The Colonic Crypt: Cellular Dynamics and Signaling Pathways in Homeostasis and Cancer
Abstract
1. Introduction
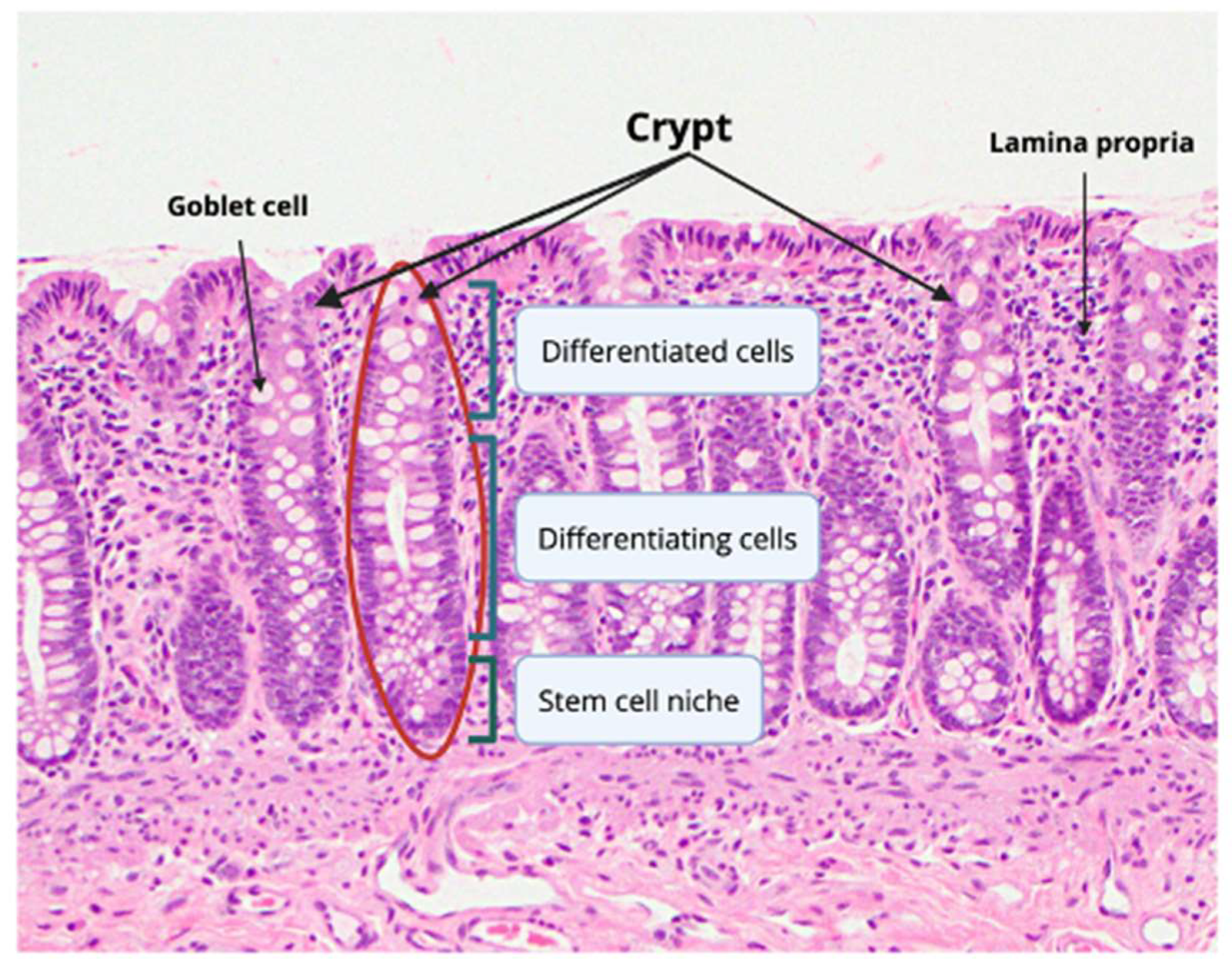
2. Normal Colonic Epithelial Cell Composition
2.1. Colonic Cells and Transit-Amplifying Cells
2.2. Colonocytes and Goblet Cells
2.3. Paneth-Like Cells
2.4. M Cells
2.5. Tuft Cells
2.6. Enteroendocrine Cells
3. Disruption of Colonic Epithelium in CRC
3.1. Stem Cells During Tumor Progression
| Cell Type | Marker | References |
|---|---|---|
| Mesenchymal cells | CD34, gp38 (podoplanin), αSMA–/low | [63] |
| Paneth-like cells | Lyz *, CD24, MMP7, REG4 | [20,21] |
| Goblet cells | Atoh1, Fcgbp, Clca1, Muc2 * | [17] |
| Stem cells | Lgr5 *, Ascl2, Smoc2, Olfm4, Bmi1 | [8,9,20] |
| Enteroendocrine cells | SYP *, ChgA | [48] |
| Tuft cells | DCKL1 *; POU2AF2 | [40,48] |
| M cells | GP2 *, PGLYRP1 | [37,38] |
| Transit Amplifying cells | MKI67 * (Ki-67) | [11] |
3.2. Transit Amplifying Cells During Tumor Progression
3.3. Goblet Cells During Tumor Progression
3.4. Paneth-Like Cells During Tumor Progression
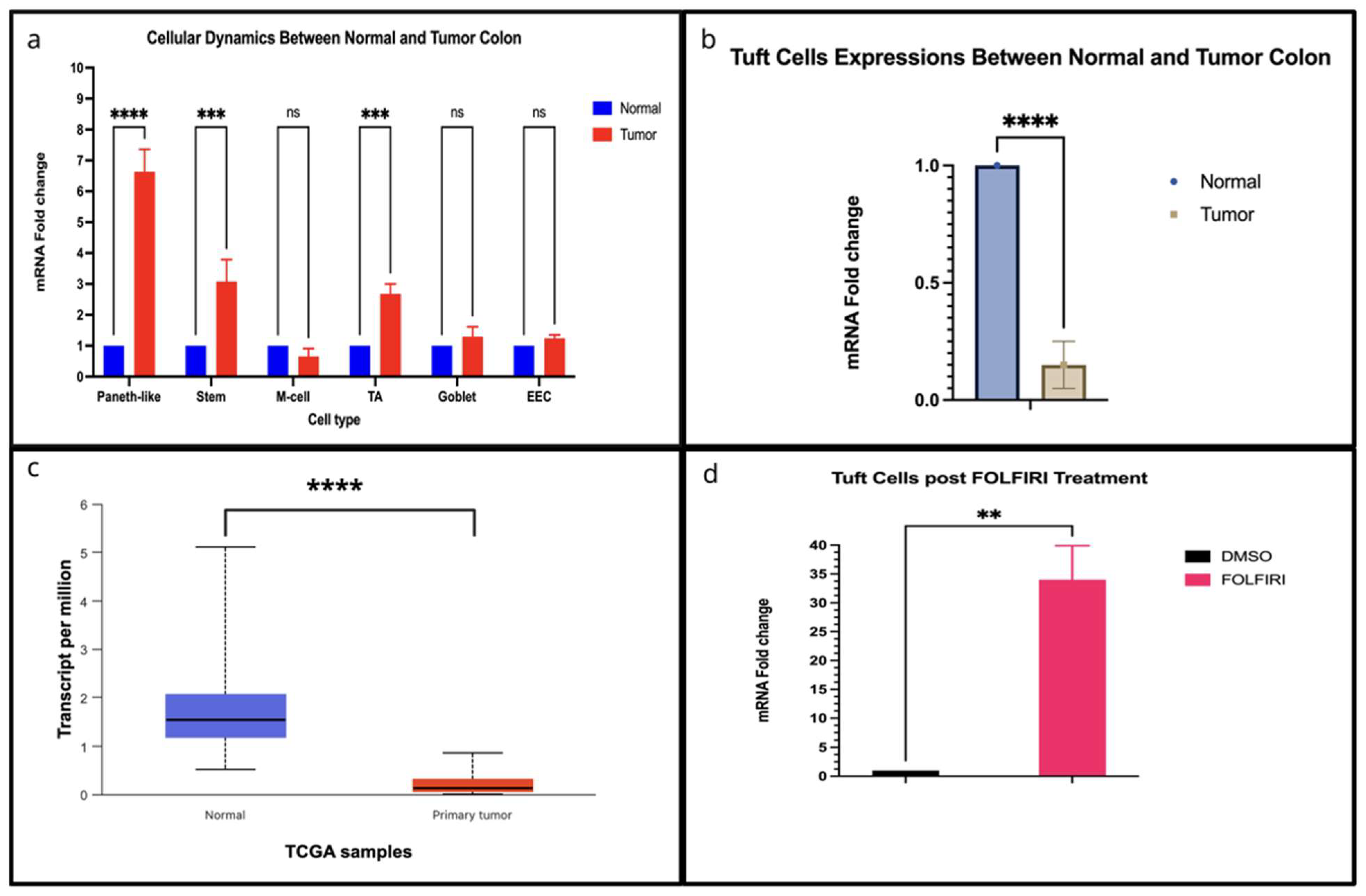
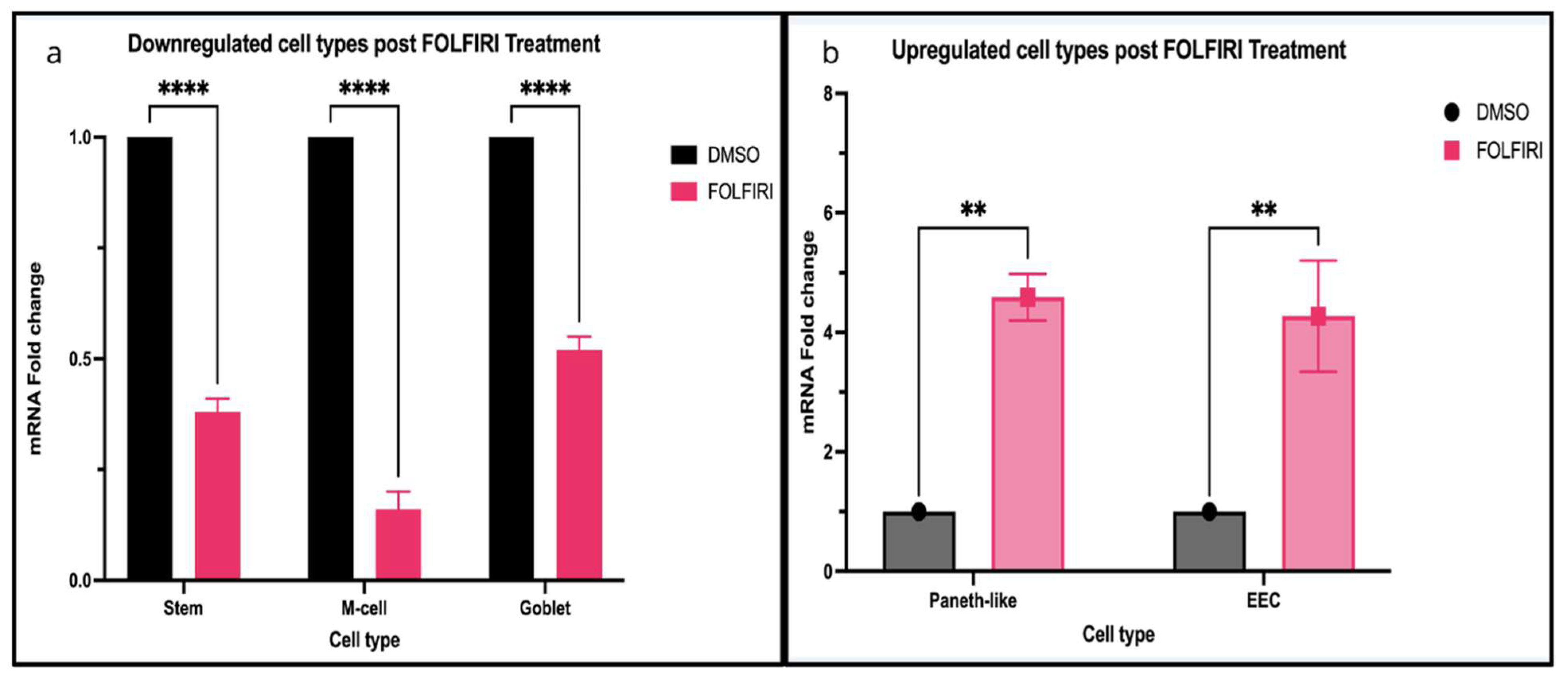
3.5. M Cells and Tuft Cells During Tumor Progression
3.6. EECs During Tumor Progression
4. Regulatory Signaling in Normal and Neoplastic Colonic Crypts
4.1. WNT Signaling in Normal Colon
4.2. WNT in CRC
4.3. Fibroblast Growth Factor Signaling in Normal Colon
4.4. FGF Signaling in CRC
4.5. Notch Signaling in Normal Colon
4.6. Notch Signaling in CRC
4.7. Bone Morphogenetic Protein Signaling in Normal Colon
4.8. BMP Signaling in CRC
4.9. EMT in CRC
5. Cellular Plasticity and EMT-Targeted Therapies in CRC
5.1. Cellular Plasticity in CRC
5.2. EMT-Targeted Therapies in CRC
| Sponsor and Clinical Trial Gov ID | Title of Study | Status of Clinical Trial | Summary | Stage |
|---|---|---|---|---|
| Istituto Clinico Humanitas NCT04323813 | High Levels of EMT-TFs for the Diagnosis of CRC | Unknown | An observational and diagnostic study that measures mRNA levels of genes involved in EMT in peripheral blood samples of CRC patients and healthy controls, to determine the presence of disease, its progression and risk of recurrence. | Observational and Diagnostic Study |
| Alethia Biotherapeutics NCT02412462 | Phase I Dose Escalation Study of AB-16B5 in Subjects with an Advanced Solid Malignancy | Completed | A clinical study to investigate the safety, pharmacokinetics and pharmacodynamics of AB-16B5 in patients with an advanced solid malignancy. AB-16B5 is a humanized monoclonal antibody that inhibits the activity of the secreted form of clusterin. Eligible subjects will have a disease that has been refractory to prior therapy and is unlikely to benefit from known therapies. No results posted at time of publication. | A Phase I Dose-Escalation Study to Evaluate the Safety, Tolerability and Pharmacokinetics of AB-16B5 in Subjects with an Advanced Solid Malignancy |
| Alethia Biotherapeutics NCT06225843 | Sotevtamab (AB-16B5) Combined With FOLFOX as Neoadjuvant Treatment Prior to Resection of CRC Liver Metastasis (EGIA-003) | Recruiting | This study will recruit 17 CRC patients with liver-dominant metastases. All recruited patients will receive Sotevtamab at a dose of 800 mg once weekly for 6 cycles combined with FOLFOX once every 2 weeks for the first 4 cycles followed by liver metastases resection surgery with or without primary cancer resection. AB-165 (Sotevtamab) a fully humanized monoclonal antibody of IgG2 isotype against tumor-associated secreted clusterin. | A Proof-of-Concept Phase II Trial to Evaluate the EMT Inhibitor Sotevtamab Combined with FOLFOX Administered as Neoadjuvant Treatment Prior to Resection of CRC Liver Metastasis. |
