Multi-Modal Profiling Reveals Contrasting Immunomodulatory Effects of Recreational Marijuana Used Alone or with Tobacco in Youth with HIV
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants and Enrollment
2.2. Study Design
2.3. Plasma Biomarker Profiling, Data Processing, Normalization and Analysis
2.4. Flow Cytometry Analysis of Lymphocyte Subsets
2.5. Peripheral Blood Cell RNA Isolation, Library Preparation, Sequencing and Data Analysis
2.6. Identification of Differentially Expressed Genes (DEGs)
2.7. Functional Enrichment and Network Analysis of Differentially Expressed Genes
3. Results
3.1. Characteristics of the Study Population Included in Bioprofiling
3.2. Marijuana Use Elevates Plasma Anti-Inflammatory Cytokine Bioprofile
3.3. Lymphocyte Subpopulations Among YWH Showed Limited Modulation by Substance Use
3.4. Distinct Differential Gene Expression Profiles Associated with Substance Use in Virally Suppressed YWH
3.5. Canonical Pathways Related to Cellular Immune Response Normalized by Marijuana
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CDC. Cannabis and Public Health; CDC: Atlanta, GA, USA, 2024.
- Russell, K. NBCWashington. 2024. Available online: https://www.nbcwashington.com/news/national-international/where-is-marijuana-legal-in-the-us-a-state-by-state-guide/3493460/# (accessed on 12 April 2025).
- Wrona, A.; Justice, A.C.; Tate, J.P.; Rentsch, C.T.; Gordon, K.S.; Kidwai-Khan, F.; Silverberg, M.J.; Satre, D.D.; Marconi, V.C.; Ingle, S.M.; et al. Cannabis Use and Self-Reported Bothersome Symptoms in People with HIV. Cannabis 2025, 8, 177–190. [Google Scholar] [CrossRef]
- Pacek, L.R.; Towe, S.L.; Hobkirk, A.L.; Nash, D.; Goodwin, R.D. Frequency of Cannabis Use and Medical Cannabis Use Among Persons Living With HIV in the United States: Findings From a Nationally Representative Sample. AIDS Educ. Prev. 2018, 30, 169–181. [Google Scholar] [CrossRef]
- Manuzak, J.A.; Granche, J.; Tassiopoulos, K.; Rower, J.E.; Knox, J.R.; Williams, D.W.; Ellis, R.J.; Goodkin, K.; Sharma, A.; Erlandson, K.M. Cannabis Use Is Associated With Decreased Antiretroviral Therapy Adherence among Older Adults With HIV. Open Forum Infect. Dis. 2023, 10, ofac699. [Google Scholar] [CrossRef]
- Azofeifa, A.; Mattson, M.E.; Schauer, G.; McAfee, T.; Grant, A.; Lyerla, R. National Estimates of Marijuana Use and Related Indicators—National Survey on Drug Use and Health, United States, 2002–2014. MMWR Surveill. Summ. 2016, 65, 1–28. [Google Scholar] [CrossRef]
- Alon, L.; Bruce, D.; Blocker, O.; Bouris, A.M.; Reirden, D.H.; Schneider, J.A. Perceptions of quality and safety in cannabis acquisition amongst young gay and bisexual men living with HIV/AIDS who use cannabis: Impact of legalisation and dispensaries. Int. J. Drug Policy 2021, 88, 103035. [Google Scholar] [CrossRef]
- Nichols, S.L.; Lowe, A.; Zhang, X.; Garvie, P.A.; Thornton, S.; Goldberger, B.A.; Hou, W.; Goodenow, M.M.; Sleasman, J.W. Concordance between self-reported substance use and toxicology among HIV-infected and uninfected at risk youth. Drug Alcohol Depend. 2014, 134, 376–382. [Google Scholar] [CrossRef]
- Gamarel, K.E.; Brown, L.; Kahler, C.W.; Fernandez, M.I.; Bruce, D.; Nichols, S. Prevalence and correlates of substance use among youth living with HIV in clinical settings. Drug Alcohol Depend. 2016, 169, 11–18. [Google Scholar] [CrossRef]
- NIDA. Reported Use of Most Drugs Among Adolescents Remained Low in 2024. Available online: https://nida.nih.gov/news-events/news-releases/2024/12/reported-use-of-most-drugs-among-adolescents-remained-low-in-2024 (accessed on 1 June 2025).
- Cabral, G.A.; Griffin-Thomas, L. Emerging role of the cannabinoid receptor CB2 in immune regulation: Therapeutic prospects for neuroinflammation. Expert Rev. Mol. Med. 2009, 11, e3. [Google Scholar] [CrossRef]
- Klein, T.W.; Cabral, G.A. Cannabinoid-induced immune suppression and modulation of antigen-presenting cells. J. Neuroimmune Pharmacol. 2006, 1, 50–64. [Google Scholar] [CrossRef]
- Zicari, S.; Sessa, L.; Cotugno, N.; Ruggiero, A.; Morrocchi, E.; Concato, C.; Rocca, S.; Zangari, P.; Manno, E.C.; Palma, P. Immune Activation, Inflammation, and Non-AIDS Co-Morbidities in HIV-Infected Patients under Long-Term ART. Viruses 2019, 11, 200. [Google Scholar] [CrossRef]
- Babu, H.; Ambikan, A.T.; Gabriel, E.E.; Svensson Akusjärvi, S.; Palaniappan, A.N.; Sundaraj, V.; Mupanni, N.R.; Sperk, M.; Cheedarla, N.; Sridhar, R.; et al. Systemic Inflammation and the Increased Risk of Inflamm-Aging and Age-Associated Diseases in People Living With HIV on Long Term Suppressive Antiretroviral Therapy. Front. Immunol. 2019, 10, 1965. [Google Scholar] [CrossRef] [PubMed]
- Mazzuti, L.; Turriziani, O.; Mezzaroma, I. The Many Faces of Immune Activation in HIV-1 Infection: A Multifactorial Interconnection. Biomedicines 2023, 11, 159. [Google Scholar] [CrossRef]
- Grabon, W.; Rheims, S.; Smith, J.; Bodennec, J.; Belmeguenai, A.; Bezin, L. CB2 receptor in the CNS: From immune and neuronal modulation to behavior. Neurosci. Biobehav. Rev. 2023, 150, 105226. [Google Scholar] [CrossRef]
- Kendall, D.A.; Yudowski, G.A. Cannabinoid Receptors in the Central Nervous System: Their Signaling and Roles in Disease. Front. Cell. Neurosci. 2016, 10, 294. [Google Scholar] [CrossRef]
- Wright, K.L.; Duncan, M.; Sharkey, K.A. Cannabinoid CB2 receptors in the gastrointestinal tract: A regulatory system in states of inflammation. Br. J. Pharmacol. 2008, 153, 263–270. [Google Scholar] [CrossRef]
- Rizzo, M.D.; Henriquez, J.E.; Blevins, L.K.; Bach, A.; Crawford, R.B.; Kaminski, N.E. Targeting Cannabinoid Receptor 2 on Peripheral Leukocytes to Attenuate Inflammatory Mechanisms Implicated in HIV-Associated Neurocognitive Disorder. J. Neuroimmune Pharmacol. 2020, 15, 780–793. [Google Scholar] [CrossRef]
- Henriquez, J.E.; Rizzo, M.D.; Schulz, M.A.; Crawford, R.B.; Gulick, P.; Kaminski, N.E. Δ9-Tetrahydrocannabinol Suppresses Secretion of IFNα by Plasmacytoid Dendritic Cells From Healthy and HIV-Infected Individuals. J. Acquir. Immune Defic. Syndr. 2017, 75, 588–596. [Google Scholar] [CrossRef]
- DeMarino, C.; Cowen, M.; Khatkar, P.; Cotto, B.; Branscome, H.; Kim, Y.; Sharif, S.A.; Agbottah, E.T.; Zhou, W.; Costiniuk, C.T.; et al. Cannabinoids Reduce Extracellular Vesicle Release from HIV-1 Infected Myeloid Cells and Inhibit Viral Transcription. Cells 2022, 11, 723. [Google Scholar] [CrossRef]
