Isolation of Porcine Umbilical Cord Cells by Mechanical Tissue Dissociation Using a Tissue Grinder
Abstract
1. Introduction
2. Materials and Methods
2.1. Pig Serum Production
2.2. Isolation and Cultivation of Umbilical Cord Cells (UCCs)
2.3. Surface Marker Analysis
2.4. CFU Assay
2.5. Metabolic Activity (WST1)
2.6. Multi-Lineage Differentiation Assay
2.7. Statistical Analyses
3. Results
3.1. Isolation of UC Cells Comparing Different Tissue-Grinding Methods
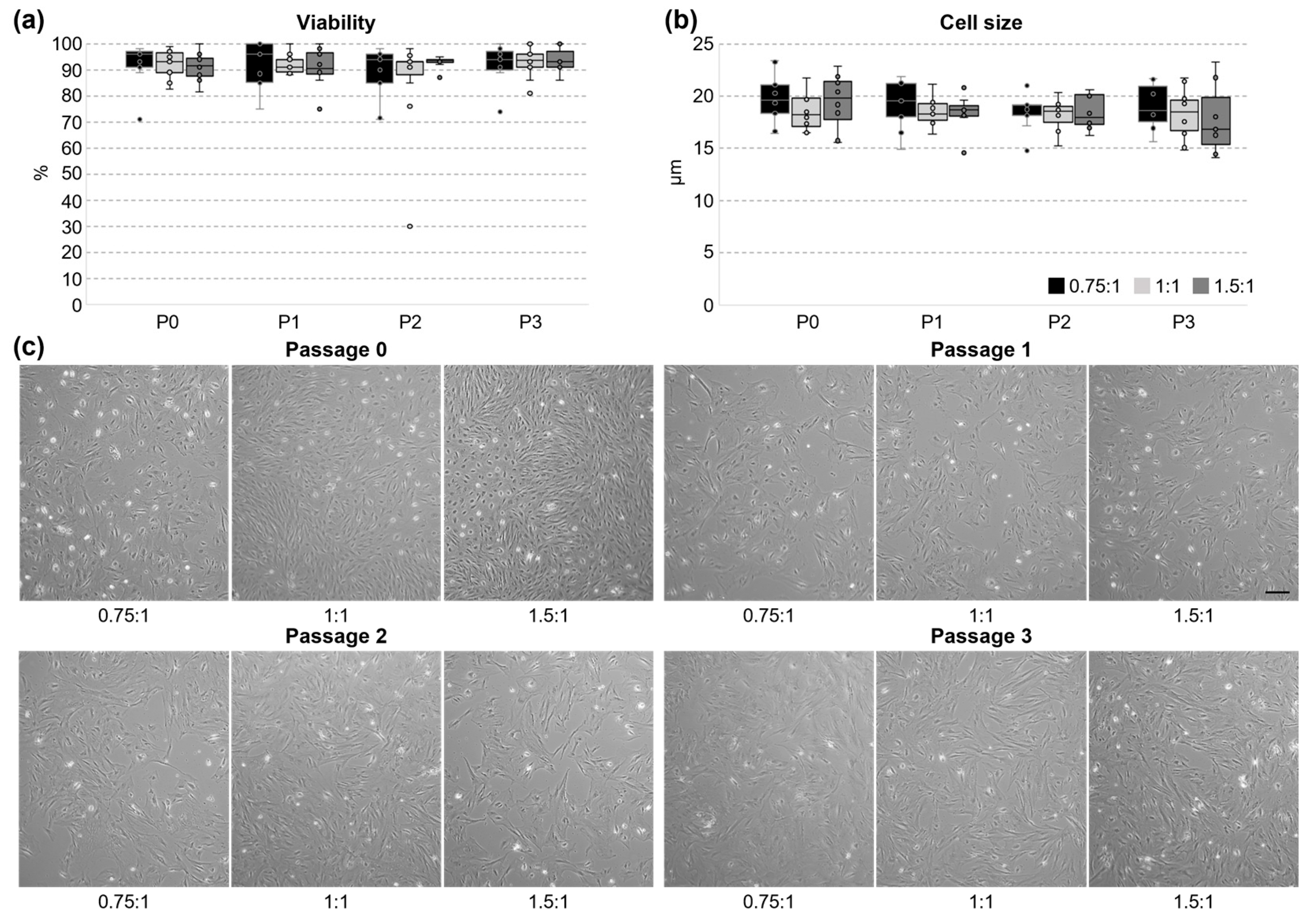
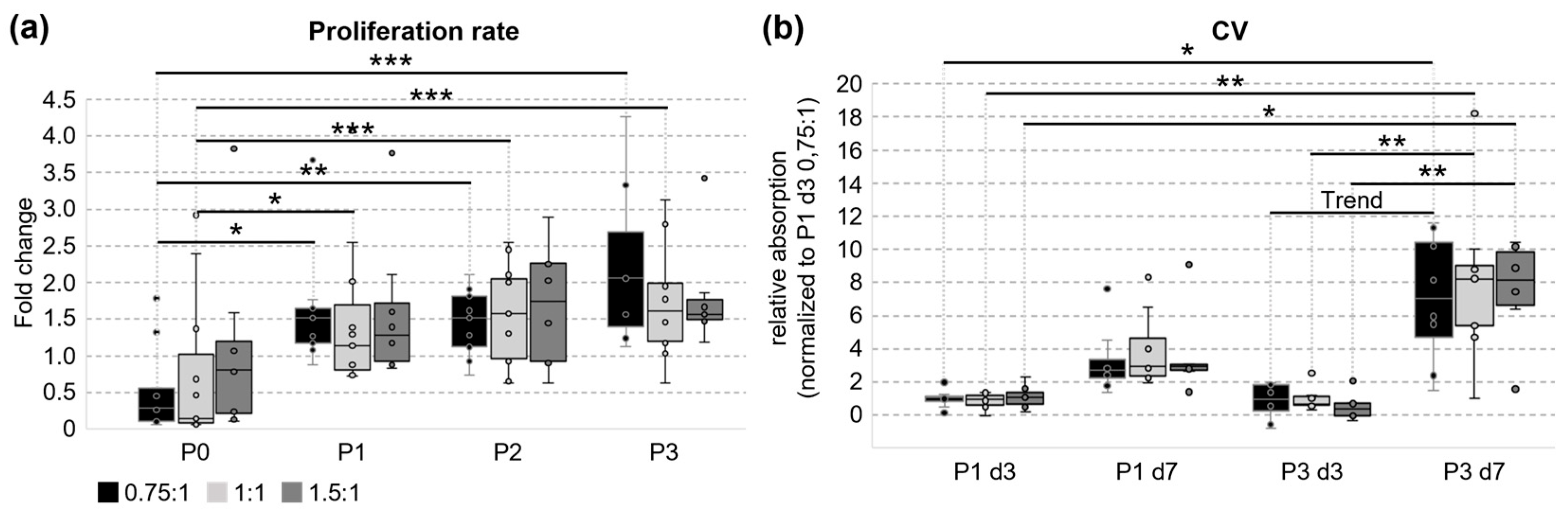
3.2. Cultivation and Proliferation of UC Cells
3.3. Metabolic Activity and Ability to Form Colonies
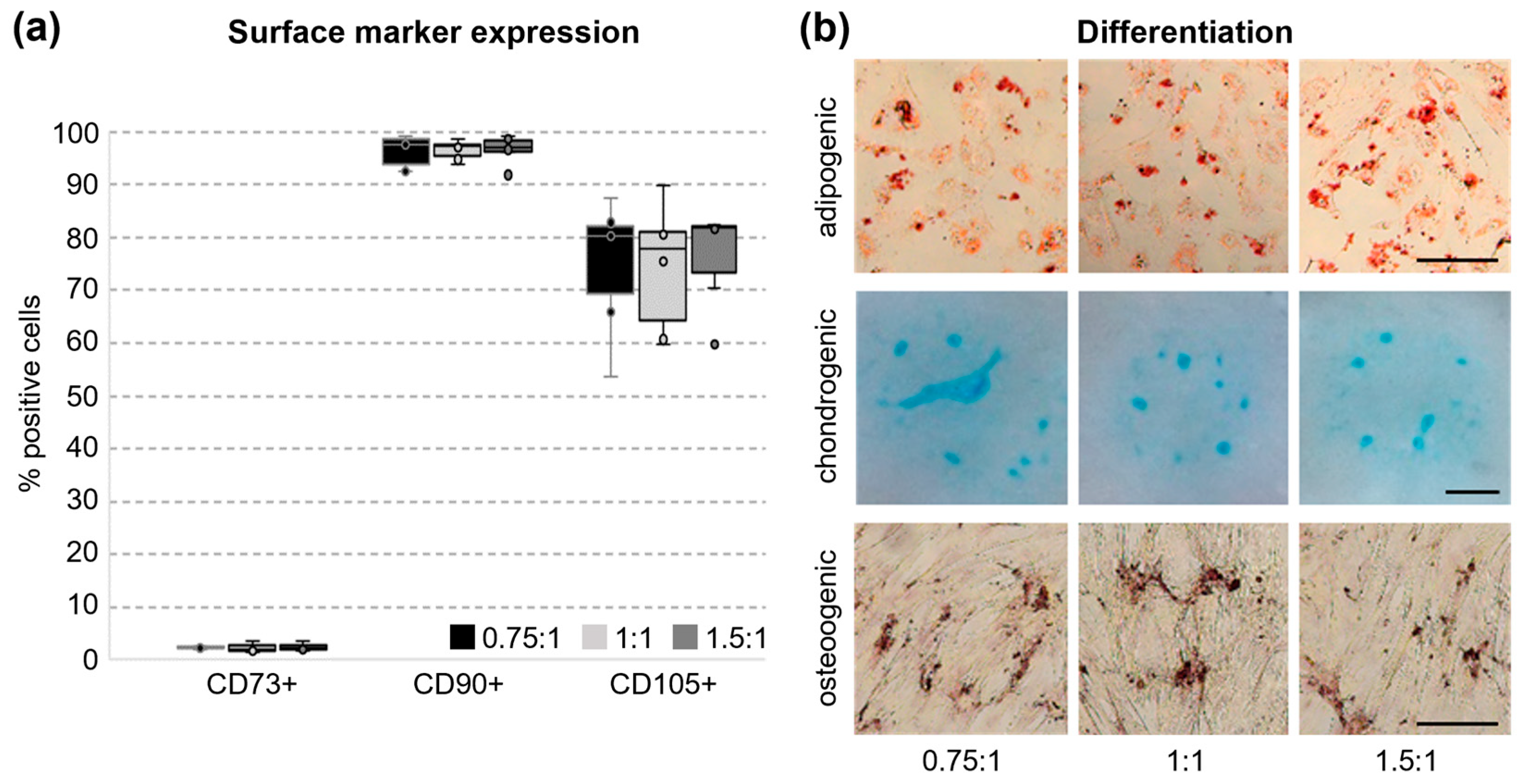
3.4. Surface Marker Expression and Tri-Lineage Differentiation of UC Cells In Vitro
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| bFGF | basic fibroblast growth factor |
| CFU | colony-forming unit |
| CHO | Chinese hamster ovary |
| CV | crystal violet |
| ECM | extracellular matrix |
| HEK | human embryonic kidney |
| MSC | mesenchymal stromal cell |
| UC | umbilical cord |
| UCC | umbilical cord cell |
| WJ | Wharton’s Jelly |
References
- Ladke, V.S.; Kumbhar, G.M.; Joshi, K.; Kheur, S.; Bhonde, R.; Raut, C. Isolation, Culture and Morphological Assessment of Primary Cell Lines from Human Primary Oral Squamous Cell Carcinoma Using Explant Technique. Asian Pac. J. Cancer Prev. 2023, 24, 257–266. [Google Scholar] [CrossRef]
- Piwocka, O.; Musielak, M.; Ampula, K.; Piotrowski, I.; Adamczyk, B.; Fundowicz, M.; Suchorska, W.M.; Malicki, J. Navigating challenges: Optimising methods for primary cell culture isolation. Cancer Cell Int. 2024, 24, 28. [Google Scholar] [CrossRef]
