Disrupted Myelination in FAHN: Insights from a Patient-Specific hiPSC Neuron–Oligodendrocyte Model
Abstract
1. Introduction
2. Materials and Methods
2.1. hiPSC Cultivation
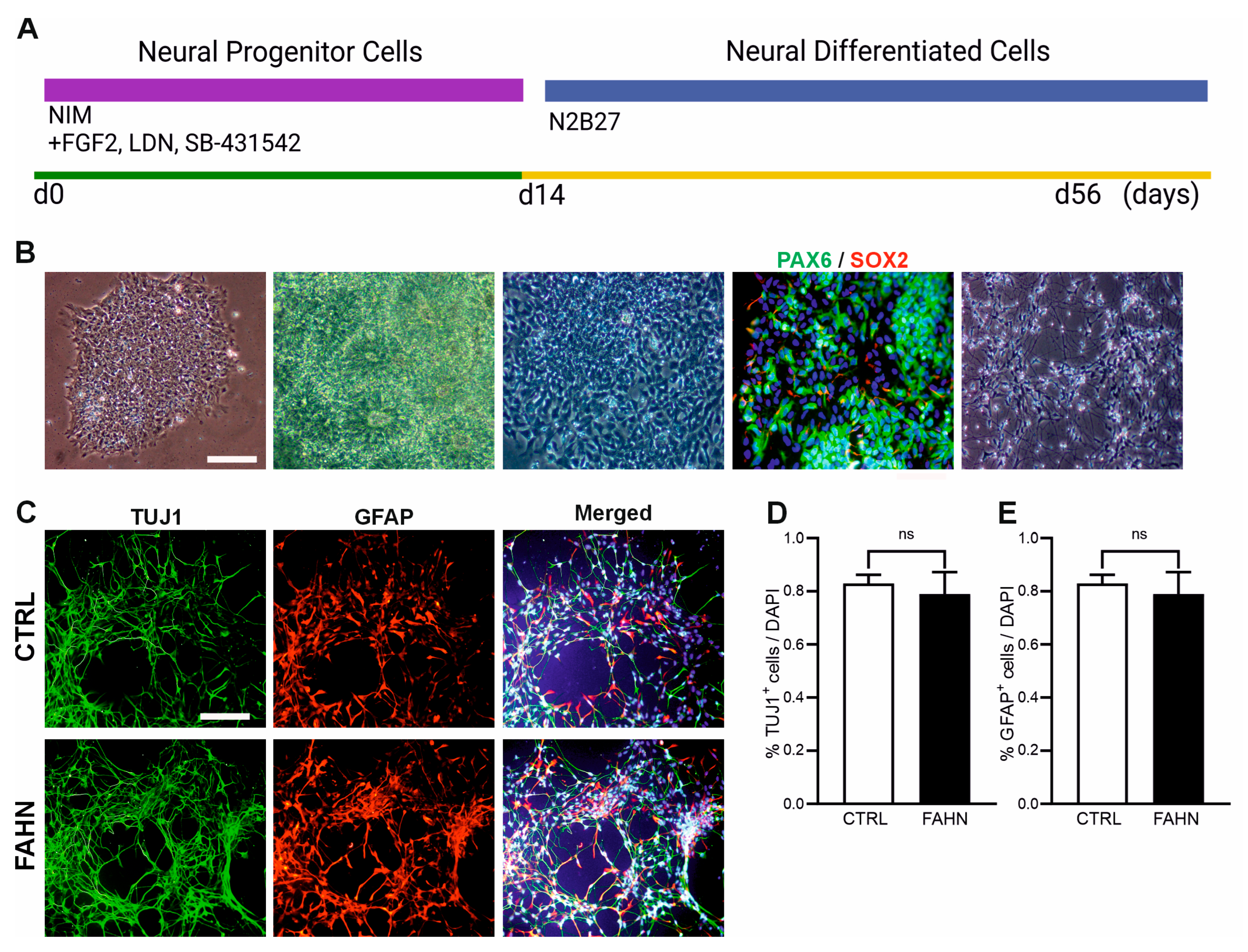
2.2. Neural Differentiation
2.3. Lentivirus Virus Generation
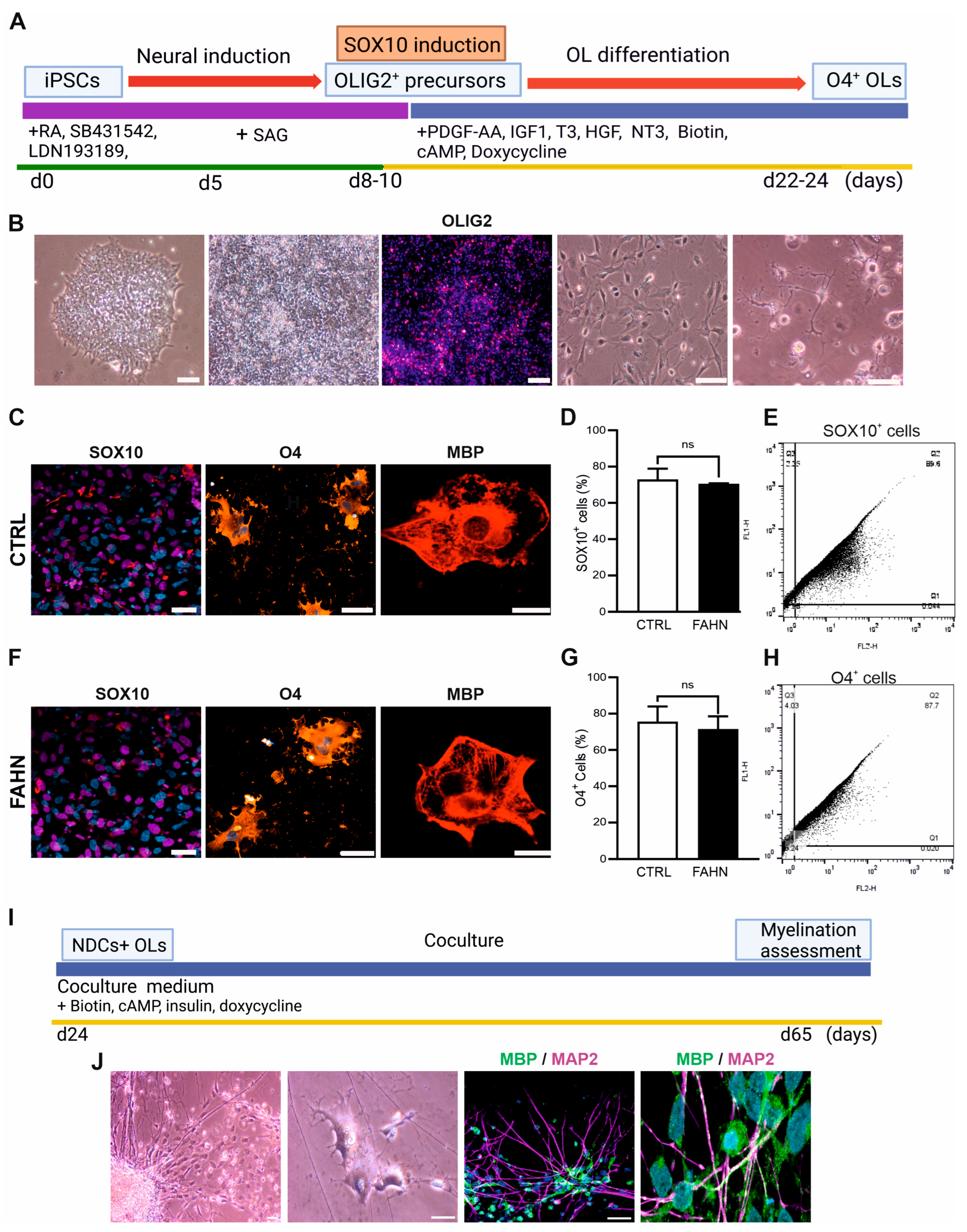
2.4. Generation of Oligodendrocyte Progenitor Cells
2.5. Flow Cytometery
2.6. Coculture Systems
2.7. Western Blot Analysis
2.8. Immunofluorescence Staining
2.9. Statistical Analyses
3. Results
3.1. Establishing the Disease Model
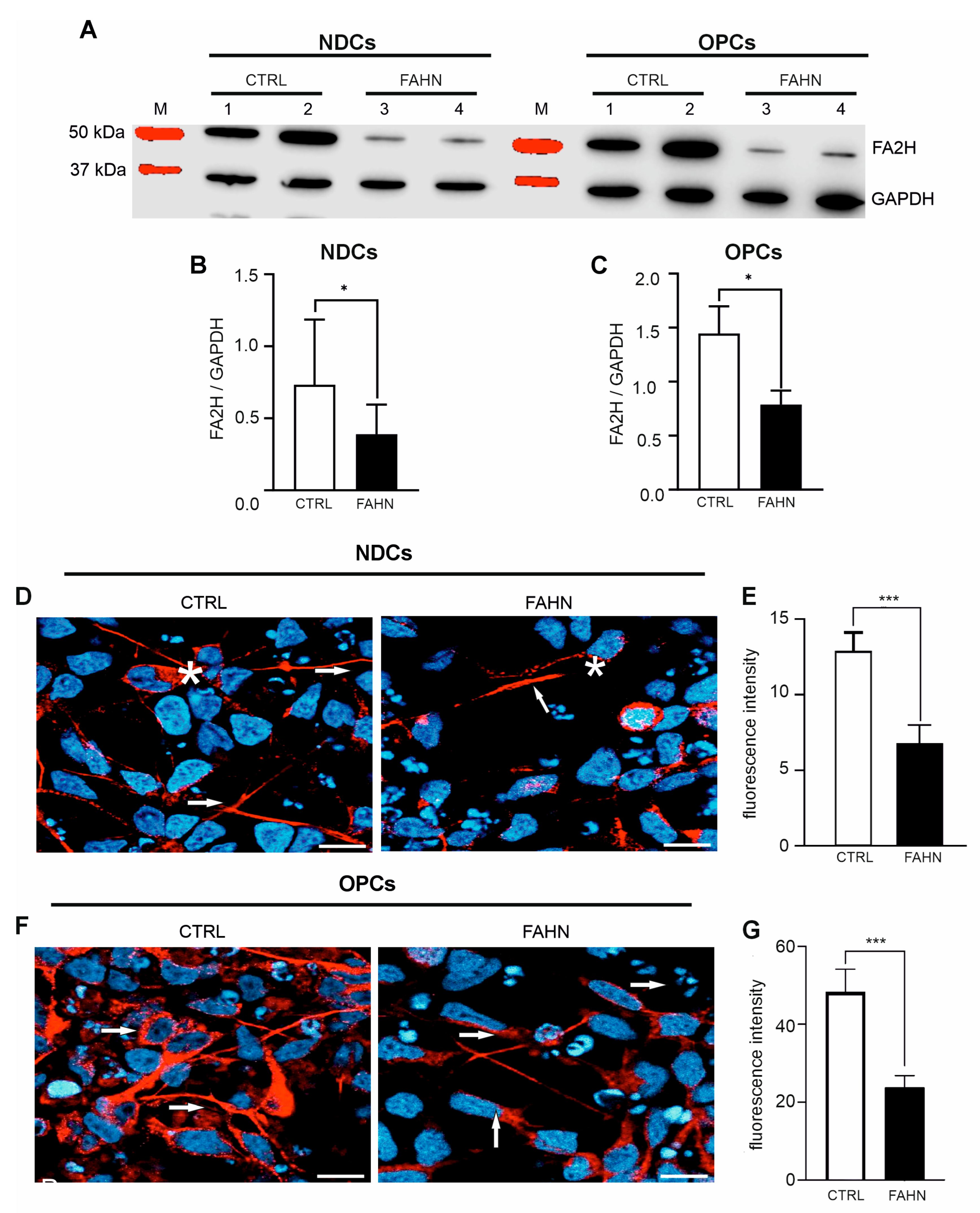
3.2. FA2H Mutant Protein Is Expressed at Reduced Levels in Patient-Derived Cells
3.3. FA2H Mutant Protein Displays Incorrect Intracellular Localization in Patient-Derived Cells
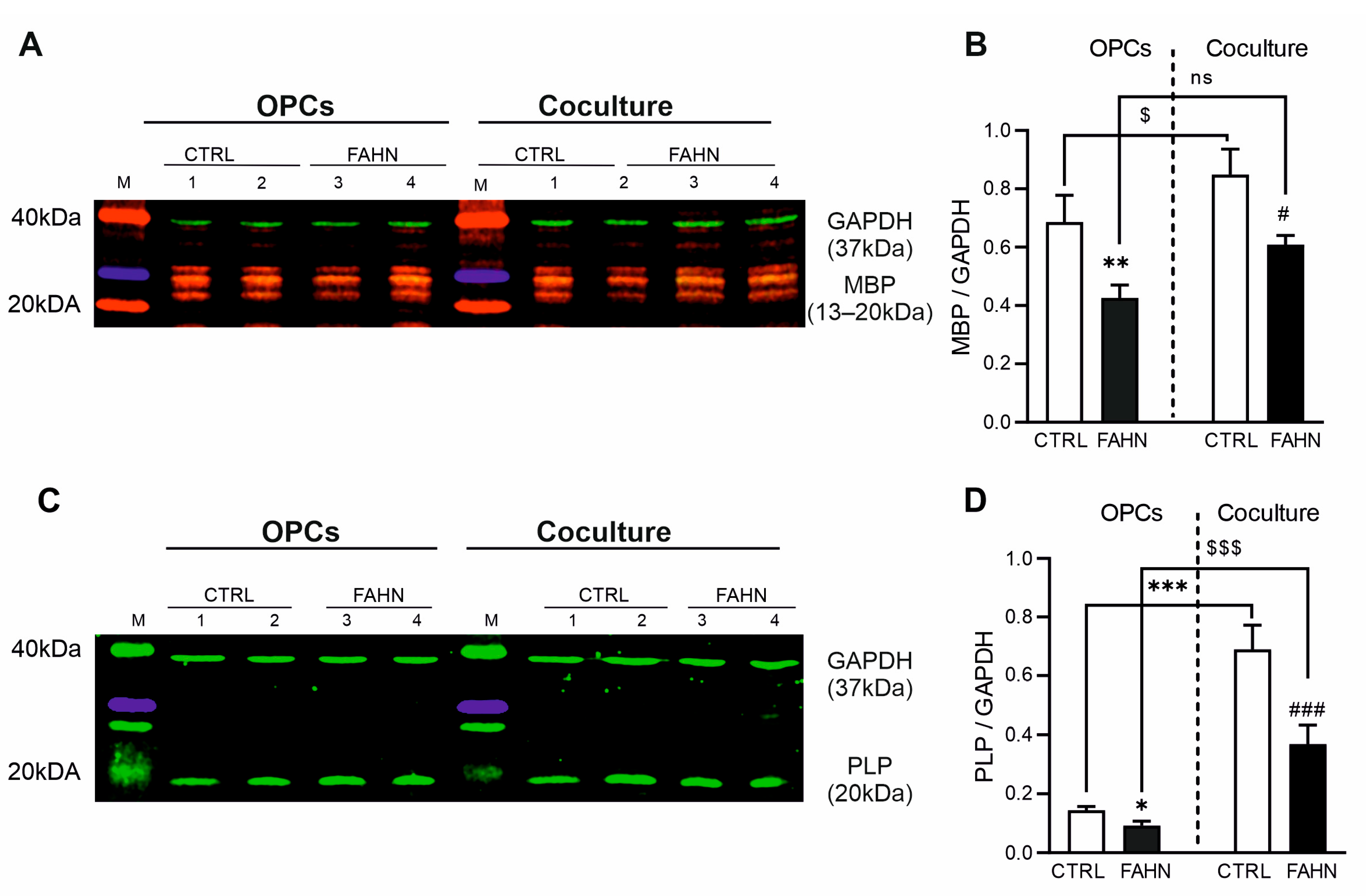
3.4. MBP and d PLP Are Expressed at Reduced Levels in FA2H-Deficient Cells
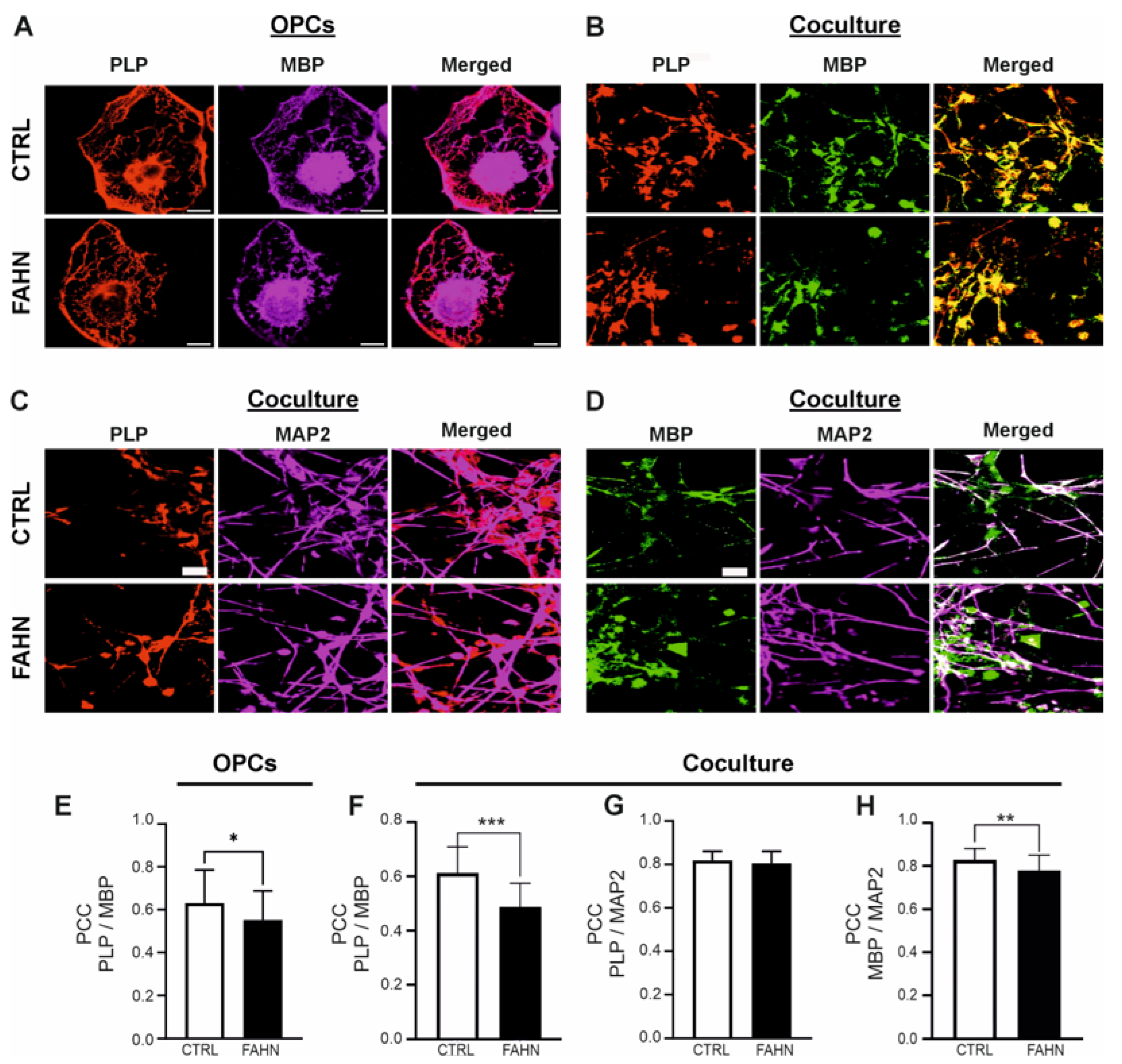
3.5. FA2H Mutant OPCs and Coculture Cells Show Impaired Localisation of MBP and PLP
3.6. Mutant FA2H Cells Line Has Reduced Quantity of Myelin Proteins
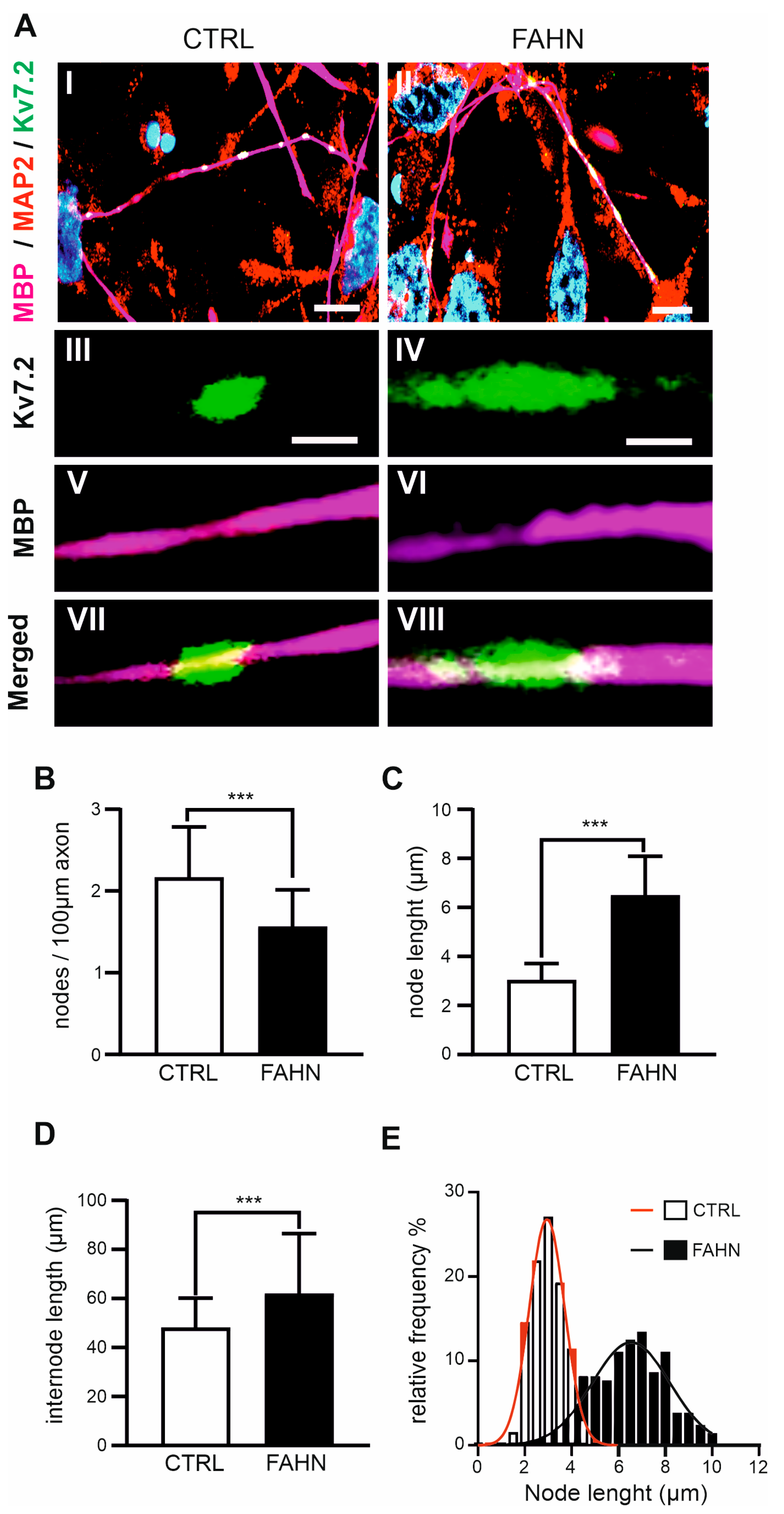
3.7. Myelin Sheath Examination and Ranvier Nodes
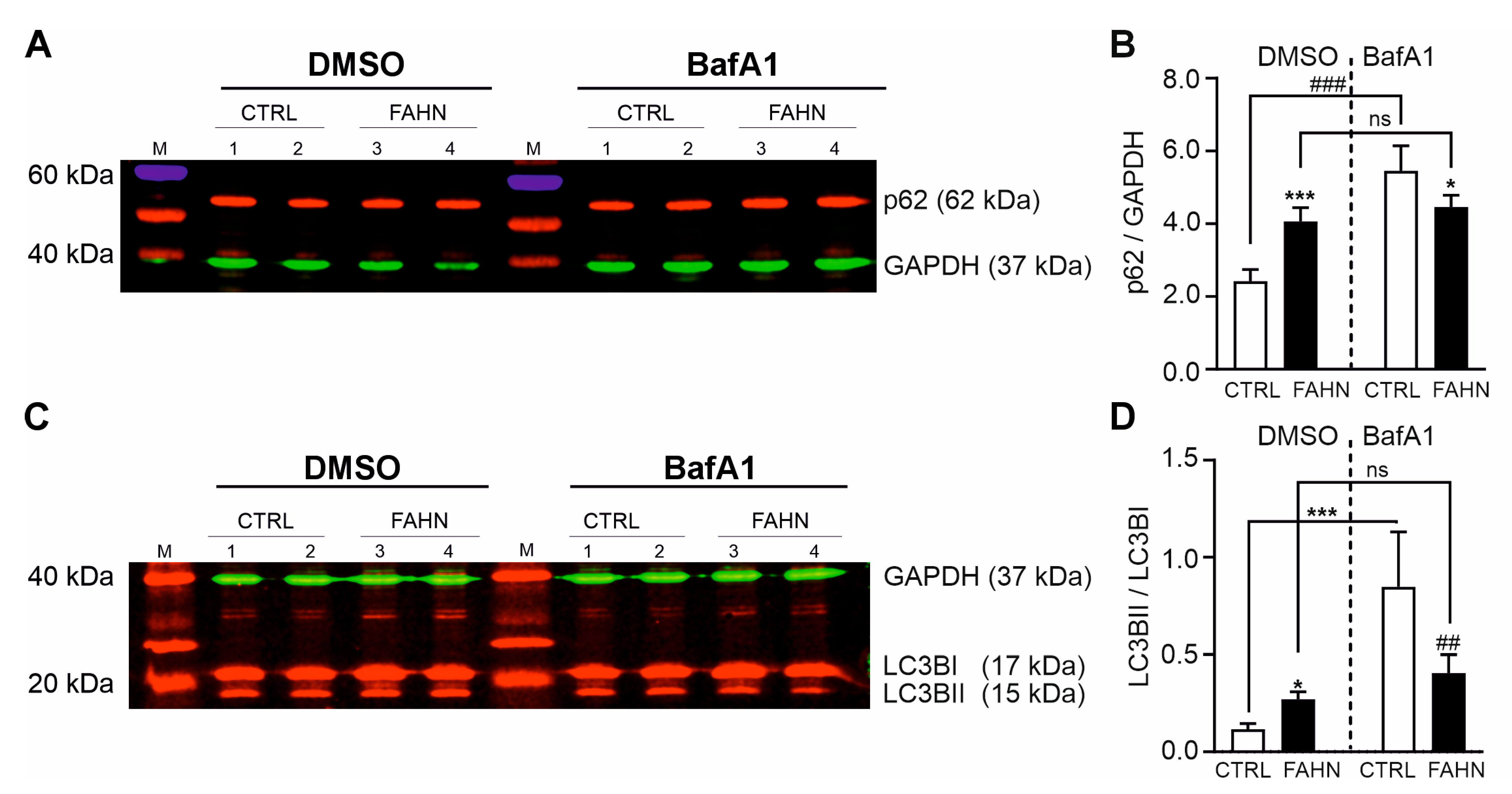
3.8. Initiation and Progression of Autophagy Is Affected
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BafA1 | Bafilomycin A1 |
| BSA | Bovine serum albumin |
| CNS | Central nervous system |
| ER | Endoplasmic reticulum |
| FAHN | Fatty-acid-hydroxylase-associated neurodegeneration |
| FA2H | Fatty acid 2-hydroxylase |
| FGF2 | Basic fibroblast growth factor |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| GFAP | Glial fibrillary acid protein |
| GSL | Glycosphingolipid |
| GlcCer | Glucosylceramide |
| hiPSC | Human induced pluripotent stem cell |
| IF | Immunofluorescence |