- Rizzo, M.D.; Crawford, R.B.; Bach, A.; Sermet, S.; Amalfitano, A.; Kaminski, N.E. Δ9-Tetrahydrocannabinol Suppresses Monocyte-Mediated Astrocyte Production of Monocyte Chemoattractant Protein 1 and Interleukin-6 in a Toll-Like Receptor 7-Stimulated Human Coculture. J. Pharmacol. Exp. Ther. 2019, 371, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.P.; Singh, N.P.; Singh, B.; Price, R.L.; Nagarkatti, M.; Nagarkatti, P.S. Cannabinoid receptor-2 (CB2) agonist ameliorates colitis in IL-10(−/−) mice by attenuating the activation of T cells and promoting their apoptosis. Toxicol. Appl. Pharmacol. 2012, 258, 256–267. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Dinasarapu, A.R.; Borkar, S.A.; Chang, K.F.; De Paris, K.; Kim-Chang, J.J.; Sleasman, J.W.; Goodenow, M.M. Anti-inflammatory effects of recreational marijuana in virally suppressed youth with HIV-1 are reversed by use of tobacco products in combination with marijuana. Retrovirology 2022, 19, 10. [Google Scholar] [CrossRef]
- Yndart Arias, A.; Kolishetti, N.; Vashist, A.; Madepalli, L.; Llaguno, L.; Nair, M. Anti-inflammatory effects of CBD in human microglial cell line infected with HIV-1. Sci. Rep. 2023, 13, 7376. [Google Scholar] [CrossRef] [PubMed]
- Shivers, S.C.; Newton, C.; Friedman, H.; Klein, T.W. delta 9-Tetrahydrocannabinol (THC) modulates IL-1 bioactivity in human monocyte/macrophage cell lines. Life Sci. 1994, 54, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.L.; Lancz, G.; Specter, S. Delta-9-tetrahydrocannabinol-(THC)-mediated inhibition of macrophage macromolecular metabolism is antagonized by human serum proteins and by cell surface proteins. Int. J. Immunopharmacol. 1993, 15, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Manuzak, J.A.; Gott, T.M.; Kirkwood, J.S.; Coronado, E.; Hensley-McBain, T.; Miller, C.; Cheu, R.K.; Collier, A.C.; Funderburg, N.T.; Martin, J.N.; et al. Heavy Cannabis Use Associated With Reduction in Activated and Inflammatory Immune Cell Frequencies in Antiretroviral Therapy-Treated Human Immunodeficiency Virus-Infected Individuals. Clin. Infect. Dis. 2018, 66, 1872–1882. [Google Scholar] [CrossRef]
- Watson, C.W.; Campbell, L.M.; Sun-Suslow, N.; Hong, S.; Umlauf, A.; Ellis, R.J.; Iudicello, J.E.; Letendre, S.; Marcotte, T.D.; Heaton, R.K.; et al. Daily Cannabis Use is Associated With Lower CNS Inflammation in People With HIV. J. Int. Neuropsychol. Soc. 2021, 27, 661–672. [Google Scholar] [CrossRef]
- Falcinelli, S.D.; Cooper-Volkheimer, A.D.; Semenova, L.; Wu, E.; Richardson, A.; Ashokkumar, M.; Margolis, D.M.; Archin, N.M.; Rudin, C.D.; Murdoch, D.; et al. Impact of Cannabis Use on Immune Cell Populations and the Viral Reservoir in People With HIV on Suppressive Antiretroviral Therapy. J. Infect. Dis. 2023, 228, 1600–1609. [Google Scholar] [CrossRef]
- Mboumba Bouassa, R.S.; Comeau, E.; Alexandrova, Y.; Pagliuzza, A.; Yero, A.; Samarani, S.; Needham, J.; Singer, J.; Lee, T.; Bobeuf, F.; et al. Effects of Oral Cannabinoids on Systemic Inflammation and Viral Reservoir Markers in People with HIV on Antiretroviral Therapy: Results of the CTN PT028 Pilot Clinical Trial. Cells 2023, 12, 1811. [Google Scholar] [CrossRef]
- Thames, A.D.; Mahmood, Z.; Burggren, A.C.; Karimian, A.; Kuhn, T.P. Combined effects of HIV and marijuana use on neurocognitive functioning and immune status. AIDS Care 2016, 28, 628–632. [Google Scholar] [CrossRef]
- Williams, J.C.; Appelberg, S.; Goldberger, B.A.; Klein, T.W.; Sleasman, J.W.; Goodenow, M.M. Δ(9)-Tetrahydrocannabinol treatment during human monocyte differentiation reduces macrophage susceptibility to HIV-1 infection. J. Neuroimmune Pharmacol. 2014, 9, 369–379. [Google Scholar] [CrossRef]
- Molina, P.E.; Winsauer, P.; Zhang, P.; Walker, E.; Birke, L.; Amedee, A.; Stouwe, C.V.; Troxclair, D.; McGoey, R.; Varner, K.; et al. Cannabinoid administration attenuates the progression of simian immunodeficiency virus. AIDS Res. Hum. Retroviruses 2011, 27, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Bonn-Miller, M.O.; Oser, M.L.; Bucossi, M.M.; Trafton, J.A. Cannabis use and HIV antiretroviral therapy adherence and HIV-related symptoms. J. Behav. Med. 2014, 37, 1–10. [Google Scholar] [CrossRef]
- Algarin, A.B.; Plazarte, G.N.; Sovich, K.R.; Seeger, S.D.; Li, Y.; Cohen, R.A.; Striley, C.W.; Goldberger, B.A.; Wang, Y.; Somboonwit, C.; et al. Marijuana Use and Health Outcomes in Persons Living with HIV: Protocol for the Marijuana Associated Planning and Long-term Effects (MAPLE) Longitudinal Cohort Study. JMIR Res. Protoc. 2022, 11, e37153. [Google Scholar] [CrossRef]
- Schouten, J.; Su, T.; Wit, F.W.; Kootstra, N.A.; Caan, M.W.; Geurtsen, G.J.; Schmand, B.A.; Stolte, I.G.; Prins, M.; Majoie, C.B.; et al. Determinants of reduced cognitive performance in HIV-1-infected middle-aged men on combination antiretroviral therapy. Aids 2016, 30, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Storck, W.; Elbaz, M.; Vindis, C.; Déguilhem, A.; Lapeyre-Mestre, M.; Jouanjus, E. Cardiovascular risk associated with the use of cannabis and cannabinoids: A systematic review and meta-analysis. Heart 2025. [Google Scholar] [CrossRef]
- Borkar, S.A.; Yin, L.; Venturi, G.M.; Shen, J.; Chang, K.F.; Fischer, B.M.; Nepal, U.; Raplee, I.D.; Sleasman, J.W.; Goodenow, M.M. Youth Who Control HIV on Antiretroviral Therapy Display Unique Plasma Biomarkers and Cellular Transcriptome Profiles Including DNA Repair and RNA Processing. Cells 2025, 14, 285. [Google Scholar] [CrossRef]
- Kim-Chang, J.J.; Donovan, K.; Loop, M.S.; Hong, S.; Fischer, B.; Venturi, G.; Garvie, P.A.; Kohn, J.; Rendina, H.J.; Woods, S.P.; et al. Higher soluble CD14 levels are associated with lower visuospatial memory performance in youth with HIV. Aids 2019, 33, 2363–2374. [Google Scholar] [CrossRef]
- Rudy, B.J.; Kapogiannis, B.G.; Worrell, C.; Squires, K.; Bethel, J.; Li, S.; Wilson, C.M.; Agwu, A.; Emmanuel, P.; Price, G.; et al. Immune Reconstitution but Persistent Activation After 48 Weeks of Antiretroviral Therapy in Youth With Pre-Therapy CD4 >350 in ATN 061. J. Acquir. Immune Defic. Syndr. 2015, 69, 52–60. [Google Scholar] [CrossRef]
- Williams, J.C.; Zhang, X.; Karki, M.; Chi, Y.Y.; Wallet, S.M.; Rudy, B.J.; Nichols, S.L.; Goodenow, M.M.; Sleasman, J.W. Soluble CD14, CD163, and CD27 biomarkers distinguish ART-suppressed youth living with HIV from healthy controls. J. Leukoc. Biol. 2018, 103, 671–680. [Google Scholar] [CrossRef]
- Roomaney, R.A.; van Wyk, B.; Pillay-van Wyk, V. Aging with HIV: Increased Risk of HIV Comorbidities in Older Adults. Int. J. Environ. Res. Public Health 2022, 19, 2359. [Google Scholar] [CrossRef] [PubMed]
- Pirrone, V.; Libon, D.J.; Sell, C.; Lerner, C.A.; Nonnemacher, M.R.; Wigdahl, B. Impact of age on markers of HIV-1 disease. Future Virol. 2013, 8, 81–101. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Diagnoses, Deaths and Prevalence of HIV in the United States and 6 Territories and Freely Associated States 2023; U.S. Department of Health and Human Services: Atlanta, GA, USA, 2025.