| Centre Leon Berard NCT05605496 | NP137 Clinical and Biological Activities Assessment in Patients With Advanced/Metastatic Solid Tumors Treated by Standard Anti PD-1/PD-L1 Immunotherapies (IMMUNONET) | Recruiting | This study aims to assess the clinical and biological impact of NP137 when added to standard PD-1/PD-L1 blockade with various sensitivity to anti-PD-1/PD-L1 NP137 is an antibody that inhibits netrin-1. NP137 has shown the ability to inhibit EMT. (Xia et al., 2024 [232]) (Cassier et al., 2023 [238]) | This study is an open-label, proof-of-concept study assessing the impact of NP137 when added to standard PD-1/PD-L1 blockade therapy of advanced or metastatic solid tumors with various sensitivity to anti-PD-1/PD-L1 |
| National Cancer Institute (NCI) NCT03030417 | Indenoisoquinoline LMP744 in Adults With Relapsed Solid Tumors and Lymphomas | Completed | LMP744 (NSC 706744) an inhibitor of topoisomerase 1 damages DNA. This causes cell death. Researchers want to see if it can treat certain kinds of cancer. Exploratory Objective: Evaluate the effect of LMP744 on markers of DNA damage (yH2AX, pNbs1, pATR, ERCC1, RAD51, Topo1cc, Top1, SLFN11) and EMT in circulating tumor cells and pre- and post-treatment tumor biopsies in patients in the expansion cohort. | A Phase I Study to establish the safety, tolerability and the maximum tolerated dose of LMP744 in patients with refractory solid tumors and lymphomas. |
5.3. Future for EMT-Targeted Therapies
6. Conclusions
7. Data Synthesis and Analysis
7.1. Gene Expression Analysis from TCGA-COAD
7.2. Single-Cell Expression Analysis from the Human Colon Cancer Atlas
7.3. RNA-Seq Expression Analysis from Mzoughi et al., 2025 [87]
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SC | Stem cell |
| CRC | Colorectal cancer |
| FGF | Fibroblast growth factor |
| BMP | Bone Morphogenetic Protein |
| CSCs | Cancer stem cells |
| EMT | Epithelial—Mesenchymal Transition |
| MET | Mesenchymal—Epithelial Transition |
| TA | Transit-amplifying |
| GI | Gastrointestinal |
| LGR5 | leucine-rich repeat-containing G protein-coupled receptor 5 |
| REG4 | regenerating family member 4 |
| sPLA2 | secretory phospholipase A2 |
| HD5 | human defensin 5 |
| PCM | Paneth-like cell metaplasia |
| GLP-1 | glucagon-like peptide-1 |
| PYY | peptide YY |
| EECs | Enteroendocrine cells |
| LEF1 | Lymphoid Enhancer Factor-1 |
| TCF | T-cell Factor |
| mCRC | mucinous CRC |
| TCGA | The Cancer Genome Atlas |
| FGFR | FGF receptor |
| NICD | NOTCH intracellular domain |
| LSD1 | lysine-specific demethylase 1A |
| IBD | inflammatory bowel disease |
References
- Zou, L.; Xiong, X.; Wang, K.; Yin, Y. MicroRNAs in the Intestine: Role in Renewal, Homeostasis, and Inflammation. Curr. Mol. Med. 2018, 18, 190–198. [Google Scholar] [CrossRef]
- Gehart, H.; Clevers, H. Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Kiela, P.R.; Ghishan, F.K. Physiology of Intestinal Absorption and Secretion. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 145–159. [Google Scholar] [CrossRef]
- van der Heijden, M.; Vermeulen, L. Stem cells in homeostasis and cancer of the gut. Mol. Cancer 2019, 18, 66. [Google Scholar] [CrossRef]
- Patil, A.Z.L. Anatomy & Histology. Available online: https://www.pathologyoutlines.com/topic/colonhistology.html (accessed on 31 July 2025).