| iPSC | Induced pluripotent stem cell |
| JXP | Juxtaparanodal junction |
| KO | Knockout |
| LC3BI/II | Microtubule-associated proteins 1A/1B light chain 3B |
| LSM | Laser scanning microscope |
| MAG | Myelin-associated glycoprotein |
| MAP2 | Microtubule-associated protein 2 |
| MBP | Myelin-binding protein |
| MOG | Myelin oligodendrocyte glycoprotein |
| NBIA | Neurodegeneration with brain iron accumulation |
| NDCs | Neuronal differentiated cells |
| NIM | Neural induction medium |
| NPCs | Neural progenitor cells |
| OLs | Oligodendrocytes |
| OMIM | Online Mendelian Inheritance in Man |
| OPCs | Oligodendrocyte progenitor cells |
| PBS | Phosphate-buffered saline |
| PDGFRα | Platelet-derived growth factor receptor alpha |
| PFA | Paraformaldehyde |
| PLP | poly-L-ornithine |
| pre-OPC | Pre-oligodendrocyte progenitor |
| PNS | Peripheral nervous system |
| PNJ | Paranodal axoglial junction |
| PSA-NCAM | Polysialylated-neural cell adhesion molecule |
| RA | Retinoic acid |
| SHH | Sonic hedgehog |
| STR | Short tandem repeat |
| SQSTM1/p62 | Sequestosome-1 |
References
- Alderson, N.L.; Rembiesa, B.M.; Walla, M.D.; Bielawska, A.; Bielawski, J.; Hama, H. The human FA2H gene encodes a fatty acid 2-hydroxylase. J. Biol. Chem. 2004, 279, 48562–48568. [Google Scholar] [CrossRef] [PubMed]
- Alderson, N.L.; Maldonado, E.N.; Kern, M.J.; Bhat, N.R.; Hama, H. FA2H-dependent fatty acid 2-hydroxylation in postnatal mouse brain. J. Lipid Res. 2006, 47, 2772–2780. [Google Scholar] [CrossRef] [PubMed]
- Edvardson, S.; Hama, H.; Shaag, A.; Gomori, J.M.; Berger, I.; Soffer, D.; Korman, S.H.; Taustein, I.; Saada, A.; Elpeleg, O. Mutations in the fatty acid 2-hydroxylase gene are associated with leukodystrophy with spastic paraparesis and dystonia. Am. J. Hum. Genet. 2008, 83, 643–648. [Google Scholar] [CrossRef]