- HIVinfo.nih.gov. HIV and Specific Populations. Available online: https://hivinfo.nih.gov/understanding-hiv/fact-sheets/hiv-and-adolescents-and-young-adults (accessed on 2 May 2025).
- Lifson, A.R.; Neuhaus, J.; Arribas, J.R.; van den Berg-Wolf, M.; Labriola, A.M.; Read, T.R. Smoking-related health risks among persons with HIV in the Strategies for Management of Antiretroviral Therapy clinical trial. Am. J. Public Health 2010, 100, 1896–1903. [Google Scholar] [CrossRef] [PubMed]
- Rahmanian, S.; Wewers, M.E.; Koletar, S.; Reynolds, N.; Ferketich, A.; Diaz, P. Cigarette smoking in the HIV-infected population. Proc. Am. Thorac. Soc. 2011, 8, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Kim-Chang, J.J.; Wilson, L.; Chan, C.; Fischer, B.; Venturi, G.; Goodenow, M.M.; Aldrovandi, G.; Weber, T.J.; Sleasman, J.W. Tenofovir Has Minimal Effect on Biomarkers of Bone Health in Youth with HIV Receiving Initial Antiretroviral Therapy. AIDS Res. Hum. Retroviruses 2019, 35, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Nichols, S.L.; Bethel, J.; Kapogiannis, B.G.; Li, T.; Woods, S.P.; Patton, E.D.; Ren, W.; Thornton, S.E.; Major-Wilson, H.O.; Puga, A.M.; et al. Antiretroviral treatment initiation does not differentially alter neurocognitive functioning over time in youth with behaviorally acquired HIV. J. Neurovirol. 2016, 22, 218–230. [Google Scholar] [CrossRef]
- Roslawski, M.J.; Remmel, R.P.; Karanam, A.; Leppik, I.E.; Marino, S.E.; Birnbaum, A.K. Simultaneous Quantification of 13 Cannabinoids and Metabolites in Human Plasma by Liquid Chromatography Tandem Mass Spectrometry in Adult Epilepsy Patients. Ther. Drug Monit. 2019, 41, 357–370. [Google Scholar] [CrossRef]
- Wall, M.A.; Johnson, J.; Jacob, P.; Benowitz, N.L. Cotinine in the serum, saliva, and urine of nonsmokers, passive smokers, and active smokers. Am. J. Public Health 1988, 78, 699–701. [Google Scholar] [CrossRef]
- WHO ASSIST Working Group. The Alcohol, Smoking and Substance Involvement Screening Test (ASSIST): Development, reliability and feasibility. Addiction 2002, 97, 1183–1194. [Google Scholar] [CrossRef]
- Schepp, R.M.; de Haan, C.A.M.; Wilkins, D.; Layman, H.; Graham, B.S.; Esser, M.T.; Berbers, G.A.M. Development and Standardization of a High-Throughput Multiplex Immunoassay for the Simultaneous Quantification of Specific Antibodies to Five Respiratory Syncytial Virus Proteins. mSphere 2019, 4, e00236-19. [Google Scholar] [CrossRef]
- FASTQC. FastQC. 2015. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 5 January 2025).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Manimaran, S.; Selby, H.M.; Okrah, K.; Ruberman, C.; Leek, J.T.; Quackenbush, J.; Haibe-Kains, B.; Bravo, H.C.; Johnson, W.E. BatchQC: Interactive software for evaluating sample and batch effects in genomic data. Bioinformatics 2016, 32, 3836–3838. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Enviroment for Statistical Computing (4.1.2); R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: http://www.R-project.org/ (accessed on 10 June 2025).
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- NCBI’s Database of Genotypes and Phenotypes. Available online: https://healthdata.gov/NIH/Database-of-Genotype-and-Phenotype-dbGaP-/th78-z3aq/about_data (accessed on 10 June 2025).
- Xie, F.; Armand, E.J.; Yao, Z.; Liu, H.; Bartlett, A.; Behrens, M.M.; Li, Y.E.; Lucero, J.D.; Luo, C.; Nery, J.R.; et al. Robust enhancer-gene regulation identified by single-cell transcriptomes and epigenomes. Cell Genom. 2023, 3, 100342. [Google Scholar] [CrossRef]
- Scott, E.N.; Wright, G.E.B.; Drögemöller, B.I.; Hasbullah, J.S.; Gunaretnam, E.P.; Miao, F.; Bhavsar, A.P.; Shen, F.; Schneider, B.P.; Carleton, B.C.; et al. Transcriptome-wide association study uncovers the role of essential genes in anthracycline-induced cardiotoxicity. NPJ Genom. Med. 2021, 6, 35. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Blighe, K.; Rana, S.; Lewis, M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2018. Available online: https://bioconductor.org/packages/devel/bioc/vignettes/EnhancedVolcano/inst/doc/EnhancedVolcano.html (accessed on 7 February 2025).
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- van der Kolk, B.W.; Muniandy, M.; Kaminska, D.; Alvarez, M.; Ko, A.; Miao, Z.; Valsesia, A.; Langin, D.; Vaittinen, M.; Pääkkönen, M.; et al. Differential Mitochondrial Gene Expression in Adipose Tissue Following Weight Loss Induced by Diet or Bariatric Surgery. J. Clin. Endocrinol. Metab. 2021, 106, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- Yoder, A.C.; Guo, K.; Dillon, S.M.; Phang, T.; Lee, E.J.; Harper, M.S.; Helm, K.; Kappes, J.C.; Ochsenbauer, C.; McCarter, M.D.; et al. The transcriptome of HIV-1 infected intestinal CD4+ T cells exposed to enteric bacteria. PLoS Pathog. 2017, 13, e1006226. [Google Scholar] [CrossRef]
- Thieurmel, B. visNetwork: Network Visualization Using ‘vis.js’ Library. Available online: https://github.com/datastorm-open/visnetwork (accessed on 25 May 2025).