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef]
- Gustafsson, J.K.; Johansson, M.E.V. The role of goblet cells and mucus in intestinal homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 785–803. [Google Scholar] [CrossRef]
- Shaker, A.; Rubin, D.C. Intestinal stem cells and epithelial–mesenchymal interactions in the crypt and stem cell niche. Transl. Res. 2010, 156, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Lee, C.; Kim, W.H.; Maeng, Y.H.; Jang, B.G. Expression profile of intestinal stem cell markers in colitis-associated carcinogenesis. Sci. Rep. 2017, 7, 6533. [Google Scholar] [CrossRef] [PubMed]
- Beumer, J.; Clevers, H. Cell fate specification and differentiation in the adult mammalian intestine. Nat. Rev. Mol. Cell Biol. 2021, 22, 39–53. [Google Scholar] [CrossRef]
- Basak, O.; van de Born, M.; Korving, J.; Beumer, J.; van der Elst, S.; van Es, J.H.; Clevers, H. Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J. 2014, 33, 2057–2068. [Google Scholar] [CrossRef]
- Wang, Y.; Song, W.; Wang, J.; Wang, T.; Xiong, X.; Qi, Z.; Fu, W.; Yang, X.; Chen, Y.-G. Single-cell transcriptome analysis reveals differential nutrient absorption functions in human intestine. J. Exp. Med. 2020, 217, e20191130. [Google Scholar] [CrossRef]
- Ordóñez-Morán, P. (Ed.) Intestinal Differentiated Cells; Springer: New York, NY, USA, 2023. [Google Scholar]
- Litvak, Y.; Byndloss, M.X.; Bäumler, A.J. Colonocyte metabolism shapes the gut microbiota. Science 2018, 362, eaat9076. [Google Scholar] [CrossRef] [PubMed]
- Raya Tonetti, F.; Eguileor, A.; Llorente, C. Goblet cells: Guardians of gut immunity and their role in gastrointestinal diseases. eGastroenterology 2024, 2, e100098. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yu, M. Role of Goblet Cells in Intestinal Barrier and Mucosal Immunity. J. Inflamm. Res. 2021, 14, 3171–3183. [Google Scholar] [CrossRef]
- Nyström, E.E.L.; Martinez-Abad, B.; Arike, L.; Birchenough, G.M.H.; Nonnecke, E.B.; Castillo, P.A.; Svensson, F.; Bevins, C.L.; Hansson, G.C.; Johansson, M.E.V. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science 2021, 372, eabb1590. [Google Scholar] [CrossRef] [PubMed]
- Coleman, O.I.; Haller, D. Microbe–Mucus Interface in the Pathogenesis of Colorectal Cancer. Cancers 2021, 13, 616. [Google Scholar] [CrossRef]
- Stolfi, C.; Maresca, C.; Monteleone, G.; Laudisi, F. Implication of Intestinal Barrier Dysfunction in Gut Dysbiosis and Diseases. Biomedicines 2022, 10, 289. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sasaki, N.; Sachs, N.; Wiebrands, K.; Ellenbroek, S.I.J.; Fumagalli, A.; Lyubimova, A.; Begthel, H.; van den Born, M.; van Es, J.H.; Karthaus, W.R.; et al. Reg4 + deep crypt secretory cells function as epithelial niche for Lgr5+ stem cells in colon. Proc. Natl. Acad. Sci. USA 2016, 113, E5399–E5407. [Google Scholar] [CrossRef]
- Clevers, H.C.; Bevins, C.L. Paneth Cells: Maestros of the Small Intestinal Crypts. Annu. Rev. Physiol. 2013, 75, 289–311. [Google Scholar] [CrossRef]
- Yu, S.; Balasubramanian, I.; Laubitz, D.; Tong, K.; Bandyopadhyay, S.; Lin, X.; Flores, J.; Singh, R.; Liu, Y.; Macazana, C.; et al. Paneth Cell-Derived Lysozyme Defines the Composition of Mucolytic Microbiota and the Inflammatory Tone of the Intestine. Immunity 2020, 53, 398–416.e8. [Google Scholar] [CrossRef]
- Abbott, A.M.; Armstrong, L.; Jensen, E.H. Small Intestine. In Shackelford’s Surgery of the Alimentary Tract; Elsevier: Amsterdam, The Netherlands, 2013; pp. 839–863. [Google Scholar]
- Ghosh, D.; Porter, E.; Shen, B.; Lee, S.K.; Wilk, D.; Drazba, J.; Yadav, S.P.; Crabb, J.W.; Ganz, T.; Bevins, C.L. Paneth cell trypsin is the processing enzyme for human defensin-5. Nat. Immunol. 2002, 3, 583–590. [Google Scholar] [CrossRef]
- Sakahara, M.; Okamoto, T.; Srivastava, U.; Natsume, Y.; Yamanaka, H.; Suzuki, Y.; Obama, K.; Nagayama, S.; Yao, R. Paneth-like cells produced from OLFM4+ stem cells support OLFM4+ stem cell growth in advanced colorectal cancer. Commun. Biol. 2024, 7, 27. [Google Scholar] [CrossRef]
- Eşrefoğlu, M. Paneth Cells: Development, Morphology, Function and Clinical Importance. Biomed. J. Sci. Tech. Res. 2024, 57, 48743–48753. [Google Scholar] [CrossRef]
- Singh, R.; Balasubramanian, I.; Zhang, L.; Gao, N. Metaplastic Paneth Cells in Extra-Intestinal Mucosal Niche Indicate a Link to Microbiome and Inflammation. Front. Physiol. 2020, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Wallaeys, C.; Garcia-Gonzalez, N.; Libert, C. Paneth cells as the cornerstones of intestinal and organismal health: A primer. EMBO Mol. Med. 2023, 15, e16427. [Google Scholar] [CrossRef]
- Lueschow, S.R.; McElroy, S.J. The Paneth Cell: The Curator and Defender of the Immature Small Intestine. Front. Immunol. 2020, 11, 587. [Google Scholar] [CrossRef]
- Simmonds, N.; Furman, M.; Karanika, E.; Phillips, A.; Bates, A.W. Paneth cell metaplasia in newly diagnosed inflammatory bowel disease in children. BMC Gastroenterol. 2014, 14, 93. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, M.A. The emerging roles of deep crypt secretory cells in colonic physiology. Am. J. Physiol. Gastrointest. Liver Physiol. 2023, 325, G493–G500. [Google Scholar] [CrossRef]
- Corr, S.C.; Gahan, C.C.G.M.; Hill, C. M-cells: Origin, morphology and role in mucosal immunity and microbial pathogenesis. FEMS Immunol. Med. Microbiol. 2008, 52, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, N.; Takahashi, D.; Takano, S.; Kimura, S.; Hase, K. The Roles of Peyer’s Patches and Microfold Cells in the Gut Immune System: Relevance to Autoimmune Diseases. Front. Immunol. 2019, 10, 2345. [Google Scholar] [CrossRef]
- Ahmed, M.; Ahmed, S. Functional, Diagnostic and Therapeutic Aspects of Gastrointestinal Hormones. Gastroenterol. Res. 2019, 12, 233–244. [Google Scholar] [CrossRef]
- Bennett, K.M.; Parnell, E.A.; Sanscartier, C.; Parks, S.; Chen, G.; Nair, M.G.; Lo, D.D. Induction of Colonic M Cells during Intestinal Inflammation. Am. J. Pathol. 2016, 186, 1166–1179. [Google Scholar] [CrossRef]
- Dillon, A.; Lo, D.D. M Cells: Intelligent Engineering of Mucosal Immune Surveillance. Front. Immunol. 2019, 10, 1499. [Google Scholar] [CrossRef] [PubMed]
- Kanaya, T.; Williams, I.R.; Ohno, H. Intestinal M cells: Tireless samplers of enteric microbiota. Traffic 2020, 21, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Mabbott, N.A.; Donaldson, D.S.; Ohno, H.; Williams, I.R.; Mahajan, A. Microfold (M) cells: Important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013, 6, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tanaka, I.; Nakahashi-Ouchida, R.; Ernst, P.B.; Kiyono, H.; Kurashima, Y. Glycoprotein 2 as a gut gate keeper for mucosal equilibrium between inflammation and immunity. Semin. Immunopathol. 2024, 45, 493–507. [Google Scholar] [CrossRef]
- Hendel, S.K.; Kellermann, L.; Hausmann, A.; Bindslev, N.; Jensen, K.B.; Nielsen, O.H. Tuft Cells and Their Role in Intestinal Diseases. Front. Immunol. 2022, 13, 822867. [Google Scholar] [CrossRef]
- Gehart, H.; van Es, J.H.; Hamer, K.; Beumer, J.; Kretzschmar, K.; Dekkers, J.F.; Rios, A.; Clevers, H. Identification of Enteroendocrine Regulators by Real-Time Single-Cell Differentiation Mapping. Cell 2019, 176, 1158–1173.e16. [Google Scholar] [CrossRef]
- Gerbe, F.; Jay, P. Intestinal tuft cells: Epithelial sentinels linking luminal cues to the immune system. Mucosal Immunol. 2016, 9, 1353–1359. [Google Scholar] [CrossRef]
- Campillo Poveda, M.; Britton, C.; Devaney, E.; McNeilly, T.N.; Gerbe, F.; Jay, P.; Maizels, R.M. Tuft Cells: Detectors, Amplifiers, Effectors and Targets in Parasite Infection. Cells 2023, 12, 2477. [Google Scholar] [CrossRef]
- Schneider, C. Tuft cell integration of luminal states and interaction modules in tissues. Pflugers Arch. 2021, 473, 1713–1722. [Google Scholar] [CrossRef] [PubMed]
- Billipp, T.E.; Nadjsombati, M.S.; von Moltke, J. Tuning tuft cells: New ligands and effector functions reveal tissue-specific function. Curr. Opin. Immunol. 2021, 68, 98–106. [Google Scholar] [CrossRef]
- McGinty, J.W.; Ting, H.-A.; Billipp, T.E.; Nadjsombati, M.S.; Khan, D.M.; Barrett, N.A.; Liang, H.-E.; Matsumoto, I.; von Moltke, J. Tuft-Cell-Derived Leukotrienes Drive Rapid Anti-helminth Immunity in the Small Intestine but Are Dispensable for Anti-protist Immunity. Immunity 2020, 52, 528–541.e7. [Google Scholar] [CrossRef]