- Richter, M.; Piwocka, O.; Musielak, M.; Piotrowski, I.; Suchorska, W.M.; Trzeciak, T. From Donor to the Lab: A Fascinating Journey of Primary Cell Lines. Front. Cell Dev. Biol. 2021, 9, 711381. [Google Scholar] [CrossRef]
- Witkowska-Zimny, M.; Wrobel, E. Perinatal sources of mesenchymal stem cells: Wharton’s jelly, amnion and chorion. Cell Mol. Biol. Lett. 2011, 16, 493–514. [Google Scholar] [CrossRef]
- Camilleri, E.T.; Gustafson, M.P.; Dudakovic, A.; Riester, S.M.; Garces, C.G.; Paradise, C.R.; Takai, H.; Karperien, M.; Cool, S.; Sampen, H.J.; et al. Identification and validation of multiple cell surface markers of clinical-grade adipose-derived mesenchymal stromal cells as novel release criteria for good manufacturing practice-compliant production. Stem Cell Res. Ther. 2016, 7, 107. [Google Scholar] [CrossRef]
- Ding, D.C.; Shyu, W.C.; Lin, S.Z. Mesenchymal stem cells. Cell Transplant. 2011, 20, 5–14. [Google Scholar] [CrossRef]
- Nagamura-Inoue, T.; He, H. Umbilical cord-derived mesenchymal stem cells: Their advantages and potential clinical utility. World J. Stem Cells 2014, 6, 195–202. [Google Scholar] [CrossRef]
- Viswanathan, S.; Shi, Y.; Galipeau, J.; Krampera, M.; Leblanc, K.; Martin, I.; Nolta, J.; Phinney, D.G.; Sensebe, L. Mesenchymal stem versus stromal cells: International Society for Cell & Gene Therapy (ISCT(R)) Mesenchymal Stromal Cell committee position statement on nomenclature. Cytotherapy 2019, 21, 1019–1024. [Google Scholar] [CrossRef]
- Alvites, R.; Branquinho, M.; Sousa, A.C.; Lopes, B.; Sousa, P.; Mauricio, A.C. Mesenchymal Stem/Stromal Cells and Their Paracrine Activity-Immunomodulation Mechanisms and How to Influence the Therapeutic Potential. Pharmaceutics 2022, 14, 381. [Google Scholar] [CrossRef]
