PA2G4 Functions as a Cofactor for MYC Family Oncoproteins in MYC-Driven Malignancies
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Cell Viability
2.3. Western Blot
2.4. siRNA Transfection
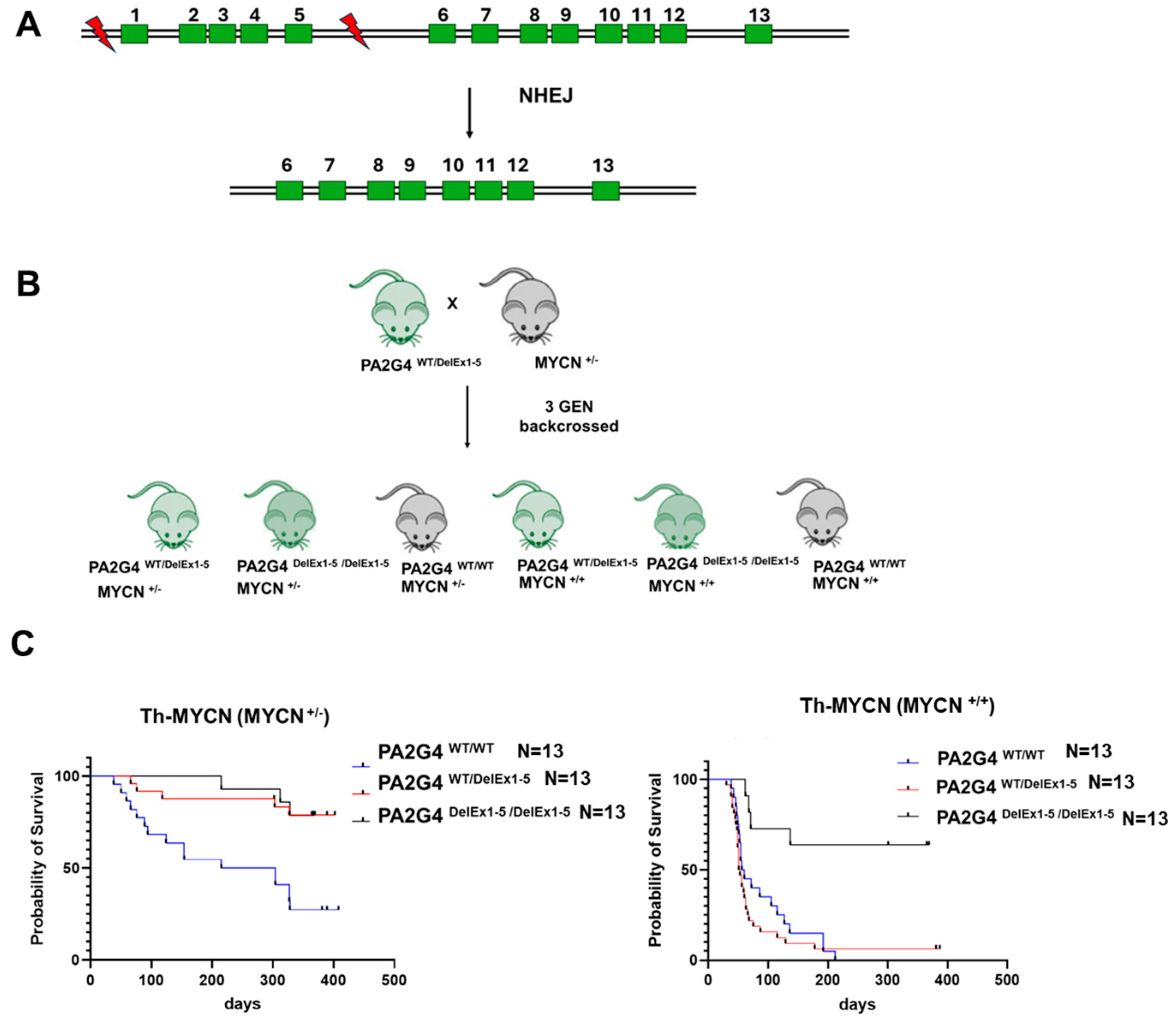
2.5. Animal Studies
2.6. Single-Cell and Bulk-RNA Sequencing Analyses of Publicly Available Datasets
2.7. Statistical Analysis
3. Results
3.1. Loss of PA2G4 Reduces MYCN-Driven Neuroblastoma Tumorigenesis In Vivo
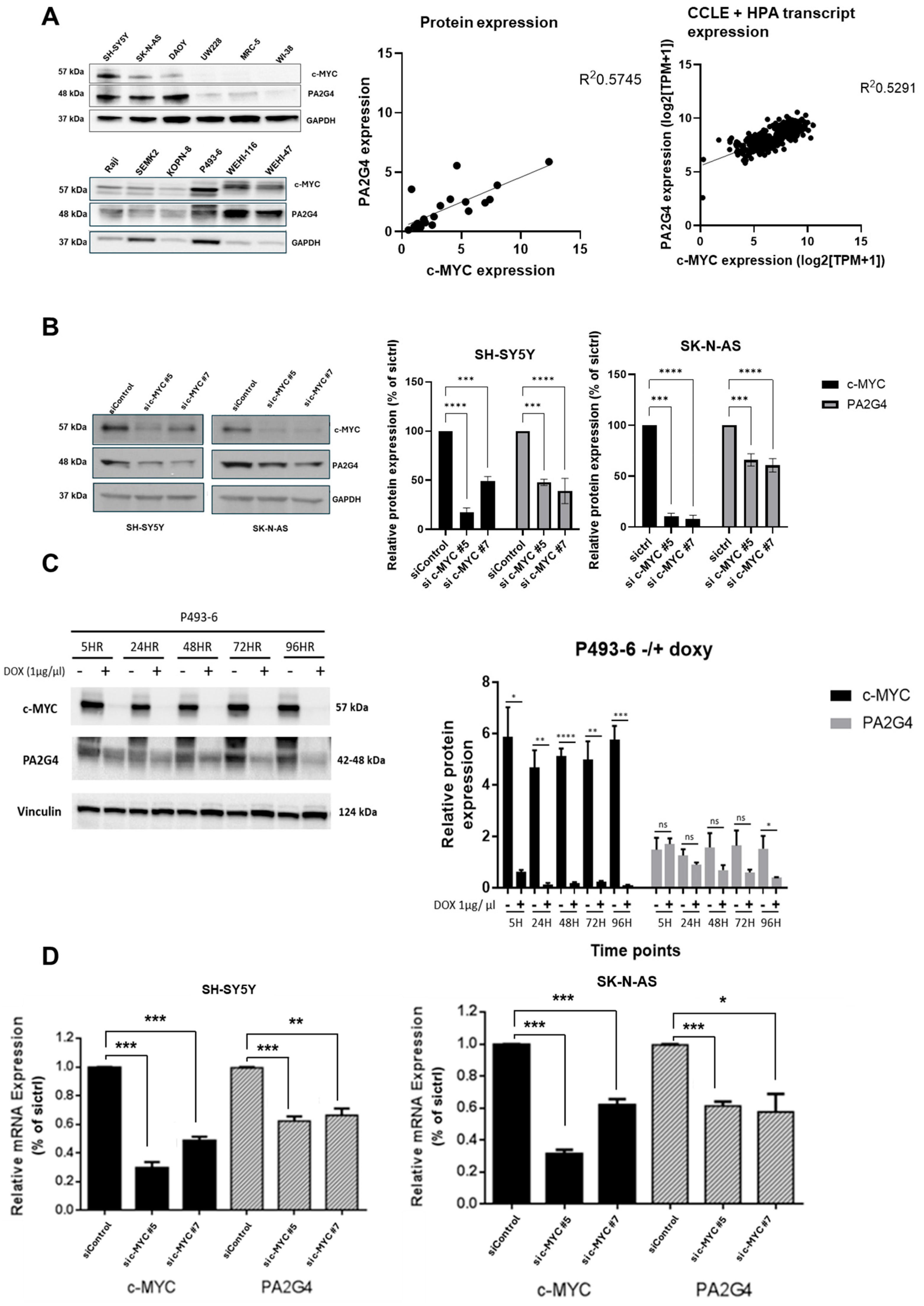
3.2. c-MYC Regulates PA2G4 Transcription
3.3. PA2G4 Stabilizes c-MYC Protein
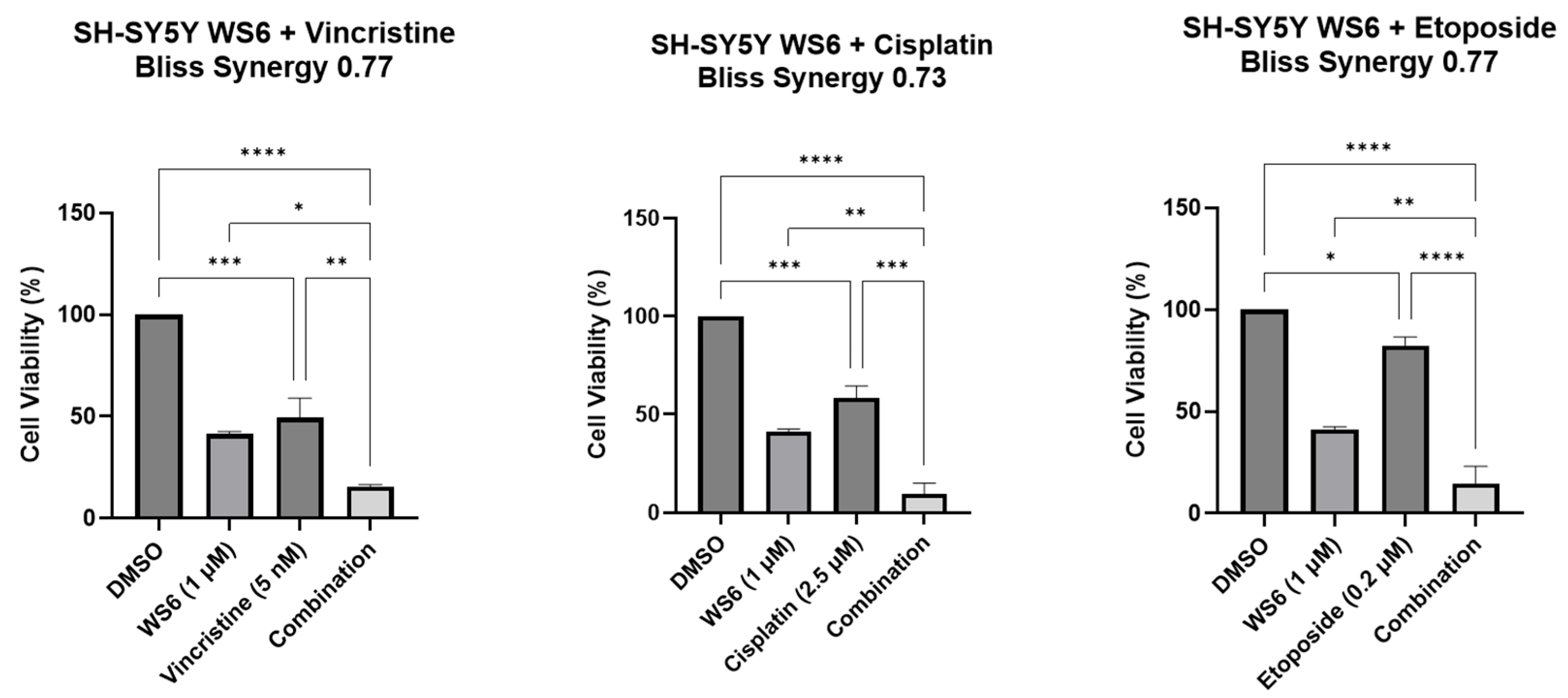
3.4. c-MYC Plays a ROLE in WS6-Induced Growth Inhibition in c-MYC-Driven Malignancies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kalkat, M.; De Melo, J.; Hickman, K.A.; Lourenco, C.; Redel, C.; Resetca, D.; Tamachi, A.; Tu, W.B.; Penn, L.Z. MYC Deregulation in Primary Human Cancers. Genes 2017, 8, 151. [Google Scholar] [CrossRef]
- Schick, M.; Habringer, S.; Nilsson, J.A.; Keller, U. Pathogenesis and therapeutic targeting of aberrant MYC expression in haematological cancers. Br. J. Haematol. 2017, 179, 724–738. [Google Scholar] [CrossRef]
- Chen, X.; Bahrami, A.; Pappo, A.; Easton, J.; Dalton, J.; Hedlund, E.; Ellison, D.; Shurtleff, S.; Wu, G.; Wei, L. Recurrent somatic structural variations contribute to tumorigenesis in pediatric osteosarcoma. Cell Rep. 2014, 7, 104–112. [Google Scholar] [CrossRef]
- Edmunds, L.R.; Sharma, L.; Kang, A.; Lu, J.; Vockley, J.; Basu, S.; Uppala, R.; Goetzman, E.S.; Beck, M.E.; Scott, D.; et al. c-Myc programs fatty acid metabolism and dictates acetyl-CoA abundance and fate. J. Biol. Chem. 2015, 290, 20100. [Google Scholar] [CrossRef]