- Varyani, F.; Löser, S.; Filbey, K.J.; Harcus, Y.; Drurey, C.; Poveda, M.C.; Rasid, O.; White, M.P.J.; Smyth, D.J.; Gerbe, F.; et al. The IL-25-dependent tuft cell circuit driven by intestinal helminths requires macrophage migration inhibitory factor (MIF). Mucosal Immunol. 2022, 15, 1243–1256. [Google Scholar] [CrossRef] [PubMed]
- Sei, Y.; Lu, X.; Liou, A.; Zhao, X.; Wank, S.A. A stem cell marker-expressing subset of enteroendocrine cells resides at the crypt base in the small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G345–G356. [Google Scholar] [CrossRef]
- Chhetri, D.; Vengadassalapathy, S.; Venkadassalapathy, S.; Balachandran, V.; Umapathy, V.R.; Veeraraghavan, V.P.; Jayaraman, S.; Patil, S.; Iyaswamy, A.; Palaniyandi, K.; et al. Pleiotropic effects of DCLK1 in cancer and cancer stem cells. Front. Mol. Biosci. 2022, 9, 965730. [Google Scholar] [CrossRef] [PubMed]
- Worthington, J.J.; Reimann, F.; Gribble, F.M. Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 2018, 11, 3–20. [Google Scholar] [CrossRef]
- Atanga, R.; Singh, V.; In, J.G. Intestinal Enteroendocrine Cells: Present and Future Druggable Targets. Int. J. Mol. Sci. 2023, 24, 8836. [Google Scholar] [CrossRef]
- Gunawardene, A.R.; Corfe, B.M.; Staton, C.A. Classification and functions of enteroendocrine cells of the lower gastrointestinal tract. Int. J. Exp. Pathol. 2011, 92, 219–231. [Google Scholar] [CrossRef]
- Sailaja, B.S.; He, X.C.; Li, L. The regulatory niche of intestinal stem cells. J. Physiol. 2016, 594, 4827–4836. [Google Scholar] [CrossRef]
- Radford, I.R.; Lobachevsky, P.N. An enteroendocrine cell-based model for a quiescent intestinal stem cell niche. Cell Prolif. 2006, 39, 403–414. [Google Scholar] [CrossRef]
- D’amico, M.A.; Ghinassi, B.; Izzicupo, P.; Manzoli, L.; Di Baldassarre, A. Biological function and clinical relevance of chromogranin A and derived peptides. Endocr. Connect. 2014, 3, R45–R54. [Google Scholar] [CrossRef]
- Beumer, J.; Gehart, H.; Clevers, H. Enteroendocrine Dynamics—New Tools Reveal Hormonal Plasticity in the Gut. Endocr. Rev. 2020, 41, bnaa018. [Google Scholar] [CrossRef]
- Haber, A.L.; Biton, M.; Rogel, N.; Herbst, R.H.; Shekhar, K.; Smillie, C.; Burgin, G.; Delorey, T.M.; Howitt, M.R.; Katz, Y.; et al. A single-cell survey of the small intestinal epithelium. Nature 2017, 551, 333–339. [Google Scholar] [CrossRef]
- Cho, H.; Lim, J. The emerging role of gut hormones. Mol. Cells 2024, 47, 100126. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Kaye, J.A.; Douglas, E.R.; Joshi, N.R.; Gribble, F.M.; Reimann, F.; Liberles, S.D. Enteroendocrine cell lineages that differentially control feeding and gut motility. elife 2023, 12, e78512. [Google Scholar] [CrossRef]
- Nwako, J.G.; McCauley, H.A. Enteroendocrine cells regulate intestinal homeostasis and epithelial function. Mol. Cell. Endocrinol. 2024, 593, 112339. [Google Scholar] [CrossRef]
- Barton, J.R.; Londregan, A.K.; Alexander, T.D.; Entezari, A.A.; Covarrubias, M.; Waldman, S.A. Enteroendocrine cell regulation of the gut-brain axis. Front. Neurosci. 2023, 17, 1272955. [Google Scholar] [CrossRef] [PubMed]
- Sesotyosari, S.L.; Kinoshita, M.; Sunardi, M.; Lihan, M.; Orii, A.; Abe, T.; Kiyonari, H.; Nakai, T.; Uesaka, T.; Kodama, Y.; et al. The long-term survival of enteroendocrine cells depends on their subtype and is linked to peripheral sensory innervation. Dev. Growth Differ. 2025, 67, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Stzepourginski, I.; Nigro, G.; Jacob, J.-M.; Dulauroy, S.; Sansonetti, P.J.; Eberl, G.; Peduto, L. CD34 + mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. USA 2017, 114, E506–E513. [Google Scholar] [CrossRef]
- Clevers, H. The Intestinal Crypt, A Prototype Stem Cell Compartment. Cell 2013, 154, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Heppert, J.K.; Davison, J.M.; Kelly, C.; Mercado, G.P.; Lickwar, C.R.; Rawls, J.F. Transcriptional programmes underlying cellular identity and microbial responsiveness in the intestinal epithelium. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 7–23. [Google Scholar] [CrossRef]
- Choi, J.; Augenlicht, L.H. Intestinal stem cells: Guardians of homeostasis in health and aging amid environmental challenges. Exp. Mol. Med. 2024, 56, 495–500. [Google Scholar] [CrossRef]
- Lo, Y.-H.; Chung, E.; Li, Z.; Wan, Y.-W.; Mahe, M.M.; Chen, M.-S.; Noah, T.K.; Bell, K.N.; Yalamanchili, H.K.; Klisch, T.J.; et al. Transcriptional Regulation by ATOH1 and its Target SPDEF in the Intestine. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 51–71. [Google Scholar] [CrossRef]
- Billingsley, J.L.; Yevdokimova, V.; Ayoub, K.; Benoit, Y.D. Colorectal Cancer Is Borrowing Blueprints from Intestinal Ontogenesis. Cancers 2023, 15, 4928. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Xu, Y.; Gao, Z.; Shi, J.; Wang, Y.; Wang, X.; Huang, W.; Li, M.; Wang, L.; Xu, J.; et al. A patient-derived organoid model captures fetal-like plasticity in colorectal cancer. Cell Res. 2025, 35, 642–655. [Google Scholar] [CrossRef]
- Moorman, A.; Benitez, E.K.; Cambulli, F.; Jiang, Q.; Mahmoud, A.; Lumish, M.; Hartner, S.; Balkaran, S.; Bermeo, J.; Asawa, S.; et al. Progressive plasticity during colorectal cancer metastasis. Nature 2025, 637, 947–954. [Google Scholar] [CrossRef]
- Lei, K.F.; Ho, Y.-C.; Huang, C.-H.; Huang, C.-H.; Pai, P.C. Characterization of stem cell-like property in cancer cells based on single-cell impedance measurement in a microfluidic platform. Talanta 2021, 229, 122259. [Google Scholar] [CrossRef]
- Posey, T.A.; Jacob, J.; Parkhurst, A.N.; Subramanian, S.; Francisco, L.E.; Liang, Z.; Carmon, K.S. Loss of LGR5 through plasticity or gene ablation is associated with therapy resistance and enhanced MET-STAT3 signaling in colorectal cancer cells. BioRxiv 2023. [Google Scholar] [CrossRef]
- Yeung, T.M.; Gandhi, S.C.; Bodmer, W.F. Hypoxia and lineage specification of cell line-derived colorectal cancer stem cells. Proc. Natl. Acad. Sci. USA 2011, 108, 4382–4387. [Google Scholar] [CrossRef] [PubMed]
- Korkaya, H.; Liu, S.; Wicha, M.S. Regulation of Cancer Stem Cells by Cytokine Networks: Attacking Cancer’s Inflammatory Roots. Clin. Cancer Res. 2011, 17, 6125–6129. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, Y.-G. Intestinal epithelial plasticity and regeneration via cell dedifferentiation. Cell Regen. 2020, 9, 14. [Google Scholar] [CrossRef]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Sato, T. Defining the role of Lgr5+ stem cells in colorectal cancer: From basic research to clinical applications. Genome Med. 2017, 9, 66. [Google Scholar] [CrossRef]
- Santos, A.J.M.; Lo, Y.-H.; Mah, A.T.; Kuo, C.J. The Intestinal Stem Cell Niche: Homeostasis and Adaptations. Trends Cell Biol. 2018, 28, 1062–1078. [Google Scholar] [CrossRef]
- Jang, B.G.; Kim, H.S.; Chang, W.Y.; Bae, J.M.; Kim, W.H.; Kang, G.H. Expression Profile of LGR5 and Its Prognostic Significance in Colorectal Cancer Progression. Am. J. Pathol. 2018, 188, 2236–2250. [Google Scholar] [CrossRef]
- Morgan, R.; Mortensson, E.; Williams, A. Targeting LGR5 in Colorectal Cancer: Therapeutic gold or too plastic? Br. J. Cancer 2018, 118, 1410–1418. [Google Scholar] [CrossRef]
- Wang, W.; Lokman, N.A.; Barry, S.C.; Oehler, M.K.; Ricciardelli, C. LGR5: An emerging therapeutic target for cancer metastasis and chemotherapy resistance. Cancer Metastasis Rev. 2025, 44, 23. [Google Scholar] [CrossRef]
- Hirsch, D.; Barker, N.; McNeil, N.; Hu, Y.; Camps, J.; McKinnon, K.; Clevers, H.; Ried, T.; Gaiser, T. LGR5 positivity defines stem-like cells in colorectal cancer. Carcinogenesis 2014, 35, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Gensbittel, V.; Kräter, M.; Harlepp, S.; Busnelli, I.; Guck, J.; Goetz, J.G. Mechanical Adaptability of Tumor Cells in Metastasis. Dev. Cell 2021, 56, 164–179. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Raghunathan, V.; Stamer, W.D.; Ganapathy, P.S.; Herberg, S. Extracellular Matrix Stiffness and TGFβ2 Regulate YAP/TAZ Activity in Human Trabecular Meshwork Cells. Front. Cell Dev. Biol. 2022, 10, 844342. [Google Scholar] [CrossRef]
- Human Colon Cancer Atlas. Available online: https://singlecell.broadinstitute.org/ (accessed on 31 July 2025).
- The Cancer Genome Atlas. Available online: https://portal.gdc.cancer.gov/ (accessed on 31 July 2025).