- Rady, D.; Abbass, M.M.S.; El-Rashidy, A.A.; El Moshy, S.; Radwan, I.A.; Dorfer, C.E.; Fawzy El-Sayed, K.M. Mesenchymal Stem/Progenitor Cells: The Prospect of Human Clinical Translation. Stem Cells Int. 2020, 2020, 8837654. [Google Scholar] [CrossRef]
- Kalamegam, G.; Sait, K.H.W.; Anfinan, N.; Kadam, R.; Ahmed, F.; Rasool, M.; Naseer, M.I.; Pushparaj, P.N.; Al-Qahtani, M. Cytokines secreted by human Wharton’s jelly stem cells inhibit the proliferation of ovarian cancer (OVCAR3) cells in vitro. Oncol. Lett. 2019, 17, 4521–4531. [Google Scholar] [CrossRef]
- Merlo, B.; Gugole, P.M.; Iacono, E. An Update on Applications of Cattle Mesenchymal Stromal Cells. Animals 2022, 12, 1956. [Google Scholar] [CrossRef]
- Kriston-Pal, E.; Haracska, L.; Cooper, P.; Kiss-Toth, E.; Szukacsov, V.; Monostori, E. A Regenerative Approach to Canine Osteoarthritis Using Allogeneic, Adipose-Derived Mesenchymal Stem Cells. Safety Results of a Long-Term Follow-Up. Front. Vet. Sci. 2020, 7, 510. [Google Scholar] [CrossRef]
- Devireddy, L.R.; Boxer, L.; Myers, M.J.; Skasko, M.; Screven, R. Questions and Challenges in the Development of Mesenchymal Stromal/Stem Cell-Based Therapies in Veterinary Medicine. Tissue Eng. Part. B Rev. 2017, 23, 462–470. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Discher, D.E.; Peault, B.M.; Phinney, D.G.; Hare, J.M.; Caplan, A.I. Mesenchymal stem cell perspective: Cell biology to clinical progress. NPJ Regen. Med. 2019, 4, 22. [Google Scholar] [CrossRef]
- Knežic, T.; Janjuševí, L.; Djisalov, M.; Yodmuang, S.; Gadjanski, I. Using vertebrate stem and progenitor cells for cellular agriculture-state-of-the-art, challenges, and future perspectives. Biomolecules 2022, 12, 699. [Google Scholar] [CrossRef]
