Efficacy of Conventional and Novel Tyrosine Kinase Inhibitors for Uncommon EGFR Mutations—An In Vitro Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection from the cBioPortal Database
2.2. Cell Lines and Reagents
2.3. Establishment of Ba/F3 Cells Harboring EGFR L861Q Mutation
2.4. Growth Inhibition Assay
2.5. Establishment of Resistant Clones to Afatinib and Osimertinib
3. Results
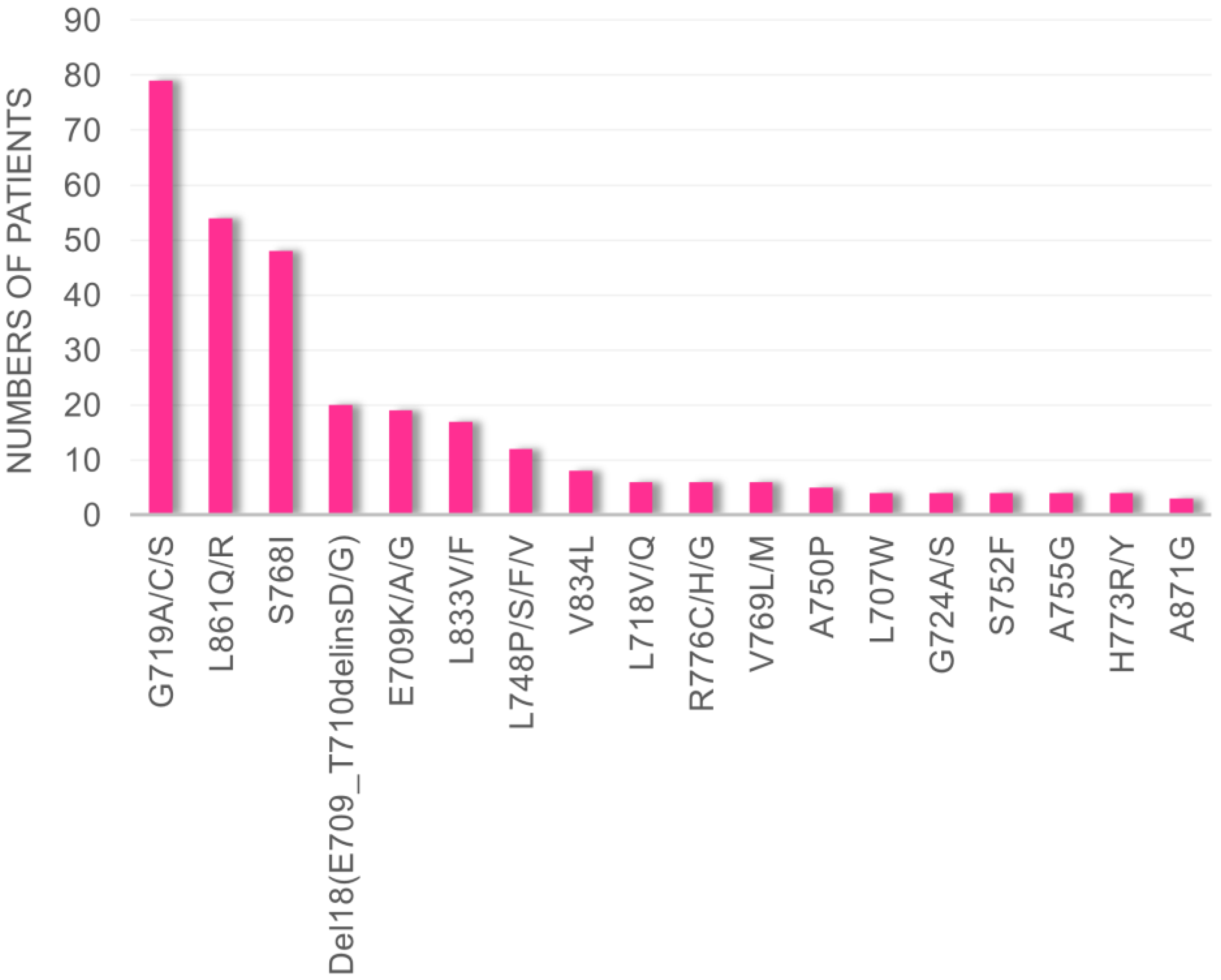
3.1. Frequency of Uncommon EGFR Mutations in cBioPortal Database
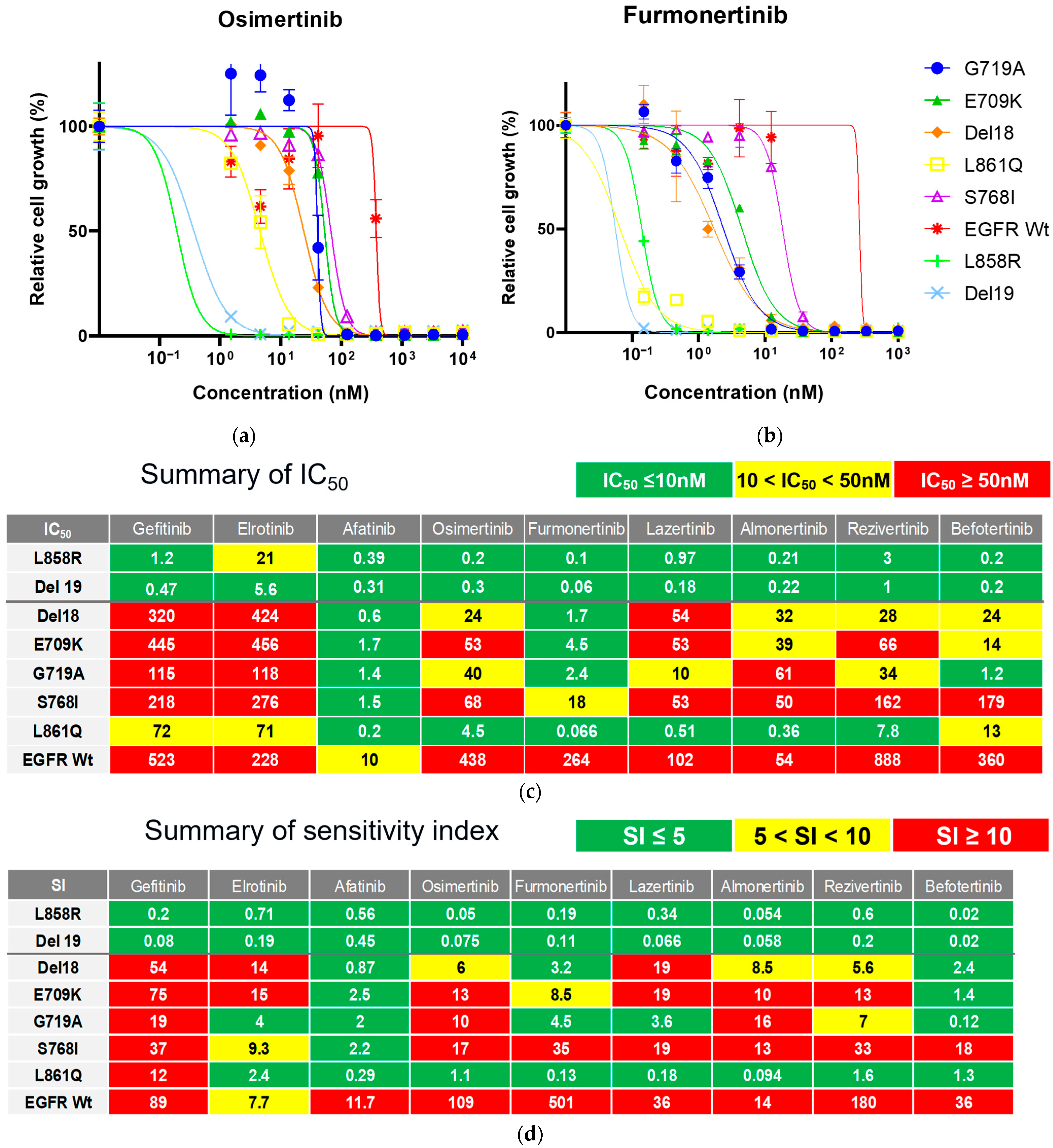
3.2. Efficacy of Novel TKIs Against Uncommon EGFR Mutations
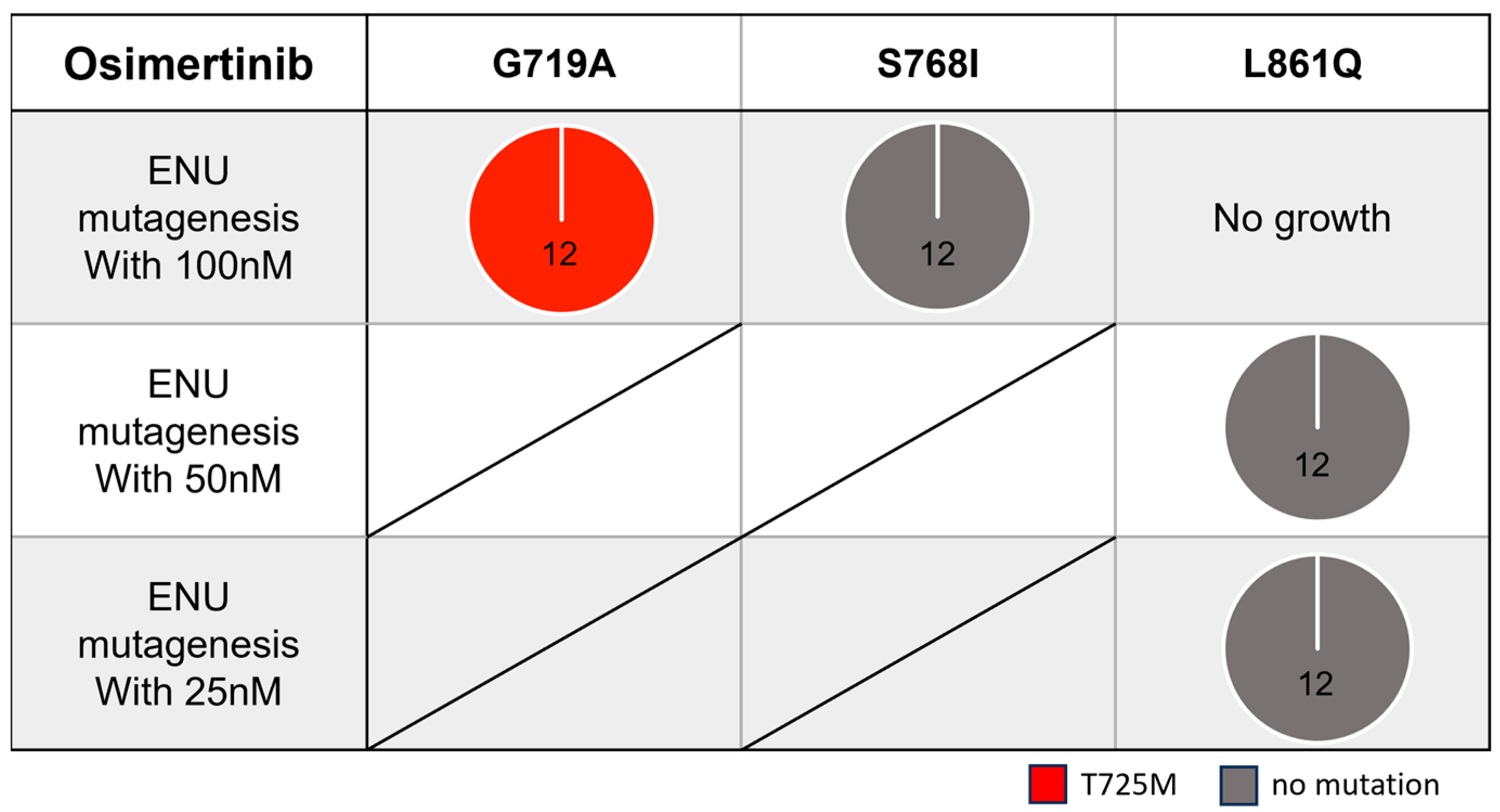
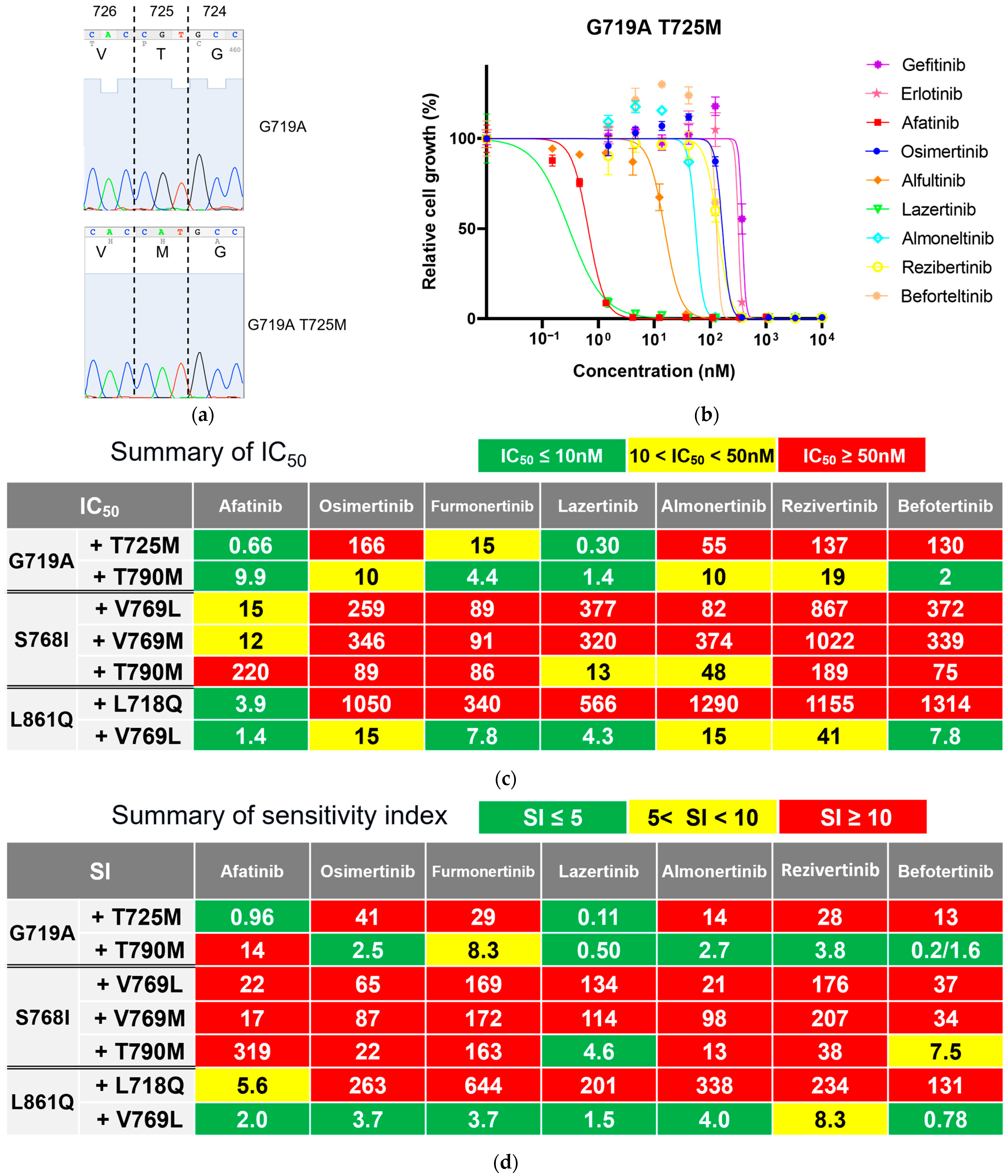
3.3. Secondary Resistance Mutations to Osimertinib and Strategies to Overcome Resistance
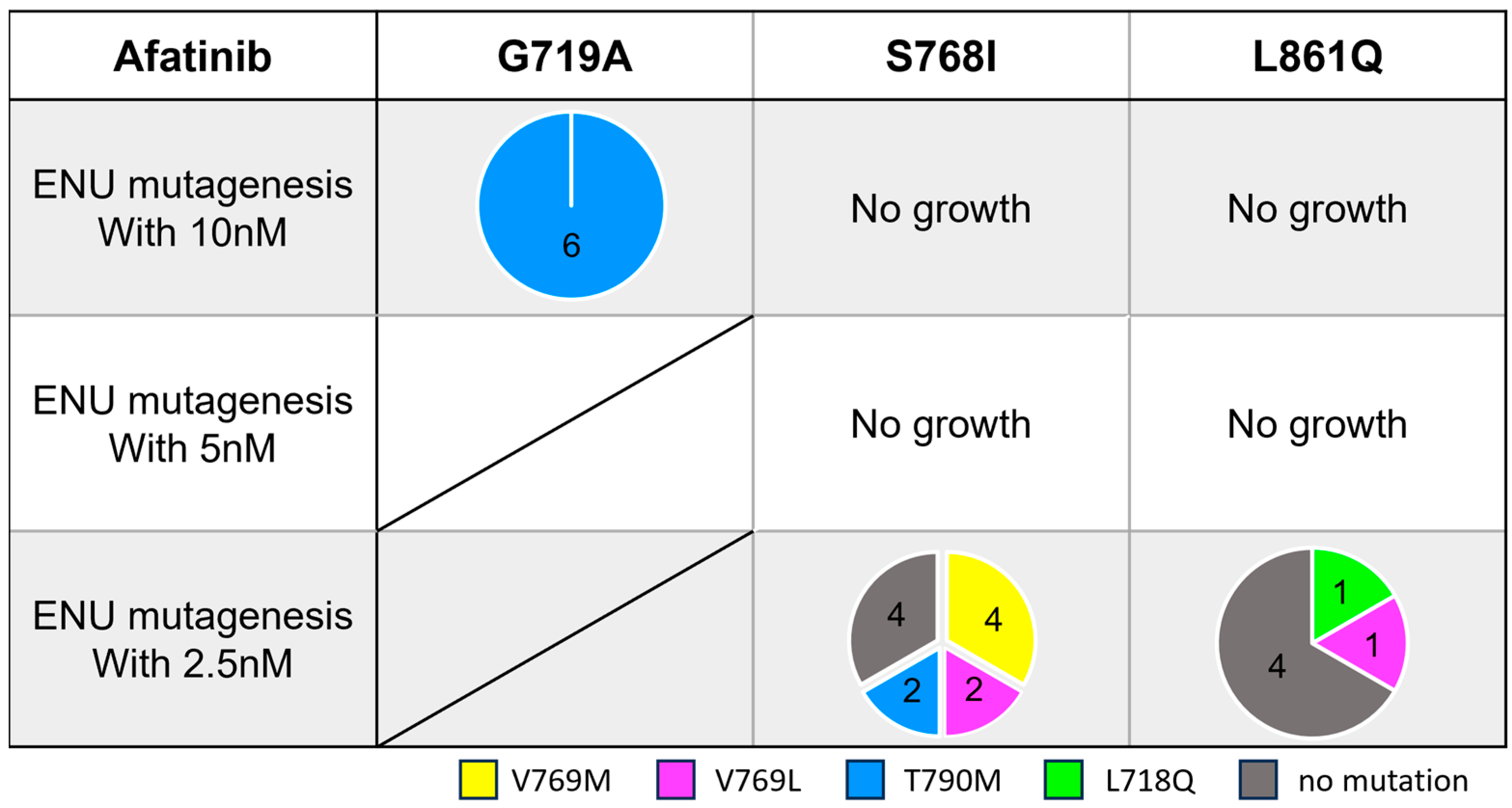
3.4. Secondary Resistance Mutations to Afatinib and Strategies to Overcome Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EGFR | Epidermal growth factor receptor |
| ENU | N-ethyl-N-nitrosourea |
| IC50 | Half maximal inhibitory concentration |
| NSCLC | Non-small cell lung cancer |
| PFS | Progression-free survival |
| SI | Sensitivity index |
| TKI | Tyrosine kinase inhibitor |
| 1G | First-generation |
| 2G | Second-generation |
| 3G | Third-generation |
References
- Shigematsu, H.; Lin, L.; Takahashi, T.; Nomura, M.; Suzuki, M.; Wistuba, I.I.; Fong, F.M.; Lee, H.; Toyooka, S.; Shimizu, N.; et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J. Natl. Cancer Inst. 2005, 97, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Sun, J.; Zhang, Y.; Hesilaiti, N.; Xia, Q.; Cui, H.; Fan, N.; Xu, X. Structure-Guided Strategies of Targeted Therapies for Patients with EGFR-Mutant Non-Small Cell Lung Cancer. Biomolecules 2023, 13, 210. [Google Scholar] [CrossRef] [PubMed]
- Suda, K.; Mitsudomi, T.; Shintani, Y.; Okami, J.; Ito, H.; Ohtsuka, T.; Toyooka, S.; Mori, T.; Watanabe, S.I.; Asamura, H.; et al. Clinical Impacts of EGFR Mutation Status: Analysis of 5780 Surgically Resected Lung Cancer Cases. Ann. Thorac. Surg. 2021, 111, 269–276. [Google Scholar] [CrossRef]
