Exploiting TCR Repertoire Analysis to Select Therapeutic TCRs for Cancer Immunotherapy
Abstract
1. Introduction
2. T Cells in Cancer Immunity
2.1. TCRs Govern T Cell Function
2.2. Cancer Targets Recognised by TCRs
3. Insights from TCR Composition and Diversity in the TME
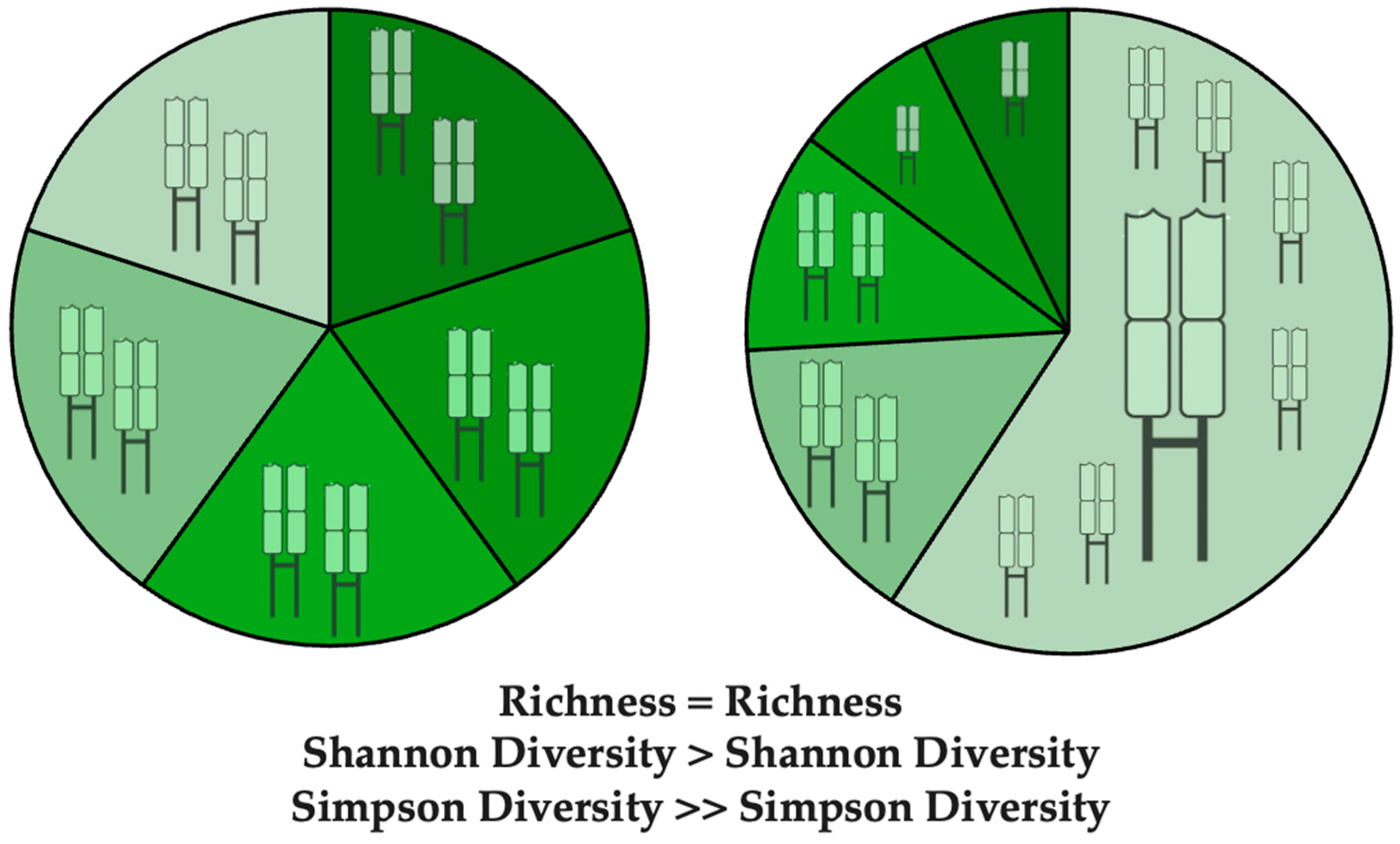
3.1. Metrics of TCR Diversity and Clonal Distribution
3.2. Predictive Power of TCR Clonality Pre and Post Treatment
3.3. TCR Sharing Between Blood and Tumour
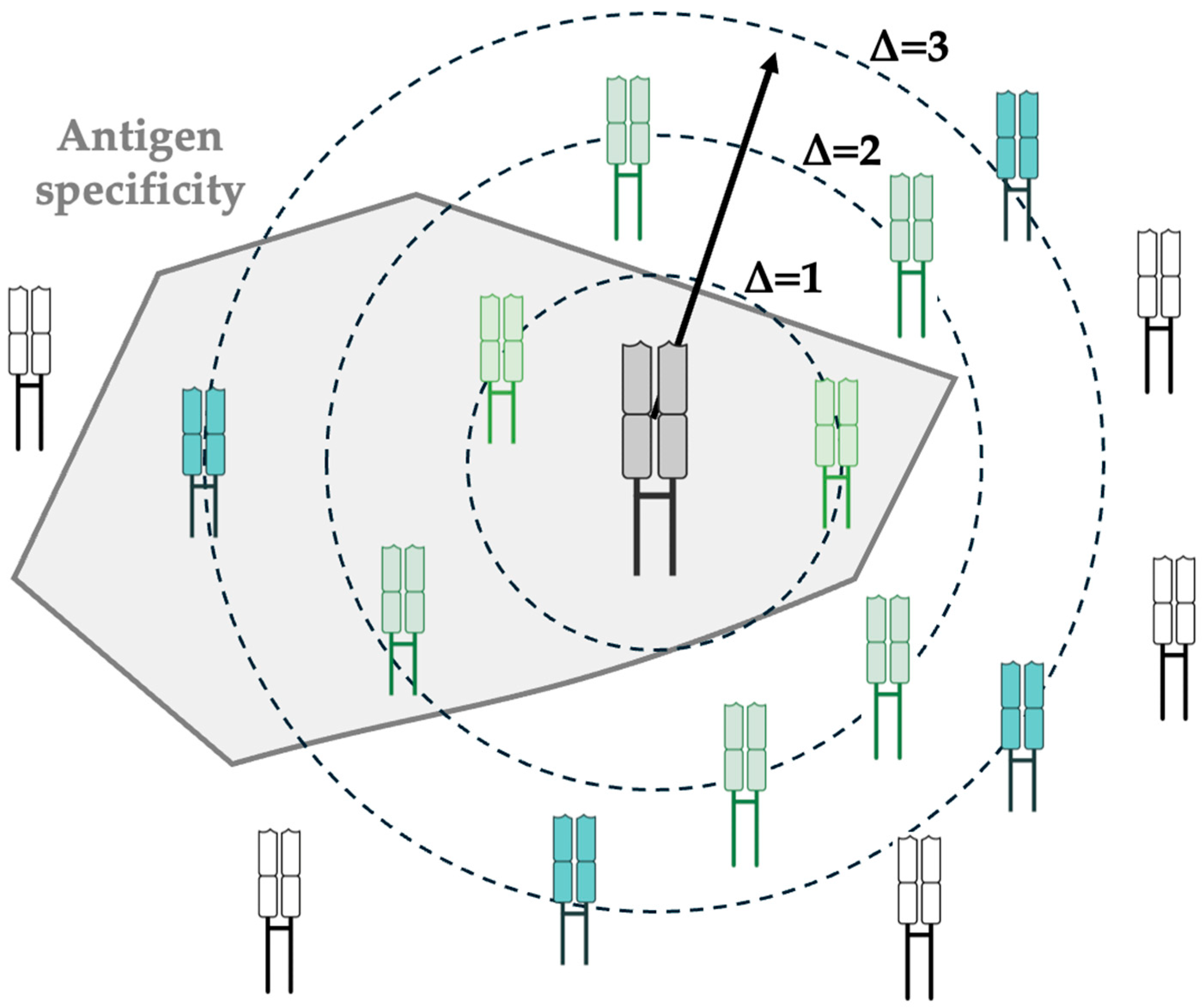