- Cesnik, A.B.; Svajger, U. The issue of heterogeneity of MSC-based advanced therapy medicinal products-a review. Front. Cell Dev. Biol. 2024, 12, 1400347. [Google Scholar] [CrossRef]
- Naji, A.; Eitoku, M.; Favier, B.; Deschaseaux, F.; Rouas-Freiss, N.; Suganuma, N. Biological functions of mesenchymal stem cells and clinical implications. Cell Mol. Life Sci. 2019, 76, 3323–3348. [Google Scholar] [CrossRef]
- Wang, Z.; Chai, C.; Wang, R.; Feng, J.; Huang, L.; Zhang, J.; Xiao, X.; Yang, S.; Zhang, J.; Zhang, X. Single-cell transcriptome atlas of human mesenchymal stem cells exploring cellular heterogeneity. Clin. Transl. Med. 2021, 11, e650. [Google Scholar] [CrossRef]
- Lim, J.; Heo, J.; Yu, H.Y.; Yun, H.D.; Lee, S.; Ju, H.; Nam, Y.J.; Jeong, S.M.; Lee, J.; Cho, Y.S.; et al. Small-sized mesenchymal stem cells with high glutathione dynamics show improved therapeutic potency in graft-versus-host disease. Clin. Transl. Med. 2021, 11, e476. [Google Scholar] [CrossRef]
- Hass, R.; Kasper, C.; Böhm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal 2011, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Carlin, R.; Davis, D.; Weiss, M.; Schultz, B.; Troyer, D. Expression of early transcription factors Oct-4, Sox-2 and Nanog by porcine umbilical cord (PUC) matrix cells. Reprod. Biol. Endocrinol. 2006, 4, 8. [Google Scholar] [CrossRef]
- Bongso, A.; Fong, C.Y.; Gauthaman, K. Taking stem cells to the clinic: Major challenges. J. Cell. Biochem. 2008, 105, 1352–1360. [Google Scholar] [CrossRef]
- Can, A.; Karahuseyinoglu, S. Concise review: Human umbilical cord stroma with regard to the source of fetus-derived stem cells. Stem Cells 2007, 25, 2886–2895. [Google Scholar] [CrossRef] [PubMed]