- Gomez-Randulfe, I.; Scanlon, L.A.; Carter, M.; Moliner, L.; Cil, E.; Califano, R.; Summers, Y.; Blackhall, F.; Lindsay, C.R.; Lewis, J.; et al. First-line osimertinib compared to earlier generation TKIs in advanced EGFR-mutant NSCLC: A real-world survival analysis. Lung Cancer 2025, 200, 108084. [Google Scholar] [CrossRef]
- Robichaux, J.P.; Le, X.; Vijayan, R.S.K.; Hicks, J.K.; Heeke, S.; Elamin, Y.Y.; Lin, H.Y.; Udagawa, H.; Skoulidis, F.; Tran, H.; et al. Structure-based classification predicts drug response in EGFR-mutant NSCLC. Nature 2021, 597, 732–737. [Google Scholar] [CrossRef]
- Yang, J.C.; Sequist, L.V.; Geater, S.L.; Tsai, C.M.; Mok, T.S.; Schuler, M.; Yamamoto, N.; Yu, C.J.; Ou, S.H.; Zhou, C.; et al. Clinical activity of afatinib in patients with advanced non-small-cell lung cancer harbouring uncommon EGFR mutations: A combined post-hoc analysis of LUX-Lung 2, LUX-Lung 3, and LUX-Lung 6. Lancet Oncol. 2015, 16, 830–838. [Google Scholar] [CrossRef]
- Miura, H.T.S.; Misumi, T. LBA66 Afatinib versus chemotherapy for treatment-naïve non-small cell lung cancer with a sensitizing uncommon epidermal growth factor receptor mutation: A phase III study (ACHILLES/TORG1834). Ann. Oncol. 2023, 34, S1310–S1311. [Google Scholar] [CrossRef]
- Okuma, Y.; Kubota, K.; Shimokawa, M.; Hashimoto, K.; Kawashima, Y.; Sakamoto, T.; Wakui, H.; Murakami, S.; Okishio, K.; Hayashihara, K.; et al. First-Line Osimertinib for Previously Untreated Patients with NSCLC and Uncommon EGFR Mutations: The UNICORN Phase 2 Nonrandomized Clinical Trial. JAMA Oncol. 2024, 10, 43–51. [Google Scholar] [CrossRef]
- Cho, J.H.; Lim, S.H.; An, H.J.; Kim, K.H.; Park, K.U.; Kang, E.J.; Choi, Y.H.; Ahn, M.S.; Lee, M.H.; Sun, J.M.; et al. Osimertinib for Patients with Non-Small-Cell Lung Cancer Harboring Uncommon EGFR Mutations: A Multicenter, Open-Label, Phase II Trial (KCSG-LU15–09). J. Clin. Oncol. 2020, 38, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Catherine, A.; Shu, K.G.; Cho, B.C.; Griesinger, F.; Yang, J.C.-H.; Felip, E.; Xie, J.; Chen, J.; Mahoney, J.; Thayu, M.; et al. CHRYSALIS-2: A phase 1/1b study of lazertinib as monotherapy and in combination with amivantamab in patients with EGFR-mutant NSCLC. J. Clin. Oncol. 2021, 39, TPS9132. [Google Scholar] [CrossRef]