- Valiathan, R.; Deeb, K.; Diamante, M.; Ashman, M.; Sachdeva, N.; Asthana, D. Reference ranges of lymphocyte subsets in healthy adults and adolescents with special mention of T cell maturation subsets in adults of South Florida. Immunobiology 2014, 219, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Castro, F.O.F.; Silva, J.M.; Dorneles, G.P.; Barros, J.B.S.; Ribeiro, C.B.; Noronha, I.; Barbosa, G.R.; Souza, L.C.S.; Guilarde, A.O.; Pereira, A.; et al. Distinct inflammatory profiles in HIV-infected individuals under antiretroviral therapy using cannabis, cocaine or cannabis plus cocaine. Aids 2019, 33, 1831–1842. [Google Scholar] [CrossRef]
- Cornwell, W.D.; Sriram, U.; Seliga, A.; Zuluaga-Ramirez, V.; Gajghate, S.; Rom, S.; Winfield, M.; Heldt, N.A.; Ambrose, D.; Rogers, T.J.; et al. Tobacco smoke and morphine alter peripheral and CNS inflammation following HIV infection in a humanized mouse model. Sci. Rep. 2020, 10, 13977. [Google Scholar] [CrossRef]
- Guo, P.; Li, R.; Piao, T.H.; Wang, C.L.; Wu, X.L.; Cai, H.Y. Pathological Mechanism and Targeted Drugs of COPD. Int. J. Chronic Obstr. Pulm. Dis. 2022, 17, 1565–1575. [Google Scholar] [CrossRef]
- AlMubarak, A.M.; Alqutub, M.N.; Javed, F.; Vohra, F.; Abduljabbar, T. Whole Salivary Cotinine Levels and Interleukin 1-β Levels among Young Adults Involuntarily Exposed to Vapor from Electronic Nicotine Delivery Systems. Oral Health Prev. Dent. 2022, 20, 127–132. [Google Scholar] [CrossRef]
- Pauwels, N.S.; Bracke, K.R.; Dupont, L.L.; Van Pottelberge, G.R.; Provoost, S.; Vanden Berghe, T.; Vandenabeele, P.; Lambrecht, B.N.; Joos, G.F.; Brusselle, G.G. Role of IL-1α and the Nlrp3/caspase-1/IL-1β axis in cigarette smoke-induced pulmonary inflammation and COPD. Eur. Respir. J. 2011, 38, 1019–1028. [Google Scholar] [CrossRef]
- Hmiel, L.; Zhang, S.; Obare, L.M.; Santana, M.A.O.; Wanjalla, C.N.; Titanji, B.K.; Hileman, C.O.; Bagchi, S. Inflammatory and Immune Mechanisms for Atherosclerotic Cardiovascular Disease in HIV. Int. J. Mol. Sci. 2024, 25, 7266. [Google Scholar] [CrossRef] [PubMed]
- Hsue, P.Y.; Li, D.; Ma, Y.; Ishai, A.; Manion, M.; Nahrendorf, M.; Ganz, P.; Ridker, P.M.; Deeks, S.G.; Tawakol, A. IL-1β Inhibition Reduces Atherosclerotic Inflammation in HIV Infection. J. Am. Coll. Cardiol. 2018, 72, 2809–2811. [Google Scholar] [CrossRef]
- Ndrepepa, G. Myeloperoxidase—A bridge linking inflammation and oxidative stress with cardiovascular disease. Clin. Chim. Acta 2019, 493, 36–51. [Google Scholar] [CrossRef] [PubMed]
- Syed, S.S.; Balluz, R.S.; Kabagambe, E.K.; Meyer, W.A., 3rd; Lukas, S.; Wilson, C.M.; Kapogiannis, B.G.; Nachman, S.A.; Sleasman, J.W. Assessment of biomarkers of cardiovascular risk among HIV type 1-infected adolescents: Role of soluble vascular cell adhesion molecule as an early indicator of endothelial inflammation. AIDS Res. Hum. Retroviruses 2013, 29, 493–500. [Google Scholar] [CrossRef]
- Cheng, D.; Talib, J.; Stanley, C.P.; Rashid, I.; Michaëlsson, E.; Lindstedt, E.L.; Croft, K.D.; Kettle, A.J.; Maghzal, G.J.; Stocker, R. Inhibition of MPO (Myeloperoxidase) Attenuates Endothelial Dysfunction in Mouse Models of Vascular Inflammation and Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1448–1457. [Google Scholar] [CrossRef]
- Mokoena, H.; Mabhida, S.E.; Choshi, J.; Dludla, P.V.; Nkambule, B.B.; McHiza, Z.J.; Ndwandwe, D.E.; Kengne, A.P.; Hanser, S. Endothelial dysfunction and cardiovascular diseases in people living with HIV on specific highly active antiretroviral therapy regimen: A systematic review of clinical studies. Atheroscler. Plus 2024, 55, 47–54. [Google Scholar] [CrossRef]
- Keane, A.M.; Swartz, T.H. The impacts of tobacco and nicotine on HIV-1 infection, inflammation, and the blood-brain barrier in the central nervous system. Front. Pharmacol. 2024, 15, 1477845. [Google Scholar] [CrossRef]
- Valiathan, R.; Miguez, M.J.; Patel, B.; Arheart, K.L.; Asthana, D. Tobacco smoking increases immune activation and impairs T-cell function in HIV infected patients on antiretrovirals: A cross-sectional pilot study. PLoS ONE 2014, 9, e97698. [Google Scholar] [CrossRef] [PubMed]
- Raplee, I.D.; Borkar, S.A.; Yin, L.; Chang, K.-F.; Nepal, U.; Shen, J.; Venturi, G.M.; Sleasman, J.W.; Goodenow, M.M. The Role of Microarray in Modern Sequencing: Statistical Approach Matters in a Comparison Between Microarray and RNA-Seq. Biotech 2025, 14, 55. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.W. Cannabinoid-based drugs as anti-inflammatory therapeutics. Nat. Rev. Immunol. 2005, 5, 400–411. [Google Scholar] [CrossRef]
- Galiègue, S.; Mary, S.; Marchand, J.; Dussossoy, D.; Carrière, D.; Carayon, P.; Bouaboula, M.; Shire, D.; Le Fur, G.; Casellas, P. Expression of central and peripheral cannabinoid receptors in human immune tissues and leukocyte subpopulations. Eur. J. Biochem. 1995, 232, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Pandey, R.; Mousawy, K.; Nagarkatti, M.; Nagarkatti, P. Endocannabinoids and immune regulation. Pharmacol. Res. 2009, 60, 85–92. [Google Scholar] [CrossRef]
- Ayoub, S.M.; Holloway, B.M.; Miranda, A.H.; Roberts, B.Z.; Young, J.W.; Minassian, A.; Ellis, R.J. The Impact of Cannabis Use on Cognition in People with HIV: Evidence of Function-Dependent Effects and Mechanisms from Clinical and Preclinical Studies. Curr. HIV/AIDS Rep. 2024, 21, 87–115. [Google Scholar] [CrossRef]
- Turcotte, C.; Blanchet, M.R.; Laviolette, M.; Flamand, N. The CB2 receptor and its role as a regulator of inflammation. Cell. Mol. Life Sci. 2016, 73, 4449–4470. [Google Scholar] [CrossRef]
- Arimilli, S.; Madahian, B.; Chen, P.; Marano, K.; Prasad, G.L. Gene expression profiles associated with cigarette smoking and moist snuff consumption. BMC Genom. 2017, 18, 156. [Google Scholar] [CrossRef]
- Hu, Y.; Ranganathan, M.; Shu, C.; Liang, X.; Ganesh, S.; Osafo-Addo, A.; Yan, C.; Zhang, X.; Aouizerat, B.E.; Krystal, J.H.; et al. Single-cell Transcriptome Mapping Identifies Common and Cell-type Specific Genes Affected by Acute Delta9-tetrahydrocannabinol in Humans. Sci. Rep. 2020, 10, 3450. [Google Scholar] [CrossRef]