- Chou, Y.T.; Lin, H.H.; Lien, Y.C.; Wang, Y.H.; Hong, C.F.; Kao, Y.R.; Lin, S.C.; Chang, Y.C.; Lin, S.Y.; Chen, S.J.; et al. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor ERF. Cancer Res. 2010, 70, 8822–8831. [Google Scholar] [CrossRef] [PubMed]
- Melnik, S.; Werth, N.; Boeuf, S.; Hahn, E.M.; Gotterbarm, T.; Anton, M.; Richter, W. Impact of c-MYC expression on proliferation, differentiation, and risk of neoplastic transformation of human mesenchymal stromal cells. Stem Cell Res. Ther. 2019, 10, 73. [Google Scholar] [CrossRef]
- Dang, C.V.; O’Donnell, K.A.; Zeller, K.I.; Nguyen, T.; Osthus, R.C.; Li, F. The c-Myc target gene network. Semin. Cancer Biol. 2006, 16, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Tavana, O.; Li, D.; Dai, C.; Lopez, G.; Banerjee, D.; Kon, N.; Chen, C.; Califano, A.; Yamashiro, D.J.; Sun, H.; et al. HAUSP deubiquitinates and stabilizes N-Myc in neuroblastoma. Nat. Med. 2016, 22, 1180–1186. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, J.R.; Soucek, L. The long journey to bring a Myc inhibitor to the clinic. J. Cell Biol. 2021, 220, e202103090. [Google Scholar] [CrossRef]
- Whitfield, J.R.; Soucek, L. MYC in cancer: From undruggable target to clinical trials. Nat. Rev. Drug Discov. 2025, 24, 445–457. [Google Scholar] [CrossRef]
- Berg, T.; Cohen, S.B.; Desharnais, J.; Sonderegger, C.; Maslyar, D.J.; Goldberg, J.; Boger, D.L.; Vogt, P.K. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc. Natl. Acad. Sci. USA 2002, 99, 3830–3835. [Google Scholar] [CrossRef] [PubMed]
- Koach, J.; Holien, J.K.; Massudi, H.; Carter, D.R.; Ciampa, O.C.; Herath, M.; Lim, T.; Seneviratne, J.A.; Milazzo, G.; Murray, J.E.; et al. Drugging MYCN Oncogenic Signaling through the MYCN-PA2G4 Binding Interface. Cancer Res. 2019, 79, 5652–5667. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, P.; Wang, Y.; Zhan, P.; Liu, C.; Mao, J.-H.; Wei, G. Distinct interactions of EBP1 isoforms with FBXW7 elicits different functions in cancer. Cancer Res. 2017, 77, 1983–1996. [Google Scholar] [CrossRef]