- Banno, E.; Togashi, Y.; Nakamura, Y.; Chiba, M.; Kobayashi, Y.; Hayashi, H.; Terashima, M.; de Velasco, M.A.; Sakai, K.; Fujita, Y.; et al. Sensitivities to various epidermal growth factor receptor-tyrosine kinase inhibitors of uncommon epidermal growth factor receptor mutations L861Q and S768I: What is the optimal epidermal growth factor receptor-tyrosine kinase inhibitor? Cancer Sci. 2016, 107, 1134–1140. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Togashi, Y.; Yatabe, Y.; Mizuuchi, H.; Jangchul, P.; Kondo, C.; Shimoji, M.; Sato, K.; Suda, K.; Tomizawa, K.; et al. EGFR Exon 18 Mutations in Lung Cancer: Molecular Predictors of Augmented Sensitivity to Afatinib or Neratinib as Compared with First- or Third-Generation TKIs. Clin. Cancer Res. 2015, 21, 5305–5313. [Google Scholar] [CrossRef]
- Koga, T.; Suda, K.; Mitsudomi, T. Utility of the Ba/F3 cell system for exploring on-target mechanisms of resistance to targeted therapies for lung cancer. Cancer Sci. 2022, 113, 815–827. [Google Scholar] [CrossRef]
- Bradeen, H.A.; Eide, C.A.; O’Hare, T.; Johnson, K.J.; Willis, S.G.; Lee, F.Y.; Druker, B.J.; Deininger, M.W. Comparison of imatinib mesylate, dasatinib (BMS-354825), and nilotinib (AMN107) in an N-ethyl-N-nitrosourea (ENU)-based mutagenesis screen: High efficacy of drug combinations. Blood 2006, 108, 2332–2338. [Google Scholar] [CrossRef]
- Suda, K.; Murakami, I.; Katayama, T.; Tomizawa, K.; Osada, H.; Sekido, Y.; Maehara, Y.; Yatabe, Y.; Mitsudomi, T. Reciprocal and complementary role of MET amplification and EGFR T790M mutation in acquired resistance to kinase inhibitors in lung cancer. Clin. Cancer Res. 2010, 16, 5489–5498. [Google Scholar] [CrossRef]
- Nakagawa, K.; Tamura, T.; Negoro, S.; Kudoh, S.; Yamamoto, N.; Yamamoto, N.; Takeda, K.; Swaisland, H.; Nakanishi, I.; Hirose, M.; et al. Phase I pharmacokinetic trial of the selective oral epidermal growth factor receptor tyrosine kinase inhibitor gefitinib (‘Iressa’, ZD1839) in Japanese patients with solid malignant tumors. Ann. Oncol. 2003, 14, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, M.; Siu, L.L.; Nemunaitis, J.; Rizzo, J.; Hammond, L.A.; Takimoto, C.; Eckhardt, S.G.; Tolcher, A.; Britten, C.D.; Denis, L.; et al. Phase I and pharmacologic study of OSI-774, an epidermal growth factor receptor tyrosine kinase inhibitor, in patients with advanced solid malignancies. J. Clin. Oncol. 2001, 19, 3267–3279. [Google Scholar] [CrossRef] [PubMed]