- Mzoughi, S.; Schwarz, M.; Wang, X.; Demircioglu, D.; Ulukaya, G.; Mohammed, K.; Zorgati, H.; Torre, D.; Tomalin, L.E.; Di Tullio, F.; et al. Oncofetal reprogramming drives phenotypic plasticity in WNT-dependent colorectal cancer. Nat. Genet. 2025, 57, 402–412. [Google Scholar] [CrossRef]
- Marles, H.; Biddle, A. Cancer stem cell plasticity and its implications in the sdevelopment of new clinical approaches for oral squamous cell carcinoma. Biochem. Pharmacol. 2022, 204, 115212. [Google Scholar] [CrossRef] [PubMed]
- Posey, T.A.; Jacob, J.; Parkhurst, A.; Subramanian, S.; Francisco, L.E.; Liang, Z.; Carmon, K.S. Loss of LGR5 through Therapy-induced Downregulation or Gene Ablation Is Associated with Resistance and Enhanced MET-STAT3 Signaling in Colorectal Cancer Cells. Mol. Cancer Ther. 2023, 22, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Asfaha, S.; Hayakawa, Y.; Muley, A.; Stokes, S.; Graham, T.A.; Ericksen, R.E.; Westphalen, C.B.; von Burstin, J.; Mastracci, T.L.; Worthley, D.L.; et al. Krt19+/Lgr5− Cells Are Radioresistant Cancer-Initiating Stem Cells in the Colon and Intestine. Cell Stem Cell 2015, 16, 627–638. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kang, D.H.; Song, G.J.; Ahn, T.S.; Son, M.W.; Lee, M.S.; Baek, M.-J. Clinical relevance of Lgr5 expression in colorectal cancer patients. Korean J. Clin. Oncol. 2018, 14, 76–82. [Google Scholar] [CrossRef]
- de Sousa e Melo, F.; Kurtova, A.V.; Harnoss, J.M.; Kljavin, N.; Hoeck, J.D.; Hung, J.; Anderson, J.E.; Storm, E.E.; Modrusan, Z.; Koeppen, H.; et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature 2017, 543, 676–680. [Google Scholar] [CrossRef]
- Zhang, C.-L.; Huang, T.; Wu, B.-L.; He, W.-X.; Liu, D. Stem cells in cancer therapy: Opportunities and challenges. Oncotarget 2017, 8, 75756–75766. [Google Scholar] [CrossRef]
- Testa, U.; Pelosi, E.; Castelli, G. Colorectal Cancer: Genetic Abnormalities, Tumor Progression, Tumor Heterogeneity, Clonal Evolution and Tumor-Initiating Cells. Med. Sci. 2018, 6, 31. [Google Scholar] [CrossRef]
- Jaladanki, R.N.; Wang, J.-Y. Regulation of Gastrointestinal Mucosal Growth. Colloq. Ser. Integr. Syst. Physiol. Mol. Funct. 2011, 3, 1–114. [Google Scholar] [CrossRef]
- Svec, J.; Onhajzer, J.; Korinek, V. Origin, development and therapy of colorectal cancer from the perspective of a biologist and an oncologist. Crit. Rev. Oncol. Hematol. 2024, 204, 104544. [Google Scholar] [CrossRef]
- Li, Q.; Geng, S.; Luo, H.; Wang, W.; Mo, Y.-Q.; Luo, Q.; Wang, L.; Song, G.-B.; Sheng, J.-P.; Xu, B. Signaling pathways involved in colorectal cancer: Pathogenesis and targeted therapy. Signal Transduct. Target. Ther. 2024, 9, 266. [Google Scholar] [CrossRef]
- Zhao, H.; Ming, T.; Tang, S.; Ren, S.; Yang, H.; Liu, M.; Tao, Q.; Xu, H. Wnt signaling in colorectal cancer: Pathogenic role and therapeutic target. Mol. Cancer 2022, 21, 144. [Google Scholar] [CrossRef]
- Loh, J.-J.; Ma, S. Hallmarks of cancer stemness. Cell Stem Cell 2024, 31, 617–639. [Google Scholar] [CrossRef]
- Becker, W.R.; Nevins, S.A.; Chen, D.C.; Chiu, R.; Horning, A.M.; Guha, T.K.; Laquindanum, R.; Mills, M.; Chaib, H.; Ladabaum, U.; et al. Single-cell analyses define a continuum of cell state and composition changes in the malignant transformation of polyps to colorectal cancer. Nat. Genet. 2022, 54, 985–995. [Google Scholar] [CrossRef]
- Suvà, M.L.; Riggi, N.; Bernstein, B.E. Epigenetic Reprogramming in Cancer. Science 2013, 339, 1567–1570. [Google Scholar] [CrossRef]
- Parmar, S.; Easwaran, H. Genetic and epigenetic dependencies in colorectal cancer development. Gastroenterol. Rep. 2022, 10, goac035. [Google Scholar] [CrossRef]
- Hammoud, S.S.; Cairns, B.R.; Jones, D.A. Epigenetic regulation of colon cancer and intestinal stem cells. Curr. Opin. Cell Biol. 2013, 25, 177–183. [Google Scholar] [CrossRef]
- Sun, J.; Chen, Y.; Xu, Z.; Wang, W.; Li, P. Notch signaling in the tumor immune microenvironment of colorectal cancer: Mechanisms and therapeutic opportunities. J. Transl. Med. 2025, 23, 315. [Google Scholar] [CrossRef]
- Burotto, M.; Chiou, V.L.; Lee, J.; Kohn, E.C. The MAPK pathway across different malignancies: A new perspective. Cancer 2014, 120, 3446–3456. [Google Scholar] [CrossRef]
- Maharati, A.; Moghbeli, M. PI3K/AKT signaling pathway as a critical regulator of epithelial-mesenchymal transition in colorectal tumor cells. Cell Commun. Signal. 2023, 21, 201. [Google Scholar] [CrossRef]
- Abdullayeva, G.; Liu, H.; Liu, T.-C.; Simmons, A.; Novelli, M.; Huseynova, I.; Lastun, V.L.; Bodmer, W. Goblet cell differentiation subgroups in colorectal cancer. Proc. Natl. Acad. Sci. USA 2024, 121, e2414213121. [Google Scholar] [CrossRef]
- Miller, S.A.; Ghobashi, A.H.; O’Hagan, H.M. Consensus molecular subtyping of colorectal cancers is influenced by goblet cell content. Cancer Genet. 2021, 254–255, 34–39. [Google Scholar] [CrossRef]
- Braga Emidio, N.; Brierley, S.M.; Schroeder, C.I.; Muttenthaler, M. Structure, Function, and Therapeutic Potential of the Trefoil Factor Family in the Gastrointestinal Tract. ACS Pharmacol. Transl. Sci. 2020, 3, 583–597. [Google Scholar] [CrossRef]
- Aihara, E.; Engevik, K.A.; Montrose, M.H. Trefoil Factor Peptides and Gastrointestinal Function. Annu. Rev. Physiol. 2017, 79, 357–380. [Google Scholar] [CrossRef]
- Larue, A.E.M.; Atlasi, Y. The epigenetic landscape in intestinal stem cells and its deregulation in colorectal cancer. Stem Cells 2024, 42, 509–525. [Google Scholar] [CrossRef]
- Tahghighi, A.; Seyedhashemi, E.; Mohammadi, J.; Moradi, A.; Esmaeili, A.; Pornour, M.; Jafarifar, K.; Ganji, S.M. Epigenetic marvels: Exploring the landscape of colorectal cancer treatment through cutting-edge epigenetic-based drug strategies. Clin. Epigenet. 2025, 17, 34. [Google Scholar] [CrossRef]
- Nakamura, Y.; Hamajima, Y.; Komori, M.; Yokota, M.; Suzuki, M.; Lin, J. The Role of Atoh1 in Mucous Cell Metaplasia. Int. J. Otolaryngol. 2012, 2012, 438609. [Google Scholar] [CrossRef]
- Jung, G.; Hernández-Illán, E.; Moreira, L.; Balaguer, F.; Goel, A. Epigenetics of colorectal cancer: Biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 111–130. [Google Scholar] [CrossRef]
- Hu, F.-J.; Li, Y.-J.; Zhang, L.; Ji, D.-B.; Liu, X.-Z.; Chen, Y.-J.; Wang, L.; Wu, A.-W. Single-cell profiling reveals differences between human classical adenocarcinoma and mucinous adenocarcinoma. Commun. Biol. 2023, 6, 85. [Google Scholar] [CrossRef]
- Schellnegger, R.; Quante, A.; Rospleszcz, S.; Schernhammer, M.; Höhl, B.; Tobiasch, M.; Pastula, A.; Brandtner, A.; Abrams, J.A.; Strauch, K.; et al. Goblet Cell Ratio in Combination with Differentiation and Stem Cell Markers in Barrett Esophagus Allow Distinction of Patients with and without Esophageal Adenocarcinoma. Cancer Prev. Res. 2017, 10, 55–66. [Google Scholar] [CrossRef]
- Luo, C.; Cen, S.; Ding, G.; Wu, W. Mucinous colorectal adenocarcinoma: Clinical pathology and treatment options. Cancer Commun. 2019, 39, 1–13. [Google Scholar] [CrossRef]
- Zhou, Q.; Lei, L.; Cheng, J.; Chen, J.; Du, Y.; Zhang, X.; Li, Q.; Li, C.; Deng, H.; Wong, C.C.; et al. Microbiota-induced S100A11-RAGE axis underlies immune evasion in right-sided colon adenomas and is a therapeutic target to boost anti-PD1 efficacy. Gut 2025, 74, 214–228. [Google Scholar] [CrossRef]
- López-Arribillaga, E.; Yan, B.; Lobo-Jarne, T.; Guillén, Y.; Menéndez, S.; Andreu, M.; Bigas, A.; Iglesias, M.; Espinosa, L. Accumulation of Paneth Cells in Early Colorectal Adenomas Is Associated with Beta-Catenin Signaling and Poor Patient Prognosis. Cells 2021, 10, 2928. [Google Scholar] [CrossRef]
- He, K.; Gan, W.-J. Wnt/β-Catenin Signaling Pathway in the Development and Progression of Colorectal Cancer. Cancer Manag. Res. 2023, 15, 435–448. [Google Scholar] [CrossRef]
- Bartley, A.N.; Parikh, N.; Hsu, C.-H.; Roe, D.J.; Buckmeier, J.A.; Corley, L.; Phipps, R.A.; Gallick, G.; Lance, P.; Thompson, P.A.; et al. Colorectal Adenoma Stem-like Cell Populations: Associations with Adenoma Characteristics and Metachronous Colorectal Neoplasia. Cancer Prev. Res. 2013, 6, 1162–1170. [Google Scholar] [CrossRef]
- Papadakos, S.P.; Petrogiannopoulos, L.; Pergaris, A.; Theocharis, S. The EPH/Ephrin System in Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 2761. [Google Scholar] [CrossRef]
- Verhagen, M.P.; Joosten, R.; Schmitt, M.; Välimäki, N.; Sacchetti, A.; Rajamäki, K.; Choi, J.; Procopio, P.; Silva, S.; van der Steen, B.; et al. Non-stem cell lineages as an alternative origin of intestinal tumorigenesis in the context of inflammation. Nat. Genet. 2024, 56, 1456–1467. [Google Scholar] [CrossRef]
- Chua, J.; Gregorieff, A.; Ayyaz, A. Bottom-up or Top-down: Inflammation Reprograms Paneth Cells to Develop Bowel Cancers. Cancer Res. 2024, 84, 3324–3326. [Google Scholar] [CrossRef]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef]
- Chen, Q.; Suzuki, K.; Sifuentes-Dominguez, L.; Miyata, N.; Song, J.; Lopez, A.; Starokadomskyy, P.; Gopal, P.; Dozmorov, I.; Tan, S.; et al. Paneth cell–derived growth factors support tumorigenesis in the small intestine. Life Sci. Alliance 2021, 4, e202000934. [Google Scholar] [CrossRef]