- Massudi, H.; Luo, J.S.; Holien, J.K.; Gadde, S.; Krishan, S.; Herath, M.; Koach, J.; Stevenson, B.W.; Gorman, M.A.; Venkat, P.; et al. Inhibitors of the Oncogenic PA2G4-MYCN Protein-Protein Interface. Cancers 2023, 15, 1822. [Google Scholar] [CrossRef]
- Zimmerman, M.W.; Liu, Y.; He, S.; Durbin, A.D.; Abraham, B.J.; Easton, J.; Shao, Y.; Xu, B.; Zhu, S.; Zhang, X.; et al. MYC Drives a Subset of High-Risk Pediatric Neuroblastomas and Is Activated through Mechanisms Including Enhancer Hijacking and Focal Enhancer Amplification. Cancer Discov. 2018, 8, 320–335. [Google Scholar] [CrossRef]
- Stanton, L.W.; Schwab, M.; Bishop, J.M. Nucleotide sequence of the human N-myc gene. Proc. Natl. Acad. Sci. USA 1986, 83, 1772–1776. [Google Scholar] [CrossRef]
- Weiss, W.A.; Aldape, K.; Mohapatra, G.; Feuerstein, B.G.; Bishop, J.M. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997, 16, 2985–2995. [Google Scholar] [CrossRef] [PubMed]
- Bonine, N.; Zanzani, V.; Van Hemelryk, A.; Vanneste, B.; Zwicker, C.; Thone, T.; Roelandt, S.; Bekaert, S.L.; Koster, J.; Janoueix-Lerosey, I.; et al. NBAtlas: A harmonized single-cell transcriptomic reference atlas of human neuroblastoma tumors. Cell Rep. 2024, 43, 114804. [Google Scholar] [CrossRef]
- Jansky, S.; Sharma, A.K.; Körber, V.; Quintero, A.; Toprak, U.H.; Wecht, E.M.; Gartlgruber, M.; Greco, A.; Chomsky, E.; Grünewald, T.G.P.; et al. Single-cell transcriptomic analyses provide insights into the developmental origins of neuroblastoma. Nat. Genet. 2021, 53, 683–693. [Google Scholar] [CrossRef]