- Yap, T.A.; Vidal, L.; Adam, J.; Stephens, P.; Spicer, J.; Shaw, H.; Ang, J.; Temple, G.; Bell, S.; Shahidi, M.; et al. Phase I trial of the irreversible EGFR and HER2 kinase inhibitor BIBW 2992 in patients with advanced solid tumors. J. Clin. Oncol. 2010, 28, 3965–3972. [Google Scholar] [CrossRef] [PubMed]
- Tsubata, Y.; Watanabe, K.; Saito, R.; Nakamura, A.; Yoshioka, H.; Morita, M.; Honda, R.; Kanaji, N.; Ohizumi, S.; Jingu, D.; et al. Osimertinib in poor performance status patients with T790M-positive advanced non-small-cell lung cancer after progression of first- and second-generation EGFR-TKI treatments (NEJ032B). Int. J. Clin. Oncol. 2022, 27, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Jänne, P.A.; Yang, J.C.; Kim, D.W.; Planchard, D.; Ohe, Y.; Ramalingam, S.S.; Ahn, M.J.; Kim, S.W.; Su, W.C.; Horn, L.; et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 1689–1699. [Google Scholar] [CrossRef] [PubMed]
- Kruithof, P.D.; Brouns, A.; Degens, J.; Hendriks, L.E.L.; Wong, D.R.; van Geel, R.; Croes, S. Osimertinib Dose Optimization Guided by Therapeutic Drug Monitoring in Patients with EGFR-Mutated Advanced Non-Small Cell Lung Cancer: A Case series. Clin. Lung Cancer 2025, in press. [Google Scholar] [CrossRef]
- Wu, Y.L.; Xue, Y.R.; Guo, Z.T.; Chen, Z.D.; Ge, X.Y.; Zhong, D.F.; Diao, X.X. Furmonertinib (Alflutinib, AST2818) is a potential positive control drug comparable to rifampin for evaluation of CYP3A4 induction in sandwich-cultured primary human hepatocytes. Acta Pharmacol. Sin. 2022, 43, 747–756. [Google Scholar] [CrossRef]
- Ahn, M.J.; Han, J.Y.; Lee, K.H.; Kim, S.W.; Kim, D.W.; Lee, Y.G.; Cho, E.K.; Kim, J.H.; Lee, G.W.; Lee, J.S.; et al. Lazertinib in patients with EGFR mutation-positive advanced non-small-cell lung cancer: Results from the dose escalation and dose expansion parts of a first-in-human, open-label, multicentre, phase 1–2 study. Lancet Oncol. 