- Kar, P.; Chatrin, C.; Đukić, N.; Suyari, O.; Schuller, M.; Zhu, K.; Prokhorova, E.; Bigot, N.; Baretić, D.; Ahel, J.; et al. PARP14 and PARP9/DTX3L regulate interferon-induced ADP-ribosylation. Embo J. 2024, 43, 2929–2953. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, T.; Luebbe, J.; Kilian, C.; Riedel, J.H.; Hiekmann, S.; Asada, N.; Ginsberg, P.; Robben, L.; Song, N.; Kaffke, A.; et al. IL-17 Receptor C Signaling Controls CD4(+) T(H)17 Immune Responses and Tissue Injury in Immune-Mediated Kidney Diseases. J. Am. Soc. Nephrol. 2021, 32, 3081–3098. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Gupta, S.C.; Kim, J.H. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood 2012, 119, 651–665. [Google Scholar] [CrossRef] [PubMed]
- Mackelprang, R.D.; Filali-Mouhim, A.; Richardson, B.; Lefebvre, F.; Katabira, E.; Ronald, A.; Gray, G.; Cohen, K.W.; Klatt, N.R.; Pecor, T.; et al. Upregulation of IFN-stimulated genes persists beyond the transitory broad immunologic changes of acute HIV-1 infection. iScience 2023, 26, 106454. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.H.; Chee, J.D.; Bradfield, C.J.; Park, E.S.; Kumar, P.; MacMicking, J.D. Interferon-induced guanylate-binding proteins in inflammasome activation and host defense. Nat. Immunol. 2016, 17, 481–489. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, Y.; Zheng, B.; Ling, X.; Wang, G.; Li, L.; Wang, W.; Pan, M.; Li, X.; Meng, Y. GBP2 upregulated in LPS-stimulated macrophages-derived exosomes accelerates septic lung injury by activating epithelial cell NLRP3 signaling. Int. Immunopharmacol. 2023, 124, 111017. [Google Scholar] [CrossRef]
- Pyrillou, K.; Burzynski, L.C.; Clarke, M.C.H. Alternative Pathways of IL-1 Activation, and Its Role in Health and Disease. Front. Immunol. 2020, 11, 613170. [Google Scholar] [CrossRef]
- Shi, L.; Song, L.; Maurer, K.; Dou, Y.; Patel, V.R.; Su, C.; Leonard, M.E.; Lu, S.; Hodge, K.M.; Torres, A.; et al. IL-1 Transcriptional Responses to Lipopolysaccharides Are Regulated by a Complex of RNA Binding Proteins. J. Immunol. 2020, 204, 1334–1344. [Google Scholar] [CrossRef]
- Pulugulla, S.H.; Packard, T.A.; Galloway, N.L.K.; Grimmett, Z.W.; Doitsh, G.; Adamik, J.; Galson, D.L.; Greene, W.C.; Auron, P.E. Distinct mechanisms regulate IL1B gene transcription in lymphoid CD4 T cells and monocytes. Cytokine 2018, 111, 373–381. [Google Scholar] [CrossRef]
- Nabatanzi, R.; Ssekamatte, P.; Castelnuovo, B.; Kambugu, A.; Nakanjako, D. Increased Levels of Caspase-1 and IL-1β Among Adults With Persistent Immune Activation After 12 Years of Suppressive Antiretroviral Therapy in the Infectious Diseases Institute HIV Treatment Cohort. Open Forum Infect. Dis. 2023, 10, ofad539. [Google Scholar] [CrossRef]
- Walsh, J.G.; Reinke, S.N.; Mamik, M.K.; McKenzie, B.A.; Maingat, F.; Branton, W.G.; Broadhurst, D.I.; Power, C. Rapid inflammasome activation in microglia contributes to brain disease in HIV/AIDS. Retrovirology 2014, 11, 35. [Google Scholar] [CrossRef]
- Ryan, S.K.; Gonzalez, M.V.; Garifallou, J.P.; Bennett, F.C.; Williams, K.S.; Sotuyo, N.P.; Mironets, E.; Cook, K.; Hakonarson, H.; Anderson, S.A.; et al. Neuroinflammation and EIF2 Signaling Persist despite Antiretroviral Treatment in an hiPSC Tri-culture Model of HIV Infection. Stem Cell Rep. 2020, 14, 991. [Google Scholar] [CrossRef]
- Mendes, E.A.; Tang, Y.; Jiang, G. The integrated stress response signaling during the persistent HIV infection. iScience 2023, 26, 108418. [Google Scholar] [CrossRef]
- Andersen, A.M.; Lei, M.K.; Beach, S.R.H.; Philibert, R.A. Inflammatory biomarker relationships with helper T cell GPR15 expression and cannabis and tobacco smoking. J. Psychosom. Res. 2021, 141, 110326. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, Y.; Shikano, S. Emerging roles of a chemoattractant receptor GPR15 and ligands in pathophysiology. Front. Immunol. 2023, 14, 1179456. [Google Scholar] [CrossRef]
- Andersen, A.M.; Lei, M.K.; Beach, S.R.H.; Philibert, R.A.; Sinha, S.; Colgan, J.D. Cigarette and Cannabis Smoking Effects on GPR15+ Helper T Cell Levels in Peripheral Blood: Relationships with Epigenetic Biomarkers. Genes 2020, 11, 149. [Google Scholar] [CrossRef] [PubMed]
- Blaak, H.; Boers, P.H.; Gruters, R.A.; Schuitemaker, H.; van der Ende, M.E.; Osterhaus, A.D. CCR5, GPR15, and CXCR6 are major coreceptors of human immunodeficiency virus type 2 variants isolated from individuals with and without plasma viremia. J. Virol. 2005, 79, 1686–1700. [Google Scholar] [CrossRef] [PubMed]
- Kiene, M.; Rethi, B.; Jansson, M.; Dillon, S.; Lee, E.; Lantto, R.; Wilson, C.; Pöhlmann, S.; Chiodi, F. Toll-like receptor 3 signalling up-regulates expression of the HIV co-receptor G-protein coupled receptor 15 on human CD4+ T cells. PLoS ONE 2014, 9, e88195. [Google Scholar] [CrossRef] [PubMed]
- Habtezion, A.; Nguyen, L.P.; Hadeiba, H.; Butcher, E.C. Leukocyte Trafficking to the Small Intestine and Colon. Gastroenterology 2016, 150, 340–354. [Google Scholar] [CrossRef]
- Wilson, N.L.; Peterson, S.N.; Ellis, R.J. Cannabis and the Gut-Brain Axis Communication in HIV Infection. Cannabis Cannabinoid Res. 2021, 6, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Borlado, L.R.; Méndez, J. CDC6: From DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 2007, 29, 237–243. [Google Scholar] [CrossRef]
- Stillman, B. Cell cycle control of DNA replication. Science 1996, 274, 1659–1664. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Graves, L.M. De novo synthesis of pyrimidine nucleotides; emerging interfaces with signal transduction pathways. Cell. Mol. Life Sci. 2003, 60, 321–336. [Google Scholar] [CrossRef]
- Ellis, R.J.; Wilson, N.; Peterson, S. Cannabis and Inflammation in HIV: A Review of Human and Animal Studies. Viruses 2021, 13, 1521. [Google Scholar] [CrossRef]
- Costiniuk, C.T.; Jenabian, M.A. Cannabinoids and inflammation: Implications for people living with HIV. Aids 2019, 33, 2273–2288. [Google Scholar] [CrossRef]
- Montgomery, L.; Bagot, K.; Brown, J.L.; Haeny, A.M. The Association Between Marijuana Use and HIV Continuum of Care Outcomes: A Systematic Review. Curr. HIV/AIDS Rep. 2019, 16, 17–28. [Google Scholar] [CrossRef]
- Badowski, M.E.; Yanful, P.K. Dronabinol oral solution in the management of anorexia and weight loss in AIDS and cancer. Ther. Clin. Risk Manag. 2018, 14, 643–651. [Google Scholar] [CrossRef]
- Benuvia, T.S.D. U.S. Food and Drug Administration; 1985. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2017/018651s029lbl.pdf (accessed on 3 April 2025).