- Janeckova, L.; Stastna, M.; Hrckulak, D.; Berkova, L.; Kubovciak, J.; Onhajzer, J.; Kriz, V.; Dostalikova, S.; Mullerova, T.; Vecerkova, K.; et al. Tcf4 regulates secretory cell fate decisions in the small intestine and colon tumors: Insights from transcriptomic, histological, and microbiome analyses. Stem. Cell Res. Ther. 2025, 16, 170. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.; Zhou, X.; Chen, H.; Song, S.; Deng, L.; Yao, Y.; Yin, X. A tunable human intestinal organoid system achieves controlled balance between self-renewal and differentiation. Nat. Commun. 2025, 16, 315. [Google Scholar] [CrossRef]
- Shin, J.H.; Park, J.; Lim, J.; Jeong, J.; Dinesh, R.K.; Maher, S.E.; Kim, J.; Park, S.; Hong, J.Y.; Wysolmerski, J.; et al. Metastasis of colon cancer requires Dickkopf-2 to generate cancer cells with Paneth cell properties. elife 2024, 13, RP97279. [Google Scholar] [CrossRef]
- Pflügler, S.; Svinka, J.; Scharf, I.; Crncec, I.; Filipits, M.; Charoentong, P.; Tschurtschenthaler, M.; Kenner, L.; Awad, M.; Stift, J.; et al. IDO1+ Paneth cells promote immune escape of colorectal cancer. Commun. Biol. 2020, 3, 252. [Google Scholar] [CrossRef]
- Bhat, A.A.; Nisar, S.; Singh, M.; Ashraf, B.; Masoodi, T.; Prasad, C.P.; Sharma, A.; Maacha, S.; Karedath, T.; Hashem, S.; et al. Cytokine- and chemokine-induced inflammatory colorectal tumor microenvironment: Emerging avenue for targeted therapy. Cancer Commun. 2022, 42, 689–715. [Google Scholar] [CrossRef]
- Molfetta, R.; Paolini, R. The Controversial Role of Intestinal Mast Cells in Colon Cancer. Cells 2023, 12, 459. [Google Scholar] [CrossRef]
- Yi, J.; Bergstrom, K.; Fu, J.; Shan, X.; McDaniel, J.M.; McGee, S.; Qu, D.; Houchen, C.W.; Liu, X.; Xia, L. Dclk1 in tuft cells promotes inflammation-driven epithelial restitution and mitigates chronic colitis. Cell Death Differ. 2019, 26, 1656–1669. [Google Scholar] [CrossRef]
- Cao, Z.; Weygant, N.; Chandrakesan, P.; Houchen, C.W.; Peng, J.; Qu, D. Tuft and Cancer Stem Cell Marker DCLK1: A New Target to Enhance Anti-Tumor Immunity in the Tumor Microenvironment. Cancers 2020, 12, 3801. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, M.; Duan, T.; Sui, X. The critical roles and therapeutic implications of tuft cells in cancer. Front. Pharmacol. 2022, 13, 1047188. [Google Scholar] [CrossRef] [PubMed]
- Harris, B.T.; Rajasekaran, V.; Blackmur, J.P.; O’Callaghan, A.; Donnelly, K.; Timofeeva, M.; Vaughan-Shaw, P.G.; Din, F.V.N.; Dunlop, M.G.; Farrington, S.M. Transcriptional dynamics of colorectal cancer risk associated variation at 11q23.1 correlate with tuft cell abundance and marker expression in silico. Sci. Rep. 2022, 12, 13609. [Google Scholar] [CrossRef]
- Sipos, F.; Műzes, G. Colonic Tuft Cells: The Less-Recognized Therapeutic Targets in Inflammatory Bowel Disease and Colorectal Cancer. Int. J. Mol. Sci. 2024, 25, 6209. [Google Scholar] [CrossRef]
- Rajasekaran, V.; Harris, B.T.; Osborn, R.T.; Smillie, C.; Donnelly, K.; Bacou, M.; Esiri-Bloom, E.; Ooi, L.-Y.; Allan, M.; Walker, M.; et al. Genetic variation at 11q23.1 confers colorectal cancer risk by dysregulation of colonic tuft cell transcriptional activator POU2AF2. Gut 2025, 74, 787–803. [Google Scholar] [CrossRef]
- Wang, Y.; Giel-Moloney, M.; Rindi, G.; Leiter, A.B. Enteroendocrine precursors differentiate independently of Wnt and form serotonin expressing adenomas in response to active β-catenin. Proc. Natl. Acad. Sci. USA 2007, 104, 11328–11333. [Google Scholar] [CrossRef] [PubMed]
- Upchurch, B.H.; Fung, B.P.; Rindi, G.; Ronco, A.; Leiter, A.B. Peptide YY expression is an early event in colonic endocrine cell differentiation: Evidence from normal and transgenic mice. Development 1996, 122, 1157–1163. [Google Scholar] [CrossRef]
- Hunt, J.E.; Holst, J.J.; Jeppesen, P.B.; Kissow, H. GLP-1 and Intestinal Diseases. Biomedicines 2021, 9, 383. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Qu, T.; Yang, J.; Dai, Q. Serotonin acts through YAP to promote cell proliferation: Mechanism and implication in colorectal cancer progression. Cell Commun. Signal. 2023, 21, 75. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Lai, Y.; Liu, C.; Teng, L.; Zhu, Y.; Lin, X.; Fu, X.; Lai, Q.; Liu, S.; Zhou, X.; et al. Comprehensively prognostic and immunological analyses of GLP-1 signaling-related genes in pan-cancer and validation in colorectal cancer. Front. Pharmacol. 2024, 15, 1387243. [Google Scholar] [CrossRef]
- Shiraishi, D.; Fujiwara, Y.; Komohara, Y.; Mizuta, H.; Takeya, M. Glucagon-like peptide-1 (GLP-1) induces M2 polarization of human macrophages via STAT3 activation. Biochem. Biophys. Res. Commun. 2012, 425, 304–308. [Google Scholar] [CrossRef]
- Ben Nasr, M.; Usuelli, V.; Dellepiane, S.; Seelam, A.J.; Fiorentino, T.V.; D’Addio, F.; Fiorina, E.; Xu, C.; Xie, Y.; Balasubramanian, H.B.; et al. Glucagon-like peptide 1 receptor is a T cell-negative costimulatory molecule. Cell Metab. 2024, 36, 1302–1319.e12. [Google Scholar] [CrossRef]
- Gahete, M.D.; Córdoba-Chacón, J.; Hergueta-Redondo, M.; Martínez-Fuentes, A.J.; Kineman, R.D.; Moreno-Bueno, G.; Luque, R.M.; Castaño, J.P. A Novel Human Ghrelin Variant (In1-Ghrelin) and Ghrelin-O-Acyltransferase Are Overexpressed in Breast Cancer: Potential Pathophysiological Relevance. PLoS ONE 2011, 6, e23302. [Google Scholar] [CrossRef]
- Kotta, A.S.; Kelling, A.S.; Corleto, K.A.; Sun, Y.; Giles, E.D. Ghrelin and Cancer: Examining the Roles of the Ghrelin Axis in Tumor Growth and Progression. Biomolecules 2022, 12, 483. [Google Scholar] [CrossRef]
- Kasprzak, A. Role of the Ghrelin System in Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 5380. [Google Scholar] [CrossRef]
- Yu, M.; Qin, K.; Fan, J.; Zhao, G.; Zhao, P.; Zeng, W.; Chen, C.; Wang, A.; Wang, Y.; Zhong, J.; et al. The evolving roles of Wnt signaling in stem cell proliferation and differentiation, the development of human diseases, and therapeutic opportunities. Genes Dis. 2024, 11, 101026. [Google Scholar] [CrossRef] [PubMed]
- Kolev, H.M.; Kaestner, K.H. Mammalian Intestinal Development and Differentiation—The State of the Art. Cell. Mol. Gastroenterol. Hepatol. 2023, 16, 809–821. [Google Scholar] [CrossRef] [PubMed]
- Demitrack, E.S.; Samuelson, L.C. Notch regulation of gastrointestinal stem cells. J. Physiol. 2016, 594, 4791–4803. [Google Scholar] [CrossRef]
- Mossahebi-Mohammadi, M.; Quan, M.; Zhang, J.-S.; Li, X. FGF Signaling Pathway: A Key Regulator of Stem Cell Pluripotency. Front. Cell Dev. Biol. 2020, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yan, K.; Xi, Q. BMP signaling in cancer stemness and differentiation. Cell Regen. 2023, 12, 37. [Google Scholar] [CrossRef]
- Kumar, V.; Vashishta, M.; Kong, L.; Wu, X.; Lu, J.J.; Guha, C.; Dwarakanath, B.S. The Role of Notch, Hedgehog, and Wnt Signaling Pathways in the Resistance of Tumors to Anticancer Therapies. Front. Cell Dev. Biol. 2021, 9, 650772. [Google Scholar] [CrossRef]
- Song, P.; Gao, Z.; Bao, Y.; Chen, L.; Huang, Y.; Liu, Y.; Dong, Q.; Wei, X. Wnt/β-catenin signaling pathway in carcinogenesis and cancer therapy. J. Hematol. Oncol. 2024, 17, 46. [Google Scholar] [CrossRef]
- Degirmenci, B.; Valenta, T.; Dimitrieva, S.; Hausmann, G.; Basler, K. GLI1-expressing mesenchymal cells form the essential Wnt-secreting niche for colon stem cells. Nature 2018, 558, 449–453. [Google Scholar] [CrossRef]
- Shoshkes-Carmel, M.; Wang, Y.J.; Wangensteen, K.J.; Tóth, B.; Kondo, A.; Massasa, E.E.; Itzkovitz, S.; Kaestner, K.H. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 2018, 557, 242–246. [Google Scholar] [CrossRef]
- Groenewald, W.; Lund, A.H.; Gay, D.M. The Role of WNT Pathway Mutations in Cancer Development and an Overview of Therapeutic Options. Cells 2023, 12, 990. [Google Scholar] [CrossRef]
- Li, J.; Li, J.; Chen, B. Oct4 was a novel target of Wnt signaling pathway. Mol. Cell Biochem. 2012, 362, 233–240. [Google Scholar] [CrossRef]
- Mann, B.; Gelos, M.; Siedow, A.; Hanski, M.L.; Gratchev, A.; Ilyas, M.; Bodmer, W.F.; Moyer, M.P.; Riecken, E.O.; Buhr, H.J.; et al. Target genes of β-catenin–T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc. Natl. Acad. Sci. USA 1999, 96, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Shtutman, M.; Zhurinsky, J.; Simcha, I.; Albanese, C.; D’Amico, M.; Pestell, R.; Ben-Ze’ev, A. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 5522–5527. [Google Scholar] [CrossRef]
- Van der Flier, L.G.; Sabates–Bellver, J.; Oving, I.; Haegebarth, A.; De Palo, M.; Anti, M.; Van Gijn, M.E.; Suijkerbuijk, S.; Van de Wetering, M.; Marra, G.; et al. The Intestinal Wnt/TCF Signature. Gastroenterology 2007, 132, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, M.; Sun, J.; Yang, X. Casein kinase 1α: Biological mechanisms and theranostic potential. Cell Commun. Signal. 2018, 16, 23. [Google Scholar] [CrossRef]