2019, 20, 1681–1690. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.C.; Han, J.Y.; Kim, S.W.; Lee, K.H.; Cho, E.K.; Lee, Y.G.; Kim, D.W.; Kim, J.H.; Lee, G.W.; Lee, J.S.; et al. A Phase 1/2 Study of Lazertinib 240 mg in Patients with Advanced EGFR T790M-Positive NSCLC After Previous EGFR Tyrosine Kinase Inhibitors. J. Thorac. Oncol. 2022, 17, 558–567. [Google Scholar] [CrossRef]
- Yang, J.C.; Camidge, D.R.; Yang, C.T.; Zhou, J.; Guo, R.; Chiu, C.H.; Chang, G.C.; Shiah, H.S.; Chen, Y.; Wang, C.C.; et al. Safety, Efficacy, and Pharmacokinetics of Almonertinib (HS-10296) in Pretreated Patients with EGFR-Mutated Advanced NSCLC: A Multicenter, Open-label, Phase 1 Trial. J. Thorac. Oncol. 2020, 15, 1907–1918. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, Y.; Yang, S.; Zhou, J.; Zhang, L.; Chen, G.; Fang, J.; Zhu, B.; Li, X.; Shu, Y.; et al. Safety, Efficacy, and Pharmacokinetics of Rezivertinib (BPI-7711) in Patients with Advanced NSCLC with EGFR T790M Mutation: A Phase 1 Dose-Escalation and Dose-Expansion Study. J. Thorac. Oncol. 2022, 17, 708–717. [Google Scholar] [CrossRef]
- Shi, Y.; Wu, S.; Wang, K.; Cang, S.; Yao, W.; Fan, Y.; Wu, L.; Huang, M.; Li, X.; Pan, Y.; et al. Efficacy and Safety of Rezivertinib (BPI-7711) in Patients with Locally Advanced or Metastatic/Recurrent EGFR T790M-Mutated NSCLC: A Phase 2b Study. J. Thorac. Oncol. 2022, 17, 1306–1317. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zhang, G.; Zhou, J.; Cang, S.; Cheng, Y.; Wu, G.; Cao, P.; Lv, D.; Jian, H.; et al. Efficacy and Safety of Befotertinib (D-0316) in Patients with EGFR T790M-Mutated NSCLC That Had Progressed After Prior EGFR Tyrosine Kinase Inhibitor Therapy: A Phase 2, Multicenter, Single-Arm, Open-Label Study. J. Thorac. Oncol. 2022, 17, 1192–1204. [Google Scholar] [CrossRef]
- Lu, S.; Zhou, J.; Jian, H.; Wu, L.; Cheng, Y.; Fan, Y.; Fang, J.; Chen, G.; Zhang, Z.; Lv, D.; et al. Befotertinib (D-0316) versus icotinib as first-line therapy for patients with EGFR-mutated locally advanced or metastatic non-small-cell lung cancer: A multicentre, open-label, randomised phase 3 study. Lancet Respir. Med. 2023, 11, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.C.; Lim, C.K.; Chang, L.Y.; Chen, K.Y.; Shih, J.Y.; Yu, C.J. Non-small cell lung cancer harbouring non-resistant uncommon EGFR mutations: Mutation patterns, effectiveness of epidermal growth factor receptor-tyrosine kinase inhibitors and prognostic factors. Eur. J. Cancer 2019, 119, 77–86. [Google Scholar] [CrossRef]
- Tanaka, I.; Morise, M.; Kodama, Y.; Matsui, A.; Ozawa, N.; Ozone, S.; Goto, D.; Miyazawa, A.; Hase, T.; Hashimoto, N.; et al. Potential for afatinib as an optimal treatment for advanced non-small cell lung carcinoma in patients with uncommon EGFR mutations. Lung Cancer 2019, 127, 169–171. [Google Scholar] [CrossRef]
- Yang, J.C.; Schuler, M.; Popat, S.; Miura, S.; Heeke, S.; Park, K.; Märten, A.; Kim, E.S. Afatinib for the Treatment of NSCLC Harboring Uncommon EGFR Mutations: A Database of 693 Cases. J. Thorac. Oncol. 2020, 15, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhao, K.; Hu, S.; Dong, W.; Gong, Y.; Xie, C. Clinical Outcomes of Afatinib Versus Osimertinib in Patients with Non-Small Cell Lung Cancer with Uncommon EGFR Mutations: A Pooled Analysis. Oncologist 2023, 28, e397–e405. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.F.; Bu, Q.; Wang, Q.; Zhao, W.; Wang, L.; Dong, X.; Chen, P.; Wen, Z.; Jia, J.; Jiang, G.; et al. 57P: Safety and efficacy of aumolertinib treatment in patients with advanced NSCLC harboring 20ins and uncommon EGFR mutations (AIM). J. Thorac. Oncol. 2025, 20 (Suppl. 1), S45–S46. [Google Scholar] [CrossRef]
- Planchard, D.; Le, X.; Yu, Y.; Zhao, Y. FURTHER (FURMO-002): A Global, Randomized Study of Firmonertinib at Two Dose Levels in TKI-Naïve, Advanced NSCLC with EGFR PACC Mutations. In Proceedings of the 2024 World Conference on Lung Cancer, San Diego, CA, USA, 7 October 2024. [Google Scholar]
- ManChon, U.; Talevich, E.; Katiyar, S.; Rasheed, K.; Kannan, N. Prediction and prioritization of rare oncogenic mutations in the cancer Kinome using novel features and multiple classifiers. PLoS Comput. Biol. 2014, 10, e1003545. [Google Scholar]