- Costiniuk, C.T.; Saneei, Z.; Routy, J.P.; Margolese, S.; Mandarino, E.; Singer, J.; Lebouche, B.; Cox, J.; Szabo, J.; Brouillette, M.J.; et al. Oral cannabinoids in people living with HIV on effective antiretroviral therapy: CTN PT028-study protocol for a pilot randomised trial to assess safety, tolerability and effect on immune activation. BMJ Open 2019, 9, e024793. [Google Scholar] [CrossRef] [PubMed]
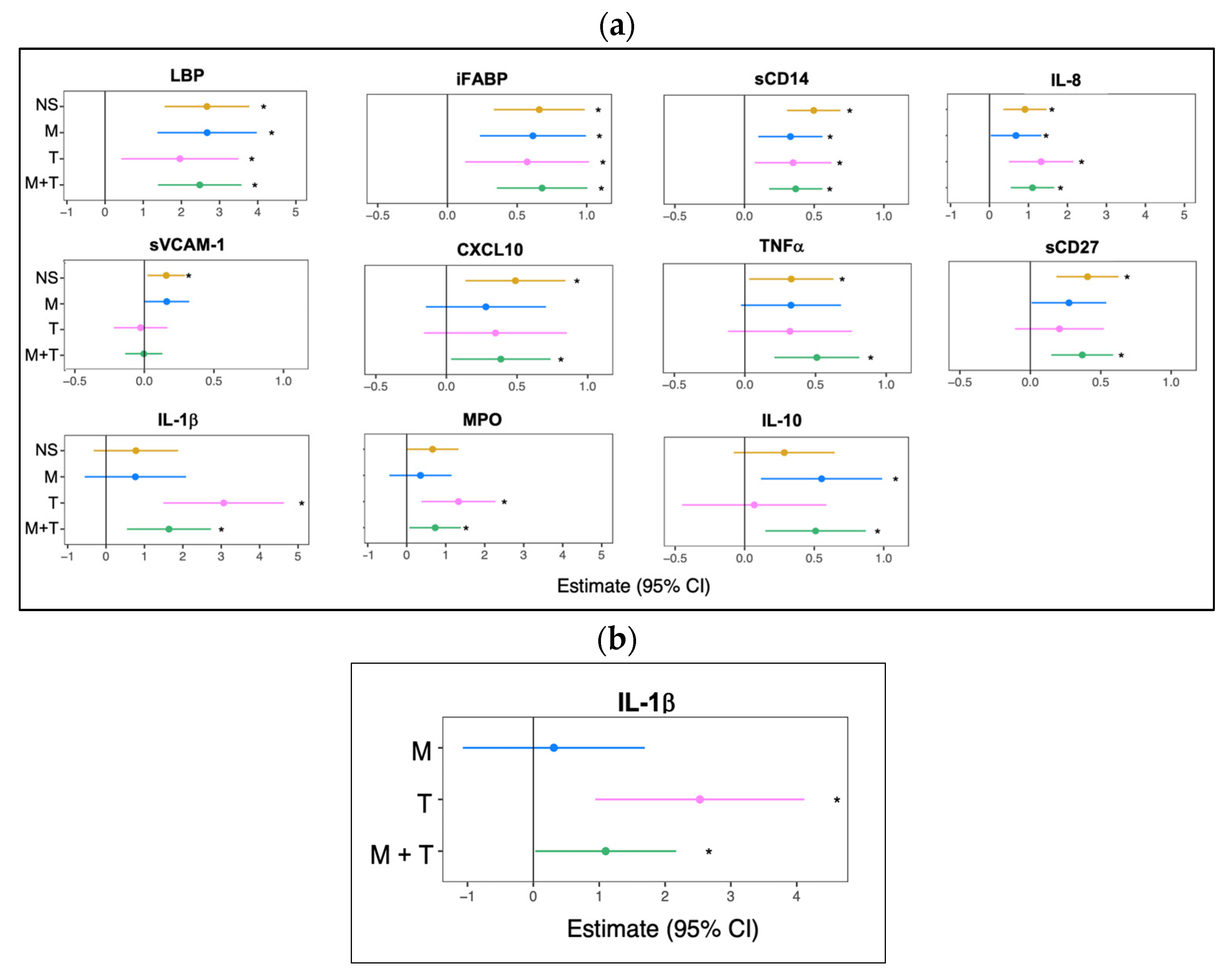
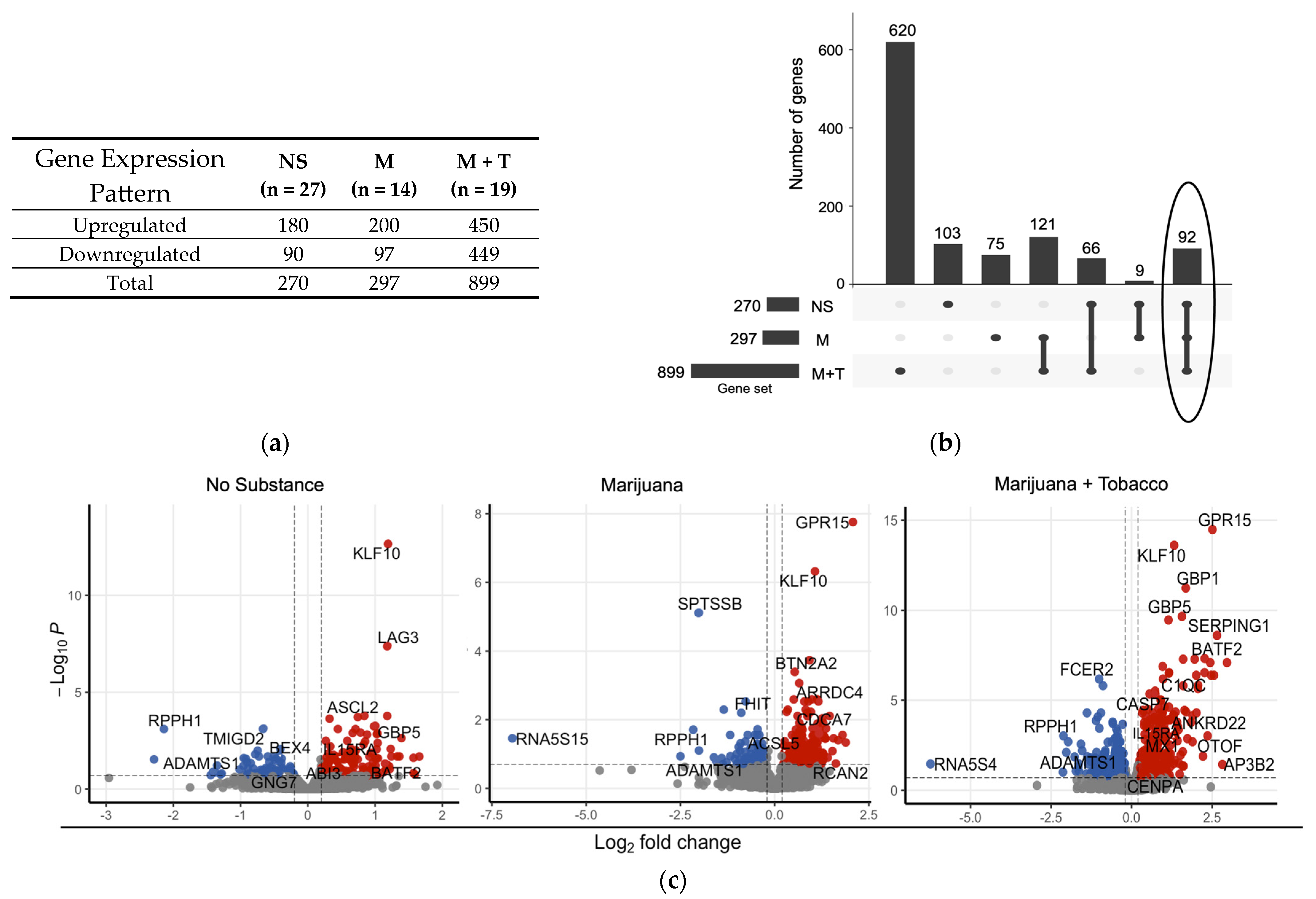
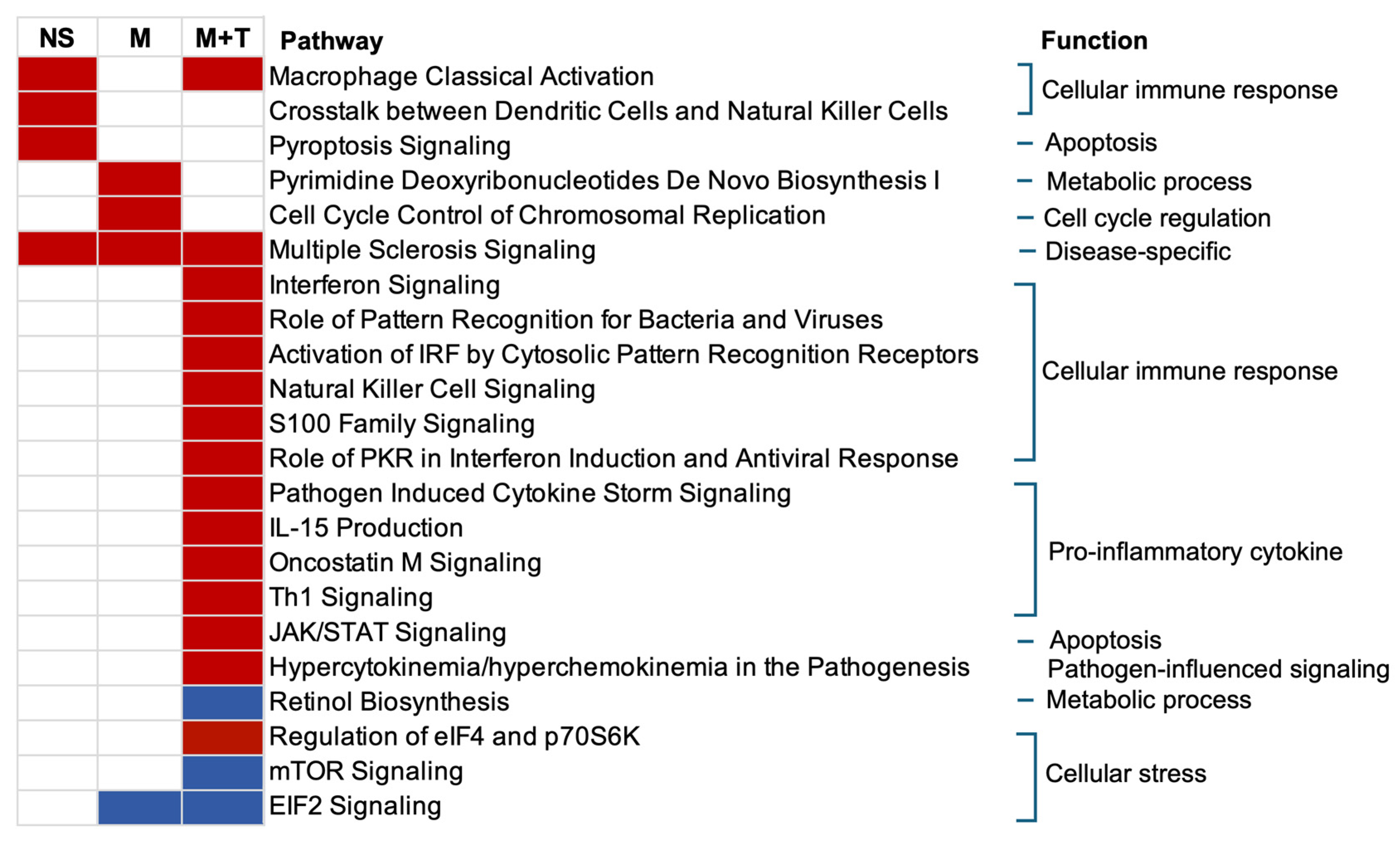
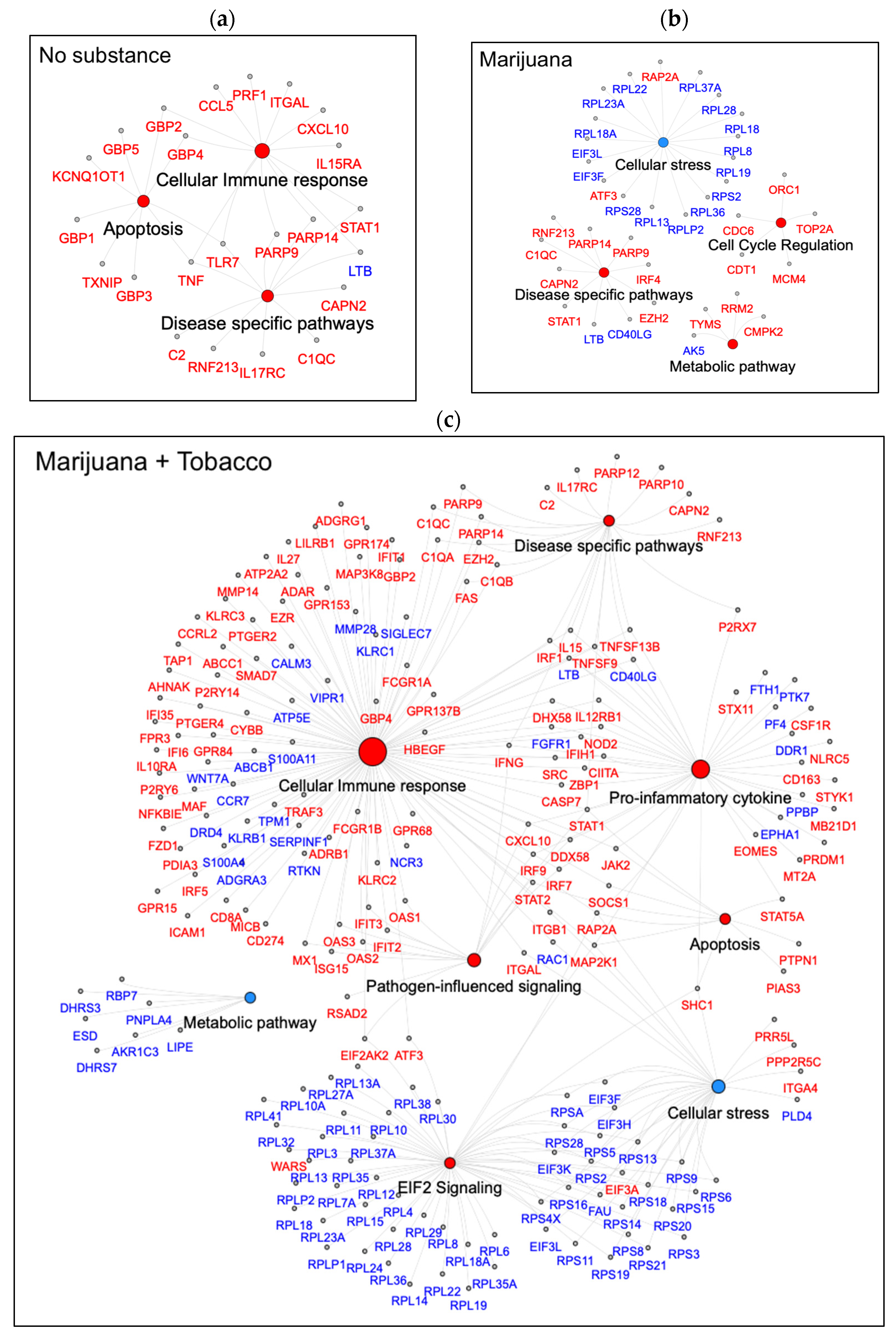
| Characteristics | Youth Without HIV | Youth with HIV (VL ≤ 50) | p-Value | ||||
|---|---|---|---|---|---|---|---|
| (n = 37) | (n = 37) | (n = 17) | (n = 10) | (n = 37) | |||
| Substance use (toxicology) | No | No | M | T | M + T | ||
| Age(years) a | 22 [20, 23] | 24 [23, 26] * | 24 [22, 25] * | 24 [23, 25] * | 24 [23, 25] * | <0.0001 * | |
| Male (%) | 68 | 84 | 87 | 70 | 86 | 0.08 | |
| African American (%) | 76 | 55 | 65 | 40 | 59 | 0.70 | |
| Days on ART a | NA | 980 [586, 1058] | 707 [358, 1018] | 1000 [736, 1056] | 973 [909, 1054] | 0.43 | |
| ART regimen (%) b | NRTI | NA | 86 | 89 | 67 | 84 | 0.72 |
| NNRTI | NA | 31 | 17 | 33 | 38 | ||
| PI | NA | 47 | 44 | 56 | 49 | ||
| INSTI | NA | 11 | 17 | 11 | 8 | ||