- Uchida, Y.; Hama, H.; Alderson, N.L.; Douangpanya, S.; Wang, Y.; Crumrine, D.A.; Elias, P.M.; Holleran, W.M. Fatty acid 2-hydroxylase, encoded by FA2H, accounts for differentiation-associated increase in 2-OH ceramides during keratinocyte differentiation. J. Biol. Chem. 2007, 282, 13211–13219. [Google Scholar] [CrossRef]
- Kruer, M.C.; Paisán-Ruiz, C.; Boddaert, N.; Yoon, M.Y.; Hama, H.; Gregory, A.; Malandrini, A.; Woltjer, R.L.; Munnich, A.; Gobin, S.; et al. Defective FA2H leads to a novel form of neurodegeneration with brain iron accumulation (NBIA). Ann. Neurol. 2010, 68, 611–618. [Google Scholar] [CrossRef]
- Dick, K.J.; Eckhardt, M.; Paisán-Ruiz, C.; Alshehhi, A.A.; Proukakis, C.; Sibtain, N.A.; Maier, H.; Sharifi, R.; Patton, M.A.; Bashir, W.; et al. Mutation of FA2H underlies a complicated form of hereditary spastic paraplegia (SPG35). Hum. Mutat. 2010, 31, E1251–E1260. [Google Scholar] [CrossRef]
- Cao, L.; Huang, X.-J.; Chen, C.-J.; Chen, S.-D. A rare family with Hereditary Spastic Paraplegia Type 35 due to novel FA2H mutations: A case report with literature review. J. Neurol. Sci. 2013, 329, 1–5. [Google Scholar] [CrossRef]
- Donkervoort, S.; Dastgir, J.; Hu, Y.; Zein, W.M.; Marks, H.; Blackstone, C.; Bönnemann, C.G. Phenotypic variability of a likely FA2H founder mutation in a family with complicated hereditary spastic paraplegia. Clin. Genet. 2014, 85, 393–395. [Google Scholar] [CrossRef]
- German, A.; Jukic, J.; Laner, A.; Arnold, P.; Socher, E.; Mennecke, A.; Schmidt, M.A.; Winkler, J.; Abicht, A.; Regensburger, M. Novel Homozygous FA2H Variant Causing the Full Spectrum of Fatty Acid Hydroxylase-Associated Neurodegeneration (SPG35). Genes 2023, 15, 14. [Google Scholar] [CrossRef]