- Jin, H.; Zhang, C.; Zwahlen, M.; von Feilitzen, K.; Karlsson, M.; Shi, M.; Yuan, M.; Song, X.; Li, X.; Yang, H.; et al. Systematic transcriptional analysis of human cell lines for gene expression landscape and tumor representation. Nat. Commun. 2023, 14, 5417. [Google Scholar] [CrossRef]
- Monaco, G.; Lee, B.; Xu, W.; Mustafah, S.; Hwang, Y.Y.; Carré, C.; Burdin, N.; Visan, L.; Ceccarelli, M.; Poidinger, M.; et al. RNA-Seq Signatures Normalized by mRNA Abundance Allow Absolute Deconvolution of Human Immune Cell Types. Cell Rep. 2019, 26, 1627–1640.e7. [Google Scholar] [CrossRef]
- Marchesini, M.; Gherli, A.; Simoncini, E.; Tor, L.M.D.; Montanaro, A.; Thongon, N.; Vento, F.; Liverani, C.; Cerretani, E.; D’Antuono, A.; et al. Orthogonal proteogenomic analysis identifies the druggable PA2G4-MYC axis in 3q26 AML. Nat. Commun. 2024, 15, 4739. [Google Scholar] [CrossRef]
- Pajic, A.; Spitkovsky, D.; Christoph, B.; Kempkes, B.; Schuhmacher, M.; Staege, M.S.; Brielmeier, M.; Ellwart, J.; Kohlhuber, F.; Bornkamm, G.W.; et al. Cell cycle activation by c-myc in a burkitt lymphoma model cell line. Int. J. Cancer 2000, 87, 787–793. [Google Scholar] [CrossRef]
- Pearson, A.D.; Pinkerton, C.R.; Lewis, I.J.; Imeson, J.; Ellershaw, C.; Machin, D.; European Neuroblastoma Study, G.; Children’s, C.; Leukaemia, G. High-dose rapid and standard induction chemotherapy for patients aged over 1 year with stage 4 neuroblastoma: A randomised trial. Lancet Oncol. 2008, 9, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Ladenstein, R.; Valteau-Couanet, D.; Brock, P.; Yaniv, I.; Castel, V.; Laureys, G.; Malis, J.; Papadakis, V.; Lacerda, A.; Ruud, E.; et al. Randomized Trial of prophylactic granulocyte colony-stimulating factor during rapid COJEC induction in pediatric patients with high-risk neuroblastoma: The European HR-NBL1/SIOPEN study. J. Clin. Oncol. 2010, 28, 3516–3524. [Google Scholar] [CrossRef] [PubMed]