3.4. Public TCR Responses Shared Across Patients
3.5. TCR Meta-Clonotypes
3.6. Which T Cell Subsets Harbour Clonal Expansion?
4. TCR Selection Criteria for TCR Based Immunotherapies
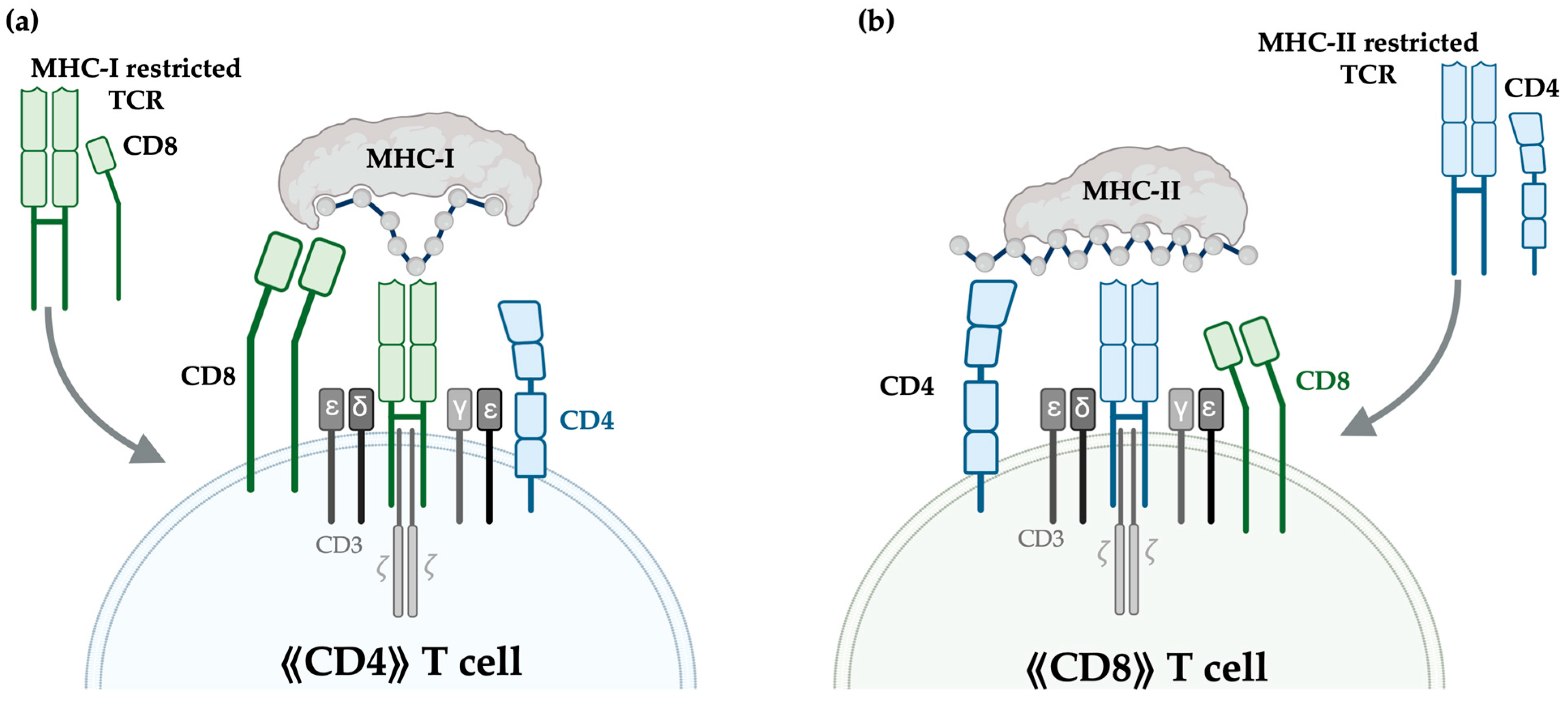