- Lin, Q.L.; Ran, C.; Ou, Q.X.; Ma, Y.T.; Bao, H.; Wu, X.; Shao, Y.; Wang, Z.X. Acquired rare recurrent EGFR mutations as mechanisms of resistance to Osimertinib in lung cancer and in silico structural modelling. Am. J. Cancer Res. 2020, 10, 4005. [Google Scholar]
- Janning, M.; Süptitz, J.; Albers-Leischner, C.; Delpy, P.; Tufman, A.; Velthaus-Rusik, J.L.; Reck, M.; Jung, A.; Kauffmann-Guerrero, D.; Bonzheim, I.; et al. Treatment outcome of atypical EGFR mutations in the German National Network Genomic Medicine Lung Cancer (nNGM). Ann. Oncol. 2022, 33, 602–615. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Hayashi, T.; Reva, B.; Yu, H.A.; Riely, G.J.; Adusumilli, P.S.; Travis, W.D.; Wilkins, O.; Bramletta, N.; Chandramohan, R.; et al. Identification and Functional Characterization of EGFR V769M, a Novel Germline Variant Associated with Multiple Lung Adenocarcinomas. JCO Precis. Oncol. 2017, 1, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Peng, M.; Chen, Q.; Yuan, M.; Chen, R.; Deng, H.; Deng, J.; Liu, O.; Weng, Y.; Chen, M.; et al. Identification of the Unique Clinical and Genetic Features of Chinese Lung Cancer Patients with EGFR Germline Mutations in a Large-Scale Retrospective Study. Front. Oncol. 2021, 11, 774156. [Google Scholar] [CrossRef]
- Huang, S.F.; Liu, H.P.; Li, L.H.; Ku, Y.C.; Fu, Y.N.; Tsai, H.Y.; Chen, Y.T.; Lin, Y.F.; Chang, W.C.; Kuo, H.P.; et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non-small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin. Cancer Res. 2004, 10, 8195–8203. [Google Scholar] [CrossRef]
- Wu, J.Y.; Yu, C.J.; Chang, Y.C.; Yang, C.H.; Shih, J.Y.; Yang, P.C. Effectiveness of tyrosine kinase inhibitors on “uncommon” epidermal growth factor receptor mutations of unknown clinical significance in non-small cell lung cancer. Clin. Cancer Res. 2011, 17, 3812–3821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shao, Y.W.; Xia, Y. Responsiveness to Full-Dose Afatinib in a Patient with Lung Adenocarcinoma Harboring EGFR S768I and V769L Mutations. J. Thorac. Oncol. 2019, 14, e25–e27. [Google Scholar] [CrossRef] [PubMed]
