol. Evol. 2023, 91, 581–597. [Google Scholar] [CrossRef]
- Sessa, G.; Carabelli, M.; Sassi, M. The Ins and Outs of Homeodomain-Leucine Zipper/Hormone Networks in the Regulation of Plant Development. Int. J. Mol. Sci. 2024, 25, 5657. [Google Scholar] [CrossRef]
- Manavella, P.A.; Dezar, C.A.; Bonaventure, G.; Baldwin, I.T.; Chan, R.L. HAHB4, a sunflower HD-Zip protein, integrates signals from the jasmonic acid and ethylene pathways during wounding and biotic stress responses. Plant J. 2008, 56, 376–388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Turner, J.G. Wound-Induced Endogenous Jasmonates Stunt Plant Growth by Inhibiting Mitosis. PLoS ONE 2008, 3, e3699. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Major, I.T.; Howe, G.A. Resolution of growth–defense conflict: Mechanistic insights from jasmonate signaling. Curr. Opin. Plant Biol. 2018, 44, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, J.-H.; Kim, J.; Kim, J.J.; Hong, S.; Kim, J.; Kim, J.H.; Woo, H.R.; Hyeon, C.; Lim, P.O.; et al. Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4930–E4939. [Google Scholar] [CrossRef]
- Cai, J.; Panda, S.; Kazachkova, Y.; Amzallag, E.; Li, Z.; Meir, S.; Rogachev, I.; Aharoni, A. A NAC triad modulates plant immunity by negatively regulating N-hydroxy pipecolic acid biosynthesis. Nat. Commun. 2024, 15, 7212. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Jing, Y.; Lin, R. The VQ Motif-Containing Protein Family of Plant-Specific Transcriptional Regulators. Plant Physiol. 2015, 169, 371–378. [Google Scholar] [CrossRef]
- Ülker, B.; Shahid Mukhtar, M.; Somssich, I.E. The WRKY70 transcription factor of Arabidopsis influences both the plant senescence and defense signaling pathways. Planta 2007, 226, 125–137. [Google Scholar] [CrossRef]
- Besseau, S.; Li, J.; Palva, E.T. WRKY54 and WRKY70 co-operate as negative regulators of leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 2667–2679. [Google Scholar] [CrossRef]


| AGI Code | Gene | log2 FC | log2 FC | Gene Annotation | |
|---|---|---|---|---|---|
| (athb1 4 h MeJA/mock)–(WT 4 h MeJA/Mock) | athb1/WT Mock 1 h | athb1/WT Mock 4 h | |||
| AT1G27730 | STZ/ZAT10 | 1.51 | −1.06 | −1.41 | SALT TOLERANCE ZINC FINGER; nucleic acid binding; transcription repressor; zinc ion binding; response to oxidative stress, response to wounding, response to salt stress |
| AT1G76650 | CML38 | 1.36 | −0.77 * | −1.6 | CALMODULIN-LIKE 38; calcium-binding EF hand family protein; response to wounding; positive regulation of brassinosteroid mediated signaling pathway, response to hypoxia |
| AT2G17040 | NAC036 | 1.27 | −1.33 | −1.46 | NAC DOMAIN CONTAINING PROTEIN 36; transcription factor; involved in leaf and inflorescence stem morphogenesis |
| AT4G39030 | EDS5 | 1.14 | −0.65 | −1.51 | ENHANCED DISEASE SUSCEPTIBILITY 5; orphan multidrug and toxin extrusion transporter; essential component of salicylic acid-dependent signaling |
| AT1G17380 | JAZ5 | 1.12 | −0.7 | −1.05 | JASMONATE-ZIM-DOMAIN PROTEIN 5; regulation of jasmonic acid mediated signaling pathway; involved in response to wounding |
| AT2G39030 | NATA1 | 1.12 | −0.22 * | −0.21 * | N-ACETYLTRANSFERASE ACTIVITY 1; ornithine metabolic process, polyamine acetylation; response to jasmonic acid |
| AT1G78410 | VQ10 | 1.1 | −0.91 | −1.55 | VQ MOTIF-CONTAINING PROTEIN 10; response to oxidative stress; protein binding, interaction with WRKY transcription factors |
| AGI Code | Gene | Annotation | Microarray (log2 FC) | RT-qPCR Validation * (log2 FC) |
|---|---|---|---|---|
| AT5G10140 | FLC | FLOWERING LOCUS C; MADS-box Transcription Factor; specific transcriptional repressor; regulation of flower development, response to abiotic stimulus. | 1.71 | 2.78 ± 0.18 |
| AT1G12010 | ACO3 | 1-Amino-Cyclopropane-1-Carboxylic Acid Oxidase 3; ethylene biosynthesis. | 0.80 | 1.27 ± 0.36 |