| CD4 T cells (number/µL) a,c | 751 [495, 885] | 670 [555, 919] | 645 [508, 818] | 756 [710, 920] | 666 [438, 837] | 0.91 | |
| CD4 T cells nadir (number/µL) a | - | 451 [341, 490] | 625 [521, 629] | 508 [445, 561] | 395 [295, 486] | 0.18 | |
| Percent of Cells at End of the Study a | Youth Without HIV | Youth with HIV (VL ≤ 50) | |||
|---|---|---|---|---|---|
| NS | M | T | M + T | ||
| Total CD4 cells | 45.9 [35.4, 53.7] | 42.6 [38.3, 48.2] | 44.6 [42.3, 48.1] | 35.7 [34.4, 44.7] | 42.9 [38.0, 45.9] |
| Naive CD4 T cells | 41.4 [36.3, 56.8] | 41.1 [38.5, 47.6] | 47.7 [35.4, 55.2] | 49.5 [46.0, 56.0] | 45.3 [38.6, 52.4] |
| Effector Memory CD4 T cells | 9.2 [7.1, 11.0] | 13.9 [9.7, 16.5] | 11.6 [10.7, 15.5] | 13.3 [10.7, 15.6] | 17.7 [15.8, 19.1] * |
| Total CD8 T cells | 24.4 [20.5, 28.7] | 32.3 [28.1, 36.7] * | 35.1 [34.3, 35.2] * | 30.8 [26.8, 33.2] | 32.4 [27.8, 37.1] * |
| HLA-DR, CD38 CD8 T cells | 0.3 [0.2, 0.5] | 1.1 [0.5, 1.9] | 0.5 [0.4, 1.0] | 0.5 [0.4, 0.6] | 0.6 [0.4, 1.5] * |
| Total CD19 B cells | 12.7 [9.5, 17.3] | 10.1 [6.8, 11.7] | 11.2 [7.4, 18.4] | 12.7 [10.3, 14.8] | 11.9 [8.3, 16.9] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borkar, S.A.; Venturi, G.M.; Chang, K.-F.; Gu, J.; Yin, L.; Shen, J.; Fischer, B.M.; Nepal, U.; Raplee, I.D.; Kim-Chang, J.J.; et al. Multi-Modal Profiling Reveals Contrasting Immunomodulatory Effects of Recreational Marijuana Used Alone or with Tobacco in Youth with HIV. Cells 2025, 14, 1267. https://doi.org/10.3390/cells14161267