- Farin, H.F.; Jordens, I.; Mosa, M.H.; Basak, O.; Korving, J.; Tauriello, D.V.F.; de Punder, K.; Angers, S.; Peters, P.J.; Maurice, M.M.; et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature 2016, 530, 340–343. [Google Scholar] [CrossRef] [PubMed]
- Mills, K.M.; Szczerkowski, J.L.A.; Habib, S.J. Wnt ligand presentation and reception: From the stem cell niche to tissue engineering. Open Biol. 2017, 7, 170140. [Google Scholar] [CrossRef]
- Dame, M.K.; Attili, D.; McClintock, S.D.; Dedhia, P.H.; Ouillette, P.; Hardt, O.; Chin, A.M.; Xue, X.; Laliberte, J.; Katz, E.L.; et al. Identification, isolation and characterization of human LGR5-positive colon adenoma cells. Development 2018, 145, dev153049. [Google Scholar] [CrossRef]
- Fevr, T.; Robine, S.; Louvard, D.; Huelsken, J. Wnt/β-Catenin Is Essential for Intestinal Homeostasis and Maintenance of Intestinal Stem Cells. Mol. Cell Biol. 2007, 27, 7551–7559. [Google Scholar] [CrossRef]
- Flanagan, D.J.; Phesse, T.J.; Barker, N.; Schwab, R.H.M.; Amin, N.; Malaterre, J.; Stange, D.E.; Nowell, C.J.; Currie, S.A.; Saw, J.T.S.; et al. Frizzled7 Functions as a Wnt Receptor in Intestinal Epithelial Lgr5+ Stem Cells. Stem Cell Rep. 2015, 4, 759–767. [Google Scholar] [CrossRef]
- Ireland, H.; Kemp, R.; Houghton, C.; Howard, L.; Clarke, A.R.; Sansom, O.J.; Winton, D.J. Inducible cre-mediated control of gene expression in the murine gastrointestinal tract: Effect of loss of β-catenin. Gastroenterology 2004, 126, 1236–1246. [Google Scholar] [CrossRef]
- Kuhnert, F.; Davis, C.R.; Wang, H.-T.; Chu, P.; Lee, M.; Yuan, J.; Nusse, R.; Kuo, C.J. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc. Natl. Acad. Sci. USA 2004, 101, 266–271. [Google Scholar] [CrossRef]
- Pinto, D.; Gregorieff, A.; Begthel, H.; Clevers, H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003, 17, 1709–1713. [Google Scholar] [CrossRef]
- Tomasetti, C.; Vogelstein, B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015, 347, 78–81. [Google Scholar] [CrossRef]
- Rao, C.V.; Yamada, H.Y. Genomic Instability and Colon Carcinogenesis: From the Perspective of Genes. Front. Oncol. 2013, 3, 130. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Ridgway, R.A.; van Es, J.H.; van de Wetering, M.; Begthel, H.; van den Born, M.; Danenberg, E.; Clarke, A.R.; Sansom, O.J.; Clevers, H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 2009, 457, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, L.; Snippert, H.J. Stem cell dynamics in homeostasis and cancer of the intestine. Nat. Rev. Cancer 2014, 14, 468–480. [Google Scholar] [CrossRef]
- Martin, M.L.; Zeng, Z.; Adileh, M.; Jacobo, A.; Li, C.; Vakiani, E.; Hua, G.; Zhang, L.; Haimovitz-Friedman, A.; Fuks, Z.; et al. Logarithmic expansion of LGR5+ cells in human colorectal cancer. Cell Signal 2018, 42, 97–105. [Google Scholar] [CrossRef]
- Ebrahimi, N.; Afshinpour, M.; Fakhr, S.S.; Kalkhoran, P.G.; Shadman-Manesh, V.; Adelian, S.; Beiranvand, S.; Rezaei-Tazangi, F.; Khorram, R.; Hamblin, M.R.; et al. Cancer stem cells in colorectal cancer: Signaling pathways involved in stemness and therapy resistance. Crit. Rev. Oncol. Hematol. 2023, 182, 103920. [Google Scholar] [CrossRef]
- De Jaime-Soguero, A.; Abreu de Oliveira, W.; Lluis, F. The Pleiotropic Effects of the Canonical Wnt Pathway in Early Development and Pluripotency. Genes 2018, 9, 93. [Google Scholar] [CrossRef]
- Sonavane, P.; Willert, K. Wnt signaling and the regulation of pluripotency. Curr. Top. Dev. Biol. 2023, 153, 95–119. [Google Scholar]
- Chen, S.; Sun, Y.-Y.; Zhang, Z.-X.; Li, Y.-H.; Xu, Z.-M.; Fu, W.-N. Transcriptional suppression of microRNA-27a contributes to laryngeal cancer differentiation via GSK-3β-involved Wnt/β-catenin pathway. Oncotarget 2017, 8, 14708–14718. [Google Scholar] [CrossRef] [PubMed]
- Divisato, G.; Piscitelli, S.; Elia, M.; Cascone, E.; Parisi, S. MicroRNAs and Stem-like Properties: The Complex Regulation Underlying Stemness Maintenance and Cancer Development. Biomolecules 2021, 11, 1074. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Fang, X.; Li, C.; Lan, P.; Guan, X. Oncofetal proteins and cancer stem cells. Essays Biochem. 2022, 66, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Ornitz, D.M.; Itoh, N. The Fibroblast Growth Factor signaling pathway. WIREs Dev. Biol. 2015, 4, 215–266. [Google Scholar] [CrossRef]
- Nguyen, A.L.; Facey, C.O.B.; Boman, B.M. The Complexity and Significance of Fibroblast Growth Factor (FGF) Signaling for FGF-Targeted Cancer Therapies. Cancers 2024, 17, 82. [Google Scholar] [CrossRef]
- Xie, Y.; Su, N.; Yang, J.; Tan, Q.; Huang, S.; Jin, M.; Ni, Z.; Zhang, B.; Zhang, D.; Luo, F.; et al. FGF/FGFR signaling in health and disease. Signal Transduct. Target. Ther. 2020, 5, 181. [Google Scholar] [CrossRef] [PubMed]
- Danopoulos, S.; Schlieve, C.R.; Grikscheit, T.C.; Al Alam, D. Fibroblast Growth Factors in the Gastrointestinal Tract: Twists and Turns. Dev. Dyn. 2017, 246, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zhang, S.; Zhang, J.; Jin, Y.; Chen, T.; Lin, R.; Lv, J.; Xu, W.; Wu, T.; Tian, S.; et al. Colonic epithelial-derived FGF1 drives intestinal stem cell commitment toward goblet cells to suppress inflammatory bowel disease. Nat. Commun. 2025, 16, 3264. [Google Scholar] [CrossRef]
- Lv, Y.-Q.; Wu, J.; Li, X.-K.; Zhang, J.-S.; Bellusci, S. Role of FGF10/FGFR2b Signaling in Mouse Digestive Tract Development, Repair and Regeneration Following Injury. Front. Cell Dev. Biol. 2019, 7, 326. [Google Scholar] [CrossRef]
- Otte, J.; Dizdar, L.; Behrens, B.; Goering, W.; Knoefel, W.T.; Wruck, W.; Stoecklein, N.H.; Adjaye, J. FGF Signalling in the Self-Renewal of Colon Cancer Organoids. Sci. Rep. 2019, 9, 17365. [Google Scholar] [CrossRef]
- Wang, Y.; Kim, R.; Gunasekara, D.B.; Reed, M.I.; DiSalvo, M.; Nguyen, D.L.; Bultman, S.J.; Sims, C.E.; Magness, S.T.; Allbritton, N.L. Formation of Human Colonic Crypt Array by Application of Chemical Gradients Across a Shaped Epithelial Monolayer. Cell. Mol. Gastroenterol Hepatol. 2017, 5, 113–130. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sala, F.G.; Curtis, J.L.; Veltmaat, J.M.; Del Moral, P.-M.; Le, L.T.; Fairbanks, T.J.; Warburton, D.; Ford, H.; Wang, K.; Burns, R.C.; et al. Fibroblast growth factor 10 is required for survival and proliferation but not differentiation of intestinal epithelial progenitor cells during murine colon development. Dev. Biol. 2006, 299, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Sphyris, N.; Hodder, M.C.; Sansom, O.J. Subversion of Niche-Signalling Pathways in Colorectal Cancer: What Makes and Breaks the Intestinal Stem Cell. Cancers 2021, 13, 1000. [Google Scholar] [CrossRef]
- Pei, J.; Song, N.; Wu, L.; Qi, J.; Xia, S.; Xu, C.; Zheng, B.; Yang, J.; Qiu, Y.; Wang, H.; et al. TCF4/β-catenin complex is directly upstream of FGF21 in mouse stomach cancer cells. Exp. Ther. Med. 2017, 15, 1041–1047. [Google Scholar] [CrossRef]
- Brodrick, B.; Vidrich, A.; Porter, E.; Bradley, L.; Buzan, J.M.; Cohn, S.M. Fibroblast Growth Factor Receptor-3 (FGFR-3) Regulates Expression of Paneth Cell Lineage-specific Genes in Intestinal Epithelial Cells through both TCF4/β-Catenin-dependent and -independent Signaling Pathways. J. Biol. Chem. 2011, 286, 18515–18525. [Google Scholar] [CrossRef]
- Raucci, A.; Laplantine, E.; Mansukhani, A.; Basilico, C. Activation of the ERK1/2 and p38 Mitogen-activated Protein Kinase Pathways Mediates Fibroblast Growth Factor-induced Growth Arrest of Chondrocytes. J. Biol. Chem. 2004, 279, 1747–1756. [Google Scholar] [CrossRef]
- Li, C.; Chen, T.; Liu, J.; Wang, Y.; Zhang, C.; Guo, L.; Shi, D.; Zhang, T.; Wang, X.; Li, J. FGF19-Induced Inflammatory CAF Promoted Neutrophil Extracellular Trap Formation in the Liver Metastasis of Colorectal Cancer. Adv. Sci. 2023, 10, e2302613. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Chen, J.; Feng, W.; Huang, W.; Wang, G.; Sun, M.; Luo, X.; Wang, Y.; Nie, Y.; Fan, D.; et al. FGF19-mediated ELF4 overexpression promotes colorectal cancer metastasis through transactivating FGFR4 and SRC. Theranostics 2023, 13, 1401–1418. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, M.; Cai, Y.; Li, X.; Zhao, C.; Cui, R. Dissecting the Role of the FGF19-FGFR4 Signaling Pathway in Cancer Development and Progression. Front. Cell Dev. Biol. 2020, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, Y.; Qin, X.; Wang, Y.; Fu, J. FGF9 promotes cisplatin resistance in colorectal cancer via regulation of Wnt/β-catenin signaling pathway. Exp. Ther. Med. 2019, 19, 1711–1718. [Google Scholar] [CrossRef]
- Sonvilla, G.; Allerstorfer, S.; Heinzle, C.; Stättner, S.; Karner, J.; Klimpfinger, M.; Wrba, F.; Fischer, H.; Gauglhofer, C.; Spiegl-Kreinecker, S.; et al. Fibroblast growth factor receptor 3-IIIc mediates colorectal cancer growth and migration. Br. J. Cancer 2010, 102, 1145–1156. [Google Scholar] [CrossRef]
- Sancho, R.; Cremona, C.A.; Behrens, A. Stem cell and progenitor fate in the mammalian intestine: Notch and lateral inhibition in homeostasis and disease. EMBO Rep. 2015, 16, 571–581. [Google Scholar] [CrossRef]