- Kushner, B.H.; Kramer, K.; Modak, S.; Qin, L.X.; Cheung, N.K. Differential impact of high-dose cyclophosphamide, topotecan, and vincristine in clinical subsets of patients with chemoresistant neuroblastoma. Cancer 2010, 116, 3054–3060. [Google Scholar] [CrossRef]
- Bliss, C.I. The Toxicity of Poisons Applied Jointly. Ann. Appl. Biol. 1939, 26, 585–615. [Google Scholar] [CrossRef]
- McKeown, M.R.; Bradner, J.E. Therapeutic strategies to inhibit MYC. Cold Spring Harb. Perspect. Med. 2014, 4, a014266. [Google Scholar] [CrossRef]
- Carabet, L.A.; Rennie, P.S.; Cherkasov, A. Therapeutic inhibition of Myc in cancer. Structural bases and computer-aided drug discovery approaches. Int. J. Mol. Sci. 2018, 20, 120. [Google Scholar] [CrossRef]
- Llombart, V.; Mansour, M.R. Therapeutic targeting of “undruggable” MYC. eBioMedicine 2022, 75, 103756. [Google Scholar] [CrossRef]
- Han, H.; Jain, A.D.; Truica, M.I.; Izquierdo-Ferrer, J.; Anker, J.F.; Lysy, B.; Sagar, V.; Luan, Y.; Chalmers, Z.R.; Unno, K.; et al. Small-Molecule MYC Inhibitors Suppress Tumor Growth and Enhance Immunotherapy. Cancer Cell 2019, 36, 483–497.e15. [Google Scholar] [CrossRef]
- Truica, M.I.; Burns, M.C.; Han, H.; Abdulkadir, S.A. Turning Up the Heat on MYC: Progress in Small-Molecule Inhibitors. Cancer Res. 2021, 81, 248–253. [Google Scholar] [CrossRef]
- Nguyen, D.Q.; Hoang, D.H.; Nguyen, T.T.V.; Ho, H.D.; Huynh, V.; Shin, J.H.; Ly, Q.T.; Thi Nguyen, D.D.; Ghoda, L.; Marcucci, G. Ebp1 p48 promotes oncogenic activities in human colon cancer cells through regulation of TIF-90-mediated ribosomal RNA synthesis. J. Cell. Physiol. 2019, 234, 17612–17621. [Google Scholar] [CrossRef]