- Yea, J.-H.; Yea, K.; Jo, C.H. Comparison of mesenchymal stem cells from bone marrow, umbilical cord blood, and umbilical cord tissue in regeneration of a full-thickness tendon defect in vitro and in vivo. Biochem. Biophys. Rep. 2023, 34, 101486. [Google Scholar] [CrossRef]
- Kulus, M.; Sibiak, R.; Stefanska, K.; Zdun, M.; Wieczorkiewicz, M.; Piotrowska-Kempisty, H.; Jaskowski, J.M.; Bukowska, D.; Ratajczak, K.; Zabel, M.; et al. Mesenchymal stem/stromal cells derived from human and animal perinatal tissues—Origins, characteristics, signaling pathways, and clinical trials. Cells 2021, 10, 3278. [Google Scholar] [CrossRef]
- Moretti, P.; Hatlapatka, T.; Marten, D.; Lavrentieva, A.; Majore, I.; Hass, R.; Kasper, C. Mesenchymal stromal cells derived from human umbilical cord tissues: Primitive cells with potential for clinical and tissue engineering applications. In Bioreactor Systems for Tissue Engineering II; Kasper, C., van Griensven, M., Pörtner, R., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2009; Volume 123, pp. 29–54. [Google Scholar] [CrossRef]
- Silini, A.R.; Di Pietro, R.; Lang-Olip, I.; Alviano, F.; Banerjee, A.; Basile, M.; Borutinskaite, V.; Eissner, G.; Gellhaus, A.; Giebel, B.; et al. Perinatal derivatives: Where do we stand? A roadmap of the human placenta and consensus for tissue and cell nomenclature. Front. Bioeng. Biotechnol. 2020, 8, 610544. [Google Scholar] [CrossRef]
- Hill, M.A. Embryology Paper—The Histology of the Umbilical Cord of the Pig. 1919. Available online: https://embryology.med.unsw.edu.au/embryology/index.php/Paper_-_The_histology_of_the_umbilical_cord_of_the_pig_(1919) (accessed on 10 September 2025).