- Garone, C.; Pippucci, T.; Cordelli, D.M.; Zuntini, R.; Castegnaro, G.; Marconi, C.; Graziano, C.; Marchiani, V.; Verrotti, A.; Seri, M.; et al. FA2H-related disorders: A novel c.270+3AT splice-site mutation leads to a complex neurodegenerative phenotype. Dev. Med. Child Neurol. 2011, 53, 958–961. [Google Scholar] [CrossRef]
- Hashemi, N.; Abadi, R.N.S.; Alavi, A.; Rohani, M.; Ghasemi, A.; Tavasoli, A.R. The first reports of FA2H-associated neurodegeneration from two unrelated Iranian families. Neurol. Sci. Off. J. Ital. Neurol. Soc. Ital. Soc. Clin. Neuro-Physiol. 2023, 44, 4359–4362. [Google Scholar] [CrossRef]
- Rupps, R.; Hukin, J.; Balicki, M.; Mercimek-Mahmutoglu, S.; Rolfs, A.; Dias, C. Novel Mutations in FA2H-Associated Neurodegeneration: An Underrecognized Condition? J. Child Neurol. 2013, 28, 1500–1504. [Google Scholar] [CrossRef]
- Eckhardt, M. Fatty Acid 2-Hydroxylase and 2-Hydroxylated Sphingolipids: Metabolism and Function in Health and Diseases. Int. J. Mol. Sci. 2023, 24, 4908. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, E.N.; Alderson, N.L.; Monje, P.V.; Wood, P.M.; Hama, H. FA2H is responsible for the formation of 2-hydroxy galactolipids in peripheral nervous system myelin. J. Lipid Res. 2008, 49, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Boiko, T.; Winckler, B. Myelin under construction—Teamwork required. J. Cell Biol. 2006, 172, 799–801. [Google Scholar] [CrossRef] [PubMed]
- Bechler, M.E.; Byrne, L.; Ffrench-Constant, C. CNS Myelin Sheath Lengths Are an Intrinsic Property of Oligodendrocytes. Curr. Biol. 2015, 25, 2411–2416. [Google Scholar] [CrossRef]
- Stadelmann, C.; Timmler, S.; Barrantes-Freer, A.; Simons, M. Myelin in the Central Nervous System: Structure, Function, and Pathology. Physiol. Rev. 2019, 99, 1381–1431. [Google Scholar] [CrossRef]