| AT5G47220 | ERF2 | Ethylene Response Factor-2; ERF/AP2 transcription factor family; transcriptional activator; ethylene-activated signaling pathway; jasmonic acid mediated signaling pathway. | −0.74 | −1.01 ± 0.61 |
| AT2G39030 | NATA1 | N-ACETYLTRANSFERASE ACTIVITY 1; ornithine metabolic process, polyamine acetylation; response to jasmonic acid. | −0.21 ** | −0.72 ± 1.08 |
| AT1G17380 | JAZ5 | Jasmonate-ZIM-domain Protein 5; regulation of jasmonic acid mediated signaling pathway; response to wounding. | −1.05 | −1.10 ± 0.41 |
| AT3G48090 | EDS1 | Enhanced Disease Susceptibility 1; component of R gene-mediated disease resistance; acts redundantly with salicylic acid to regulate resistance gene-mediated signaling; lipase; signal transducer. | −1.10 | −1.04 ± 0.05 |
| AT4G21090 | MFDX2 | Mitochondrial Ferredoxin 2; metal ion binding. | −1.29 | −1.00 ± 0.39 |
| AT1G27730 | ZAT10 | STZ (salt tolerance zinc finger); nucleic acid binding; transcriptional repressor involved in abiotic stress responses; probably involved in jasmonate (JA) early signaling response; response to oxidative stress, response to wounding, response to salt stress; zinc ion binding. | −1.41 | −1.57 ± 0.14 |
| AT2G17040 | NAC36 | Arabidopsis NAC domain containing protein 36; transcription factor; involved in leaf and inflorescence stem morphogenesis. | −1.46 | −1.39 ± 0.05 |
| AT1G80840 | WRKY40 | Pathogen-induced transcription factor; binds W-box sequences in vitro; regulation of defense response; response to salicylic acid; response to wounding. | −1.50 | −1.83 ± 0.59 |
| AT4G39030 | EDS5 | Enhanced Disease Susceptibility 5; multidrug and toxin extrusion transporter; component of salicylic acid-dependent signaling. | −1.51 | −1.68 ± 0.08 |
| AT1G78410 | VQ10 | VQ motif-containing protein, response to oxidative stress; protein binding, interaction with WRKY transcription factors. | −1.55 | −2.26 ± 0.48 |
| AT1G76650 | CML38 | CALMODULIN-LIKE 38; calcium-binding EF hand family protein; response to wounding; positive regulation of brassinosteroid mediated signaling pathway, response to hypoxia. | −1.6 | −1.76 ± 0.26 |
| AT5G22380 | NAC90 | NAC domain containing protein 90; Transcription Factor. | −1.70 | −4.61 ± 0.94 |
| AT3G56400 | WRKY70 | Transcription factor involved in senescence, biotic and abiotic stress responses by modulating various phytohormones signaling pathways; function as activator of SA-dependent defence genes and a repressor of JA-regulated genes; ethylene signaling pathway; BR signaling pathway. | −1.97 | −2.49 ± 0.59 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forte, V.; Lucchetti, S.; Ciolfi, A.; Felici, B.; Possenti, M.; D’Orso, F.; Morelli, G.; Baima, S. ATHB1 Interacts with Hormone-Related Gene Regulatory Networks Involved in Biotic and Abiotic Stress Responses in Arabidopsis. Cells 2025, 14, 1456. https://doi.org/10.3390/cells14181456
Forte V, Lucchetti S, Ciolfi A, Felici B, Possenti M, D’Orso F, Morelli G, Baima S. ATHB1 Interacts with Hormone-Related Gene Regulatory Networks Involved in Biotic and Abiotic Stress Responses in Arabidopsis. Cells. 2025; 14(18):1456. https://doi.org/10.3390/cells14181456
Chicago/Turabian StyleForte, Valentina, Sabrina Lucchetti, Andrea Ciolfi, Barbara Felici, Marco Possenti, Fabio D’Orso, Giorgio Morelli, and Simona Baima. 2025. "ATHB1 Interacts with Hormone-Related Gene Regulatory Networks Involved in Biotic and Abiotic Stress Responses in Arabidopsis" Cells 14, no. 18: 1456. https://doi.org/10.3390/cells14181456
APA StyleForte, V., Lucchetti, S., Ciolfi, A., Felici, B., Possenti, M., D’Orso, F., Morelli, G., & Baima, S. (2025). ATHB1 Interacts with Hormone-Related Gene Regulatory Networks Involved in Biotic and Abiotic Stress Responses in Arabidopsis. Cells, 14(18), 1456. https://doi.org/10.3390/cells14181456