- Barrios-Arpi, L.M.; Rodríguez Gutiérrez, J.-L.; Lopez-Torres, B. Histological characterization of umbilical cord in alpaca (Vicugna pacos). Anat. Histol. Embryol. 2017, 46, 533–538. [Google Scholar] [CrossRef]
- Majore, I.; Moretti, P.; Hass, R.; Kasper, C. Identification of subpopulations in mesenchymal stem cell-like cultures from human umbilical cord. Cell Comm. Signal 2009, 7, 6. [Google Scholar] [CrossRef]
- Mishra, S.; Sevak, J.K.; Das, A.; Arimbasseri, G.A.; Bhatnagar, S.; Gopinath, S.D. Umbilical cord tissue is a robust source for mesenchymal stem cells with enhanced myogenic differentiation potential compared to cord blood. Sci. Rep. 2020, 10, 18978. [Google Scholar] [CrossRef] [PubMed]
- Toupadakis, C.A.; Wong, A.; Genetos, D.C.; Cheung, W.K.; Borjesson, D.L.; Ferraro, G.L.; Galuppo, L.D.; Leach, J.K.; Owens, S.D.; Clare, E.; et al. Comparison of the osteogenic potential of equine mesenchymal stem cells from bone marrow, adipose tissue, umbilical cord blood, and umbilical cord tissue. Am. J. Vet. Res. 2010, 71, 1237–1245. [Google Scholar] [CrossRef]
- Cardoso, T.C.; Ferrari, H.F.; Garcia, A.F.; Novais, J.B.; Silva-Frade, C.; Ferrarezi, M.C.; Andrade, A.L.; Gameiro, R. Isolation and characterization of Wharton’s jelly-derived multipotent mesenchymal stromal cells obtained from bovine umbilical cord and maintained in a defined serum-free three-dimensional system. BMC Biotechnol. 2012, 12, 18. [Google Scholar] [CrossRef]
- Singh, J.; Mann, A.; Kumar, D.; Duhan, J.S.; Yadav, P.S. Cultured buffalo umbilical cord matrix cells exhibit characteristics of multipotent mesenchymal stem cells. Vitr. Cell. Dev. Biol. Anim. 2013, 49, 408–416. [Google Scholar] [CrossRef]
- Koutonin, B.O.M.; Zhang, F.; Jiang, Y.; Jia, C.; Saeed, H.A.; Fu, Y.; Liu, H.; Adoligbe, C.M.; Li, J. Isolation, characterization, and transcriptome profiling of umbilical cord mesenchymal stem cells in pigs. Anim. Adv. 2024, 1, e008. [Google Scholar] [CrossRef]
- Kitala, D.; Klama-Baryla, A.; Labus, W.; Kraut, M.; Glik, J.; Kawecki, M.; Kuzma, A. Amniotic and Umbilical Cord of Transgenic Pigs as an Alternative Source of Stem Cells. Transplant. Proc. 2020, 52, 2193–2197. [Google Scholar] [CrossRef]
- Bharti, D.; Shivakumar, S.B.; Subbarao, R.B.; Rho, G.-J. Research advancements in porcine derived mesenchymal stem cells. Curr. Stem Cell Res. Therapy 2016, 11, 1–16. [Google Scholar] [CrossRef]
- Gan, M.; Liu, L.; Zhang, S.; Guo, Z.; Tan, Y.; Luo, J.; Yang, Q.; Pan, H.; Li, X.; Wang, J.; et al. Expression Characteristics of microRNA in Pig Umbilical Venous Blood and Umbilical Arterial Blood. Animals 2021, 11, 1563. [Google Scholar] [CrossRef]
- Griffith, B.P.; Grazioli, A.; Singh, A.K.; Tully, A.; Galindo, J.; Saharia, K.K.; Shah, A.; Strauss, E.R.; Odonkor, P.N.; Williams, B.; et al. Transplantation of a genetically modified porcine heart into a live human. Nat. Med. 2025, 31, 589–598. [Google Scholar] [CrossRef]
- Kawai, T.; Williams, W.W.; Elias, N.; Fishman, J.A.; Crisalli, K.; Longchamp, A.; Rosales, I.A.; Duggan, M.; Kimura, S.; Morena, L.; et al. Xenotransplantation of a Porcine Kidney for End-Stage Kidney Disease. N. Engl. J. Med. 2025, 392, 1933–1940. [Google Scholar] [CrossRef]
- Chatzistamatiou, T.K.; Papassavas, A.C.; Michalopoulos, E.; Gamaloutsos, C.; Mallis, P.; Gontika, I.; Panagouli, E.; Koussoulakos, S.L.; Stavropoulos-Giokas, C. Optimizing isolation culture and freezing methods to preserve Wharton’s jelly’s mesenchymal stem cell (MSC) properties: An MSC banking protocol validation for the Hellenic Cord Blood Bank. Transfusion 2014, 54, 3108–3120. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, B.; Tao, Y.; Cheng, M.; Hu, J.; Xu, M.; Chen, H. Isolation and characterization of mesenchymal stem cells from whole human umbilical cord applying a single enzyme approach. Cell Biochem. Funct. 2012, 30, 643–649. [Google Scholar] [CrossRef]