- Kota, V.; Hama, H. 2’-Hydroxy ceramide in membrane homeostasis and cell signaling. Adv. Biol. Regul. 2014, 54, 223–230. [Google Scholar] [CrossRef]
- Zöller, I.; Meixner, M.; Hartmann, D.; Büssow, H.; Meyer, R.; Gieselmann, V.; Eckhardt, M. Absence of 2-hydroxylated sphingolipids is compatible with normal neural development but causes late-onset axon and myelin sheath degeneration. J. Neurosci. Off. J. Soc. Neurosci. 2008, 28, 9741–9754. [Google Scholar] [CrossRef]
- Potter, K.A.; Kern, M.J.; Fullbright, G.; Bielawski, J.; Scherer, S.S.; Yum, S.W.; Li, J.J.; Cheng, H.; Han, X.; Venkata, J.K.; et al. Central nervous system dysfunction in a mouse model of FA2H deficiency. Glia 2011, 59, 1009–1021. [Google Scholar] [CrossRef]
- Mandik, F.; Kanana, Y.; Rody, J.; Misera, S.; Wilken, B.; Laabs von Holt, B.-H.; Klein, C.; Vos, M. A new model for fatty acid hydroxylase-associated neurodegeneration reveals mitochondrial and autophagy abnormalities. Front. Cell Dev. Biol. 2022, 10, 1000553. [Google Scholar] [CrossRef]
- Völkner, C.; Liedtke, M.; Petters, J.; Huth, K.; Knuebel, G.; Murua Escobar, H.; Bullerdiek, J.; Lukas, J.; Hermann, A.; Frech, M.J. Generation of an iPSC line (AKOSi006-A) from fibroblasts of an NPC1 patient, carrying the homozygous mutation p.I1061T (c.3182 T C) and a control iPSC line (AKOSi007-A) using a non-integrating Sendai virus system. Stem Cell Res. 2020, 49, 102056. [Google Scholar] [CrossRef]
- Efendic, F.; Völkner, C.; Krohn, S.; Murua Escobar, H.; Venkateswaran, S.; Bennett, S.; Hermann, A.; Frech, M.J. Generation of the human iPSC line AKOSi010-A from fibroblasts of a female FAHN patient, carrying the compound heterozygous mutation p.Gly45Arg/p.His319Arg. Stem Cell Res. 2022, 63, 102863. [Google Scholar] [CrossRef]