Borkar SA, Venturi GM, Chang K-F, Gu J, Yin L, Shen J, Fischer BM, Nepal U, Raplee ID, Kim-Chang JJ, et al. Multi-Modal Profiling Reveals Contrasting Immunomodulatory Effects of Recreational Marijuana Used Alone or with Tobacco in Youth with HIV. Cells. 2025; 14(16):1267. https://doi.org/10.3390/cells14161267
Chicago/Turabian StyleBorkar, Samiksha A., Guglielmo M. Venturi, Kai-Fen Chang, Jingwen Gu, Li Yin, Jerry Shen, Bernard M. Fischer, Upasana Nepal, Isaac D. Raplee, Julie J. Kim-Chang, and et al. 2025. "Multi-Modal Profiling Reveals Contrasting Immunomodulatory Effects of Recreational Marijuana Used Alone or with Tobacco in Youth with HIV" Cells 14, no. 16: 1267. https://doi.org/10.3390/cells14161267
APA StyleBorkar, S. A., Venturi, G. M., Chang, K.-F., Gu, J., Yin, L., Shen, J., Fischer, B. M., Nepal, U., Raplee, I. D., Kim-Chang, J. J., Murdoch, D. M., Nichols, S. L., Hightow-Weidman, L. B., Somboonwit, C., Sleasman, J. W., & Goodenow, M. M. (2025). Multi-Modal Profiling Reveals Contrasting Immunomodulatory Effects of Recreational Marijuana Used Alone or with Tobacco in Youth with HIV. Cells, 14(16), 1267. https://doi.org/10.3390/cells14161267