- Scheuermann, S.; Lehmann, J.M.; Ramani Mohan, R.; Reissfelder, C.; Ruckert, F.; Langejurgen, J.; Pallavi, P. TissueGrinder, a novel technology for rapid generation of patient-derived single cell suspensions from solid tumors by mechanical tissue dissociation. Front. Med. 2022, 9, 721639. [Google Scholar] [CrossRef]
- Yoon, J.H.; Roh, E.Y.; Shin, S.; Jung, N.H.; Song, E.Y.; Chang, J.Y.; Kim, B.J.; Jeon, H.W. Comparison of explant-derived and enzymatic digestion-derived MSCs and the growth factors from Wharton’s Jelly. BioMed Res. Int. 2013, 2013, 428726. [Google Scholar] [CrossRef]
- Skiles, M.L.; Marzan, A.J.; Brown, K.S.; Shamonki, J.M. Comparison of umbilical cord tissue-derived mesenchymal stromal cells isolated from cryopreserved material and extracted by explantation and digestion methods utilizing a split manufacturing model. Cytotherapy 2020, 22, 581–591. [Google Scholar] [CrossRef]
- Duarte, A.C.; Costa, E.C.; Filipe, H.A.L.; Saraiva, S.M.; Jacinto, T.; Miguel, S.P.; Ribeiro, M.P.; Coutinho, P. Animal-derived products in science and current alternatives. Biomater. Adv. 2023, 151, 213428. [Google Scholar] [CrossRef]
- Marcus-Sekura, C.; Richardson, J.C.; Harston, R.K.; Sane, N.; Sheets, R.L. Evaluation of the human host range of bovine and porcine viruses that may contaminate bovine serum and porcine trypsin used in the manufacture of biological products. Biologicals 2011, 39, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Burden, D.W. Guide to the Disruption of Biological Samples; OPS Diagnostics: Lebanon, NJ, USA, 2012; pp. 1–25. [Google Scholar]
- Soteriou, D.; Kubankova, M.; Schweitzer, C.; Lopez-Posadas, R.; Pradhan, R.; Thoma, O.M.; Gyorfi, A.H.; Matei, A.E.; Waldner, M.; Distler, J.H.W.; et al. Rapid single-cell physical phenotyping of mechanically dissociated tissue biopsies. Nat. Biomed. Eng. 2023, 7, 1392–1403. [Google Scholar] [CrossRef] [PubMed]
- Schellenberg, A.; Stiehl, T.; Horn, P.; Joussen, S.; Pallua, N.; Ho, A.D.; Wagner, W. Population dynamics of mesenchymal stromal cells during culture expansion. Cytotherapy 2012, 14, 401–411. [Google Scholar] [CrossRef] [PubMed]
- Samsonraj, R.M.; Rai, B.; Sathiyanathan, P.; Puan, K.J.; Rotzschke, O.; Hui, J.H.; Raghunath, M.; Stanton, L.W.; Nurcombe, V.; Cool, S.M. Establishing criteria for human mesenchymal stem cell potency. Stem Cells 2015, 33, 1878–1891. [Google Scholar] [CrossRef]
- Digirolamo, C.M.; Stokes, D.; Colter, D.; Phinney, D.G.; Class, R.; Prockop, D.J. Propagation and senescence of human marrow stromal cells in culture: A simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br. J. Haematol. 1999, 107, 275–281. [Google Scholar] [CrossRef]
- Peters, R.; Wolf, M.J.; van den Broek, M.; Nuvolone, M.; Dannenmann, S.; Stieger, B.; Rapold, R.; Konrad, D.; Rubin, A.; Bertino, J.R.; et al. Efficient generation of multipotent mesenchymal stem cells from umbilical cord blood in stroma-free liquid culture. PLoS ONE 2010, 5, e15689. [Google Scholar] [CrossRef] [PubMed]
- Sarugaser, R.; Hanoun, L.; Keating, A.; Stanford, W.L.; Davies, J.E. Human mesenchymal stem cells self-renew and differentiate according to a deterministic hierarchy. PLoS ONE 2009, 4, e6498. [Google Scholar] [CrossRef]
- Mojsilović, S.; Krstić, A.; Ilić, V.; Okić-Đorđević, I.; Kocić, J.; Trivanović, D.; Santibañez, J.F.; Jovčić, G.; Bugarski, D. IL-17 and FGF signaling involved in mouse mesenchymal stem cell proliferation. Cell Tissue Res. 2011, 346, 305–316. [Google Scholar] [CrossRef]