- Mei, Y.; Zhang, P.; Zuo, H.; Clark, D.; Xia, R.; Li, J.; Liu, Z.; Mao, L. Ebp1 activates podoplanin expression and contributes to oral tumorigenesis. Oncogene 2014, 33, 3839–3850. [Google Scholar] [CrossRef] [PubMed]
- Squatrito, M.; Mancino, M.; Donzelli, M.; Areces, L.B.; Draetta, G.F. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene 2004, 23, 4454–4465. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, B.W.; Gorman, M.A.; Koach, J.; Cheung, B.B.; Marshall, G.M.; Parker, M.W.; Holien, J.K. A structural view of PA2G4 isoforms with opposing functions in cancer. J. Biol. Chem. 2020, 295, 16100–16112. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krishan, S.; Koach, J.; Lim, T.; Yeo, K.; Cheong, F.; Luo, J.-S.; Massudi, H.; Gao, X.; Heangsarath, S.; Kueh, A.J.; et al. PA2G4 Functions as a Cofactor for MYC Family Oncoproteins in MYC-Driven Malignancies. Cells 2025, 14, 1422. https://doi.org/10.3390/cells14181422
Krishan S, Koach J, Lim T, Yeo K, Cheong F, Luo J-S, Massudi H, Gao X, Heangsarath S, Kueh AJ, et al. PA2G4 Functions as a Cofactor for MYC Family Oncoproteins in MYC-Driven Malignancies. Cells. 2025; 14(18):1422. https://doi.org/10.3390/cells14181422
Chicago/Turabian StyleKrishan, Sukriti, Jessica Koach, Taylor Lim, Kenny Yeo, Faith Cheong, Jie-Si Luo, Hassina Massudi, Xiaomian Gao, Sopheakwealthy Heangsarath, Andrew J. Kueh, and et al. 2025. "PA2G4 Functions as a Cofactor for MYC Family Oncoproteins in MYC-Driven Malignancies" Cells 14, no. 18: 1422. https://doi.org/10.3390/cells14181422
APA StyleKrishan, S., Koach, J., Lim, T., Yeo, K., Cheong, F., Luo, J.-S., Massudi, H., Gao, X., Heangsarath, S., Kueh, A. J., Herold, M. J., Cheung, B. B., & Marshall, G. M. (2025). PA2G4 Functions as a Cofactor for MYC Family Oncoproteins in MYC-Driven Malignancies. Cells, 14(18), 1422. https://doi.org/10.3390/cells14181422