- Liang, S.; Li, X.; Wang, X. Notch Signaling in Mammalian Intestinal Stem Cells: Determining Cell Fate and Maintaining Homeostasis. Curr. Stem Cell Res. Ther. 2019, 14, 583–590. [Google Scholar] [CrossRef]
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.M. Notch signaling and Notch signaling modifiers. Int. J. Biochem. Cell Biol. 2011, 43, 1550–1562. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R. Notch Signaling. Cold Spring Harb. Perspect. Biol. 2012, 4, a011213. [Google Scholar] [CrossRef]
- Troll, J.V.; Hamilton, M.K.; Abel, M.L.; Ganz, J.; Bates, J.M.; Stephens, W.Z.; Melancon, E.; van der Vaart, M.; Meijer, A.H.; Distel, M.; et al. Microbiota promote secretory cell determination in the intestinal epithelium by modulating host Notch signaling. Development 2018, 145, dev155317. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Biehs, B.; Chiu, C.; Siebel, C.W.; Wu, Y.; Costa, M.; de Sauvage, F.J.; Klein, O.D. Opposing Activities of Notch and Wnt Signaling Regulate Intestinal Stem Cells and Gut Homeostasis. Cell Rep. 2015, 11, 33–42. [Google Scholar] [CrossRef]
- Shi, Q.; Xue, C.; Zeng, Y.; Yuan, X.; Chu, Q.; Jiang, S.; Wang, J.; Zhang, Y.; Zhu, D.; Li, L. Notch signaling pathway in cancer: From mechanistic insights to targeted therapies. Signal Transduct. Target. Ther. 2024, 9, 128. [Google Scholar] [CrossRef]
- Rajakulendran, N.; Rowland, K.J.; Selvadurai, H.J.; Ahmadi, M.; Park, N.I.; Naumenko, S.; Dolma, S.; Ward, R.J.; So, M.; Lee, L.; et al. Wnt and Notch signaling govern self-renewal and differentiation in a subset of human glioblastoma stem cells. Genes Dev. 2019, 33, 498–510. [Google Scholar] [CrossRef]
- López-Nieva, P.; González-Sánchez, L.; Cobos-Fernández, M.Á.; Córdoba, R.; Santos, J.; Fernández-Piqueras, J. More Insights on the Use of γ-Secretase Inhibitors in Cancer Treatment. Oncologist 2021, 26, e298–e305. [Google Scholar] [CrossRef]
- Beederman, M.; Lamplot, J.D.; Nan, G.; Wang, J.; Liu, X.; Yin, L.; Li, R.; Shui, W.; Zhang, H.; Kim, S.H.; et al. BMP signaling in mesenchymal stem cell differentiation and bone formation. J. Biomed. Sci. Eng. 2013, 06, 32–52. [Google Scholar] [CrossRef]
- Wang, R.N.; Green, J.; Wang, Z.; Deng, Y.; Qiao, M.; Peabody, M.; Zhang, Q.; Ye, J.; Yan, Z.; Denduluri, S.; et al. Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 2014, 1, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Li, Y.; Zhao, B.; Xu, C.; Liu, Y.; Li, H.; Zhang, B.; Wang, X.; Yang, X.; Xie, W.; et al. BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat. Commun. 2017, 8, 13824. [Google Scholar] [CrossRef] [PubMed]
- Kraiczy, J.; McCarthy, N.; Malagola, E.; Tie, G.; Madha, S.; Boffelli, D.; Wagner, D.E.; Wang, T.C.; Shivdasani, R.A. Graded BMP signaling within intestinal crypt architecture directs self-organization of the Wnt-secreting stem cell niche. Cell Stem Cell 2023, 30, 433–449.e8. [Google Scholar] [CrossRef]
- Malijauskaite, S.; Connolly, S.; Newport, D.; McGourty, K. Gradients in the in vivo intestinal stem cell compartment and their in vitro recapitulation in mimetic platforms. Cytokine Growth Factor Rev. 2021, 60, 76–88. [Google Scholar] [CrossRef]
- Voorneveld, P.W.; Kodach, L.L.; Jacobs, R.J.; Liv, N.; Zonnevylle, A.C.; Hoogenboom, J.P.; Biemond, I.; Verspaget, H.W.; Hommes, D.W.; de Rooij, K.; et al. Loss of SMAD4 Alters BMP Signaling to Promote Colorectal Cancer Cell Metastasis via Activation of Rho and ROCK. Gastroenterology 2014, 147, 196–208.e13. [Google Scholar] [CrossRef]
- Clarkson, E.; Lewis, A. BMP signalling in colorectal cancer: Losing the yin to WNTs yang. J. Pathol. 2025, 266, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, Y.; Watanabe, T.; Tamura, Y.; Hashizume, Y.; Miyazono, K.; Ehata, S. Autocrine BMP-4 Signaling Is a Therapeutic Target in Colorectal Cancer. Cancer Res. 2017, 77, 4026–4038. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Kornmann, M.; Traub, B. Role of Epithelial to Mesenchymal Transition in Colorectal Cancer. Int. J. Mol. Sci. 2023, 24, 14815. [Google Scholar] [CrossRef]
- Nie, F.; Sun, X.; Sun, J.; Zhang, J.; Wang, Y. Epithelial-mesenchymal transition in colorectal cancer metastasis and progression: Molecular mechanisms and therapeutic strategies. Cell Death Discov. 2025, 11, 336. [Google Scholar] [CrossRef]
- Tan, Z.; Sun, W.; Li, Y.; Jiao, X.; Zhu, M.; Zhang, J.; Qing, C.; Jia, Y. Current Progress of EMT: A New Direction of Targeted Therapy for Colorectal Cancer with Invasion and Metastasis. Biomolecules 2022, 12, 1723. [Google Scholar] [CrossRef]
- Huang, Y.; Hong, W.; Wei, X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol. Oncol. 2022, 15, 129. [Google Scholar] [CrossRef]
- Zhang, N.; Ng, A.S.; Cai, S.; Li, Q.; Yang, L.; Kerr, D. Novel therapeutic strategies: Targeting epithelial–mesenchymal transition in colorectal cancer. Lancet Oncol. 2021, 22, e358–e368. [Google Scholar] [CrossRef] [PubMed]
- Ogden, S.; Metic, N.; Leylek, O.; Smith, E.A.; Berner, A.M.; Baker, A.-M.; Uddin, I.; Buzzetti, M.; Gerlinger, M.; Graham, T.; et al. Phenotypic heterogeneity and plasticity in colorectal cancer metastasis. Cell Genom. 2025, 5, 100881. [Google Scholar] [CrossRef]
- Ladaika, C.A.; Ghobashi, A.H.; Boulton, W.C.; Miller, S.A.; O’Hagan, H.M. Single-cell multi-omics reveals insights into differentiation of rare cell types in mucinous colorectal cancer. BioRxiv 2024. [Google Scholar] [CrossRef]
- Ladaika, C.A.; Ghobashi, A.H.; Boulton, W.C.; Miller, S.A.; O’Hagan, H.M. LSD1 and CoREST2 Potentiate STAT3 Activity to Promote Enteroendocrine Cell Differentiation in Mucinous Colorectal Cancer. Cancer Res. 2025, 85, 52–68. [Google Scholar] [CrossRef]
- Ladaika, C.A.; Chakraborty, A.; Masood, A.; Hostetter, G.; Yi, J.M.; O’Hagan, H.M. LSD1 inhibition attenuates targeted therapy-induced lineage plasticity in BRAF mutant colorectal cancer. Mol. Cancer 2025, 24, 122. [Google Scholar] [CrossRef]
- Thankamony, A.P.; Subbalakshmi, A.R.; Jolly, M.K.; Nair, R. Lineage Plasticity in Cancer: The Tale of a Skin-Walker. Cancers 2021, 13, 3602. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.; Datta, P. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Lengrand, J.; Pastushenko, I.; Vanuytven, S.; Song, Y.; Venet, D.; Sarate, R.M.; Bellina, M.; Moers, V.; Boinet, A.; Sifrim, A.; et al. Pharmacological targeting of netrin-1 inhibits EMT in cancer. Nature 2023, 620, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Yin, K.; Wang, S. Targeting of netrin-1 by monoclonal antibody NP137 inhibits the EMT in cancer. J. Immunother. Cancer 2024, 12, e008937. [Google Scholar] [CrossRef]
- Shiota, M.; Zardan, A.; Takeuchi, A.; Kumano, M.; Beraldi, E.; Naito, S.; Zoubeidi, A.; Gleave, M.E. Clusterin Mediates TGF-β–Induced Epithelial–Mesenchymal Transition and Metastasis via Twist1 in Prostate Cancer Cells. Cancer Res. 2012, 72, 5261–5272. [Google Scholar] [CrossRef]
- Peng, M.; Deng, J.; Zhou, S.; Tao, T.; Su, Q.; Xue, Y.; Yang, X. The role of Clusterin in cancer metastasis. Cancer Manag. Res. 2019, 11, 2405–2414. [Google Scholar] [CrossRef]
- Miranda, I.; Jahan, N.; Shevde, L.A. The metastatic cascade through the lens of therapeutic inhibition. Cell Rep. Med. 2025, 6, 101872. [Google Scholar] [CrossRef]
- Wu, F.; Yang, J.; Liu, J.; Wang, Y.; Mu, J.; Zeng, Q.; Deng, S.; Zhou, H. Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer. Signal Transduct. Target. Ther. 2021, 6, 218. [Google Scholar] [CrossRef] [PubMed]
- Martín-García, D.; García-Aranda, M.; Redondo, M. Therapeutic Potential of Clusterin Inhibition in Human Cancer. Cells. 2024, 13, 665. [Google Scholar] [CrossRef] [PubMed]
- Cassier, P.A.; Navaridas, R.; Bellina, M.; Rama, N.; Ducarouge, B.; Hernandez-Vargas, H.; Delord, J.P.; Lengrand, J.; Paradisi, A.; Fattet, L.; et al. Netrin-1 blockade inhibits tumour growth and EMT features in endometrial cancer. Nature 2023, 620, 409–416. [Google Scholar] [CrossRef]
- Jonckheere, S.; Adams, J.; De Groote, D.; Campbell, K.; Berx, G.; Goossens, S. Epithelial-Mesenchymal Transition (EMT) as a Therapeutic Target. Cells Tissues Organs 2022, 211, 157–182. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, A.L.; Lausten, M.A.; Boman, B.M. The Colonic Crypt: Cellular Dynamics and Signaling Pathways in Homeostasis and Cancer. Cells 2025, 14, 1428. https://doi.org/10.3390/cells14181428
Nguyen AL, Lausten MA, Boman BM. The Colonic Crypt: Cellular Dynamics and Signaling Pathways in Homeostasis and Cancer. Cells. 2025; 14(18):1428. https://doi.org/10.3390/cells14181428
Chicago/Turabian StyleNguyen, Anh L., Molly A. Lausten, and Bruce M. Boman. 2025. "The Colonic Crypt: Cellular Dynamics and Signaling Pathways in Homeostasis and Cancer" Cells 14, no. 18: 1428. https://doi.org/10.3390/cells14181428
APA StyleNguyen, A. L., Lausten, M. A., & Boman, B. M. (2025). The Colonic Crypt: Cellular Dynamics and Signaling Pathways in Homeostasis and Cancer. Cells, 14(18), 1428. https://doi.org/10.3390/cells14181428