- Lv, F.J.; Tuan, R.S.; Cheung, K.M.; Leung, V.Y. Concise review: The surface markers and identity of human mesenchymal stem cells. Stem Cells 2014, 32, 1408–1419. [Google Scholar] [CrossRef]
- Swamynathan, P.; Venugopal, P.; Kannan, S.; Thej, C.; Kolkundar, U.; Bhagwat, S.; Ta, M.; Majumdar, A.S.; Balasubramanian, S. Are serum-free and xeno-free culture conditions ideal for large scale clinical grade expansion of Wharton’s jelly derived mesenchymal stem cells? A comparative study. Stem Cell Res. Ther. 2014, 5, 88. [Google Scholar] [CrossRef]
- Cao, Y.; Boss, A.L.; Bolam, S.M.; Munro, J.T.; Crawford, H.; Dalbeth, N.; Poulsen, R.C.; Matthews, B.G. In Vitro Cell Surface Marker Expression on Mesenchymal Stem Cell Cultures does not Reflect Their Ex Vivo Phenotype. Stem Cell Rev. Rep. 2024, 20, 1656–1666. [Google Scholar] [CrossRef]
- Sanz-Rodriguez, F.; Guerrero-Esteo, M.; Botella, L.-M.; Banville, D.; Vary, C.P.H.; Bernabe, C. Endoglin Regulates Cytoskeletal Organization through Binding to ZRP-1, a Member of the Lim Family of Proteins. J. Biol. Chem. 2004, 279, 32858–32868. [Google Scholar] [CrossRef]
- Duff, S.E.; Li, C.; Garland, J.M.; Kumar, S. CD105 is important for angiogenesis: Evidence and potential applications. FASEB J. 2003, 17, 984–992. [Google Scholar] [CrossRef]
- Weiss, M.L.; Medicetty, S.; Bledsoe, A.R.; Rachakatla, R.S.; Choi, M.; Merchav, S.; Luo, Y.; Rao, M.S.; Velagaleti, G.; Troyer, D. Human umbilical cord matrix stem cells: Preliminary characterization and effect of transplantation in a rodent model of Parkinson’s disease. Stem Cells 2006, 24, 781–792. [Google Scholar] [CrossRef]
- Cardoso, T.C.; Okamura, L.H.; Baptistella, J.C.; Gameiro, R.; Ferreira, H.L.; Marinho, M.; Flores, E.F. Isolation, characterization and immunomodulatory-associated gene transcription of Wharton’s jelly-derived multipotent mesenchymal stromal cells at different trimesters of cow pregnancy. Cell Tissue Res. 2017, 367, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Corotchi, M.C.; Popa, M.A.; Remes, A.; Sima, L.E.; Gussi, I.; Lupu Plesu, M. Isolation method and xeno-free culture conditions influence multipotent differentiation capacity of human Wharton’s jelly-derived mesenchymal stem cells. Stem Cell Res. Ther. 2013, 4, 81. [Google Scholar] [CrossRef]
- Weiss, M.L.; Troyer, D.L. Stem cells in the umbilical cord. Stem Cell Rev. 2006, 2, 155–162. [Google Scholar] [CrossRef]
- Alatyyat, S.M.; Alasmari, H.M.; Aleid, O.A.; Abdel-Maksoud, M.S.; Elsherbiny, N. Umbilical cord stem cells: Background, processing and applications. Tissue Cell 2020, 65, 101351. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stange, K.; Wolter, T.; Fu, Z.; Burdeos, G.; Mideksa, Y.; Friese, A.; Röntgen, M. Isolation of Porcine Umbilical Cord Cells by Mechanical Tissue Dissociation Using a Tissue Grinder. Cells 2025, 14, 1425. https://doi.org/10.3390/cells14181425
Stange K, Wolter T, Fu Z, Burdeos G, Mideksa Y, Friese A, Röntgen M. Isolation of Porcine Umbilical Cord Cells by Mechanical Tissue Dissociation Using a Tissue Grinder. Cells. 2025; 14(18):1425. https://doi.org/10.3390/cells14181425
Chicago/Turabian StyleStange, Katja, Tessa Wolter, Zhenpei Fu, Gregor Burdeos, Yonatan Mideksa, Andreas Friese, and Monika Röntgen. 2025. "Isolation of Porcine Umbilical Cord Cells by Mechanical Tissue Dissociation Using a Tissue Grinder" Cells 14, no. 18: 1425. https://doi.org/10.3390/cells14181425
APA StyleStange, K., Wolter, T., Fu, Z., Burdeos, G., Mideksa, Y., Friese, A., & Röntgen, M. (2025). Isolation of Porcine Umbilical Cord Cells by Mechanical Tissue Dissociation Using a Tissue Grinder. Cells, 14(18), 1425. https://doi.org/10.3390/cells14181425