- García-León, J.A.; García-Díaz, B.; Eggermont, K.; Cáceres-Palomo, L.; Neyrinck, K.; Da Madeiro Costa, R.; Dávila, J.C.; Baron-Van Evercooren, A.; Gutiérrez, A.; Verfaillie, C.M. Generation of oligodendrocytes and establishment of an all-human myelinating platform from human pluripotent stem cells. Nat. Protoc. 2020, 15, 3716–3744. [Google Scholar] [CrossRef] [PubMed]
- Hama, H. Fatty acid 2-Hydroxylation in mammalian sphingolipid biology. Biochim. Biophys. Acta 2010, 1801, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, M.; Yaghootfam, A.; Fewou, S.N.; Zöller, I.; Gieselmann, V. A mammalian fatty acid hydroxylase responsible for the formation of alpha-hydroxylated galactosylceramide in myelin. Biochem. J. 2005, 388, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Maier, H.; Meixner, M.; Hartmann, D.; Sandhoff, R.; Wang-Eckhardt, L.; Zöller, I.; Gieselmann, V.; Eckhardt, M. Normal fur development and sebum production depends on fatty acid 2-hydroxylase expression in sebaceous glands. J. Biol. Chem. 2011, 286, 25922–25934. [Google Scholar] [CrossRef]
- Jordans, S.; Hardt, R.; Becker, I.; Winter, D.; Wang-Eckhardt, L.; Eckhardt, M. Age-Dependent Increase in Schmidt-Lanterman Incisures and a Cadm4-Associated Membrane Skeletal Complex in Fatty Acid 2-hydroxylase Deficient Mice: A Mouse Model of Spastic Paraplegia SPG35. Mol. Neurobiol. 2022, 59, 3969–3979. [Google Scholar] [CrossRef]
- Fitzner, D.; Schneider, A.; Kippert, A.; Möbius, W.; Willig, K.I.; Hell, S.W.; Bunt, G.; Gaus, K.; Simons, M. Myelin basic protein-dependent plasma membrane reorganization in the formation of myelin. EMBO J. 2006, 25, 5037–5048. [Google Scholar] [CrossRef]