- Asahina, H.; Yamazaki, K.; Kinoshita, I.; Yokouchi, H.; Dosaka-Akita, H.; Nishimura, M. Non-responsiveness to gefitinib in a patient with lung adenocarcinoma having rare EGFR mutations S768I and V769L. Lung Cancer 2006, 54, 419–422. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Niogret, J.; Coudert, B.; Boidot, R. Primary Resistance to Afatinib in a Patient with Lung Adenocarcinoma Harboring Uncommon EGFR Mutations: S768I and V769L. J. Thorac. Oncol. 2018, 13, e113. [Google Scholar] [CrossRef]
- Deng, Q.; Xie, B.; Wu, L.; Ji, X.; Li, C.; Feng, L.; Fang, Q.; Bao, Y.; Li, J.; Jin, S.; et al. Competitive evolution of NSCLC tumor clones and the drug resistance mechanism of first-generation EGFR-TKIs in Chinese NSCLC patients. Heliyon 2018, 4, e01031. [Google Scholar] [CrossRef]
- Simionato, F.; Calvetti, L.; Cosci, M.; Scarparo, S.; Aprile, G. Case Report: A Metabolic Complete Response to Upfront Osimertinib in a Smoker Non-Small Cell Lung Cancer Patient Harbouring EGFR G719A/V769M Complex Mutation. Onco Targets Ther. 2020, 13, 12027–12031. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oiki, H.; Suda, K.; Hamada, A.; Fujino, T.; Obata, K.; Kobayashi, Y.; Sakai, K.; Fukuda, S.; Ohara, S.; Ito, M.; et al. Efficacy of Conventional and Novel Tyrosine Kinase Inhibitors for Uncommon EGFR Mutations—An In Vitro Study. Cells 2025, 14, 1386. https://doi.org/10.3390/cells14171386
Oiki H, Suda K, Hamada A, Fujino T, Obata K, Kobayashi Y, Sakai K, Fukuda S, Ohara S, Ito M, et al. Efficacy of Conventional and Novel Tyrosine Kinase Inhibitors for Uncommon EGFR Mutations—An In Vitro Study. Cells. 2025; 14(17):1386. https://doi.org/10.3390/cells14171386
Chicago/Turabian StyleOiki, Hana, Kenichi Suda, Akira Hamada, Toshio Fujino, Keiko Obata, Yoshihisa Kobayashi, Kazuko Sakai, Shota Fukuda, Shuta Ohara, Masaoki Ito, and et al. 2025. "Efficacy of Conventional and Novel Tyrosine Kinase Inhibitors for Uncommon EGFR Mutations—An In Vitro Study" Cells 14, no. 17: 1386. https://doi.org/10.3390/cells14171386
APA StyleOiki, H., Suda, K., Hamada, A., Fujino, T., Obata, K., Kobayashi, Y., Sakai, K., Fukuda, S., Ohara, S., Ito, M., Soh, J., Nishio, K., Mitsudomi, T., & Tsutani, Y. (2025). Efficacy of Conventional and Novel Tyrosine Kinase Inhibitors for Uncommon EGFR Mutations—An In Vitro Study. Cells, 14(17), 1386. https://doi.org/10.3390/cells14171386