- Lappe-Siefke, C.; Goebbels, S.; Gravel, M.; Nicksch, E.; Lee, J.; Braun, P.E.; Griffiths, I.R.; Nave, K.-A. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat. Genet. 2003, 33, 366–374. [Google Scholar] [CrossRef]
- Windschiegl, B.; Steinem, C. Influence of alpha-hydroxylation of glycolipids on domain formation in lipid monolayers. Langmuir 2006, 22, 7454–7457. [Google Scholar] [CrossRef]
- Edgar, J.M.; McLaughlin, M.; Yool, D.; Zhang, S.-C.; Fowler, J.H.; Montague, P.; Barrie, J.A.; McCulloch, M.C.; Duncan, I.D.; Garbern, J.; et al. Oligodendroglial modulation of fast axonal transport in a mouse model of hereditary spastic paraplegia. J. Cell Biol. 2004, 166, 121–131. [Google Scholar] [CrossRef]
- Honke, K.; Hirahara, Y.; Dupree, J.; Suzuki, K.; Popko, B.; Fukushima, K.; Fukushima, J.; Nagasawa, T.; Yoshida, N.; Wada, Y.; et al. Paranodal junction formation and spermatogenesis require sulfoglycolipids. Proc. Natl. Acad. Sci. USA 2002, 99, 4227–4232. [Google Scholar] [CrossRef] [PubMed]
- Arber, C.E.; Li, A.; Houlden, H.; Wray, S. Review: Insights into molecular mechanisms of disease in neurodegeneration with brain iron accumulation: Unifying theories. Neuropathol. Appl. Neurobiol. 2016, 42, 220–241. [Google Scholar] [CrossRef]
- Maruyama, T.; Noda, N.N. Autophagy-regulating protease Atg4: Structure, function, regulation and inhibition. J. Antibiot. 2017, 71, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Fernández, Á.F.; López-Otín, C. The functional and pathologic relevance of autophagy proteases. J. Clin. Invest. 2015, 125, 33–41. [Google Scholar] [CrossRef]
- Yu, Z.-Q.; Ni, T.; Hong, B.; Wang, H.-Y.; Jiang, F.-J.; Zou, S.; Chen, Y.; Zheng, X.-L.; Klionsky, D.J.; Liang, Y.; et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 2012, 8, 883–892. [Google Scholar] [CrossRef]
| Name | Host | Dilution | Company | Reference No# |
|---|---|---|---|---|
| Anti-O4-APC FACS | Mouse | 1:50 | Miltenyi Biotec | 130-095-891 |
| BETA III TUBULIN | Mouse IgG | 1:100 | Santa Cruz | SC51670 |
| FA2H | Rabbit IgG | 1:250 | Invitrogen | PA5-24728 |
| GAPDH | Mouse IgG | 1:10,000 | Abcam | AB8245 |
| GFAP | Rabbit IgG | 1:500 | Agilent | Z0334 |
| KCNQ2/KV7.2 | Mouse IgG | 1:100 | Abcam | AB22897 |
| LC3B (E7X4S) | Rabbit IgG | 1:1000 | Cell Signaling | 43566S |
| MAP2 | Chicken IgY | 1:2000 | Abcam | AB5392 |
| MBP | Chicken IgY | 1:100 | ThermoFisher | AB9348 |
| MBP aa 82–87 | Rat IgG | 1:75 | EMD Millipore | MAB386 |
| O4 | Mouse IgM | 1:100 | R&D Systems | MAB1326 |
| PAX6 | Rabbit IgG | 1:1000 | Abcam | AB5790 |
| PLP | Rabbit IgG | 1:100 | Abcam | AB_9311 |
| SOX2 | Rabbit IgG | 1:500 | Abcam | AB92494 |
| SOX10 | Goat IgG | 1:100 | R&D Systems | AF2864 |
| SQSTML/P62 | Rabbit IgG | 1:500 | Abcam | ABl09012 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Efendic, F.; Hermann, A.; Frech, M.J. Disrupted Myelination in FAHN: Insights from a Patient-Specific hiPSC Neuron–Oligodendrocyte Model. Cells 2025, 14, 1261. https://doi.org/10.3390/cells14161261
Efendic F, Hermann A, Frech MJ. Disrupted Myelination in FAHN: Insights from a Patient-Specific hiPSC Neuron–Oligodendrocyte Model. Cells. 2025; 14(16):1261. https://doi.org/10.3390/cells14161261
Chicago/Turabian StyleEfendic, Fatima, Andreas Hermann, and Moritz J. Frech. 2025. "Disrupted Myelination in FAHN: Insights from a Patient-Specific hiPSC Neuron–Oligodendrocyte Model" Cells 14, no. 16: 1261. https://doi.org/10.3390/cells14161261
APA StyleEfendic, F., Hermann, A., & Frech, M. J. (2025). Disrupted Myelination in FAHN: Insights from a Patient-Specific hiPSC Neuron–Oligodendrocyte Model. Cells, 14(16), 1261. https://doi.org/10.3390/cells14161261