4.1. MHC-I- vs. MHC-II-Restricted TCRs
4.2. Selecting TCRs by Motif Search and Clonal Expansion
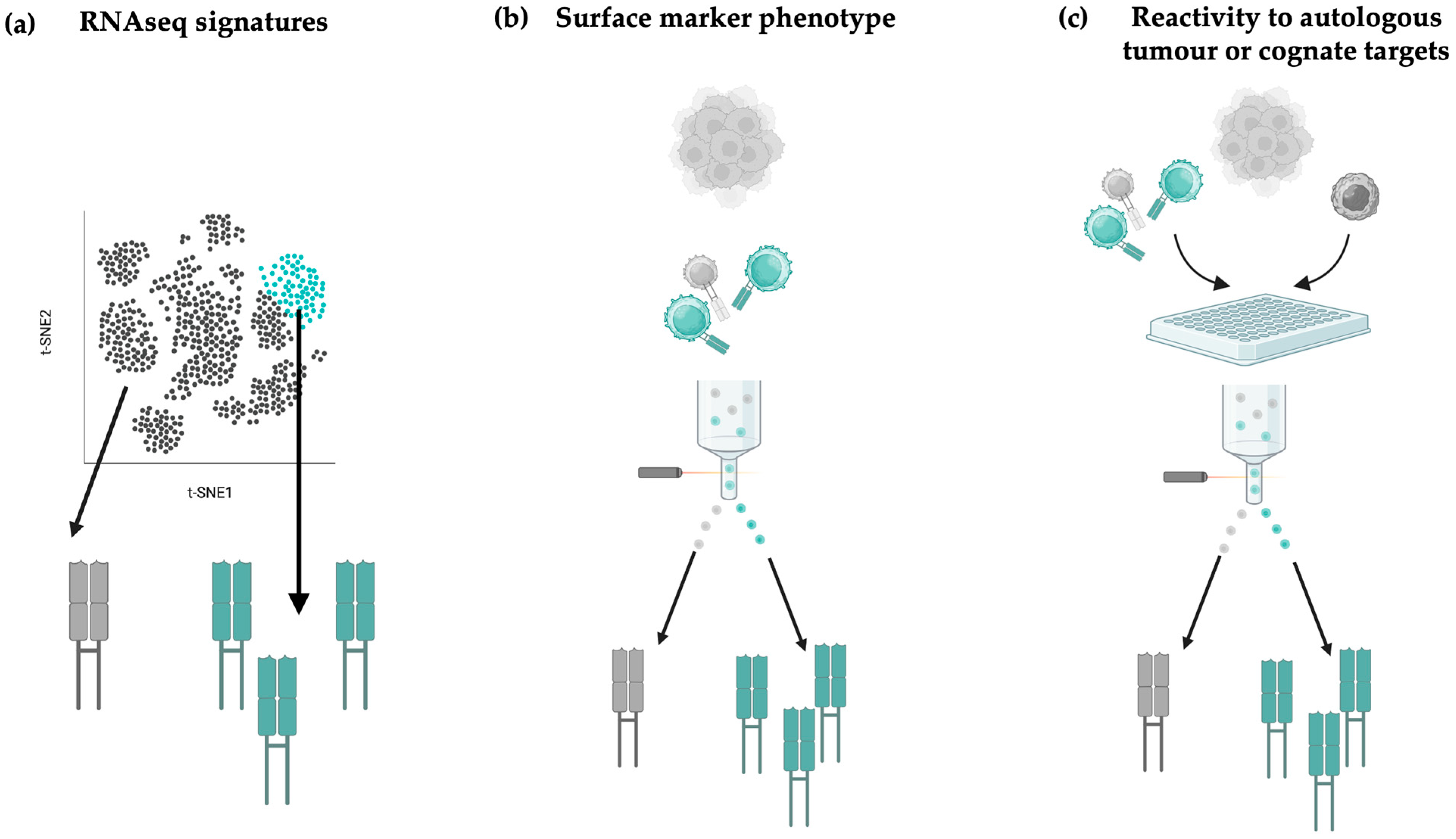
4.3. Prioritising TCRs Based on Gene-Expression Signatures
4.4. Prioritising TCRs Based on Surface Marker Activation Phenotypes
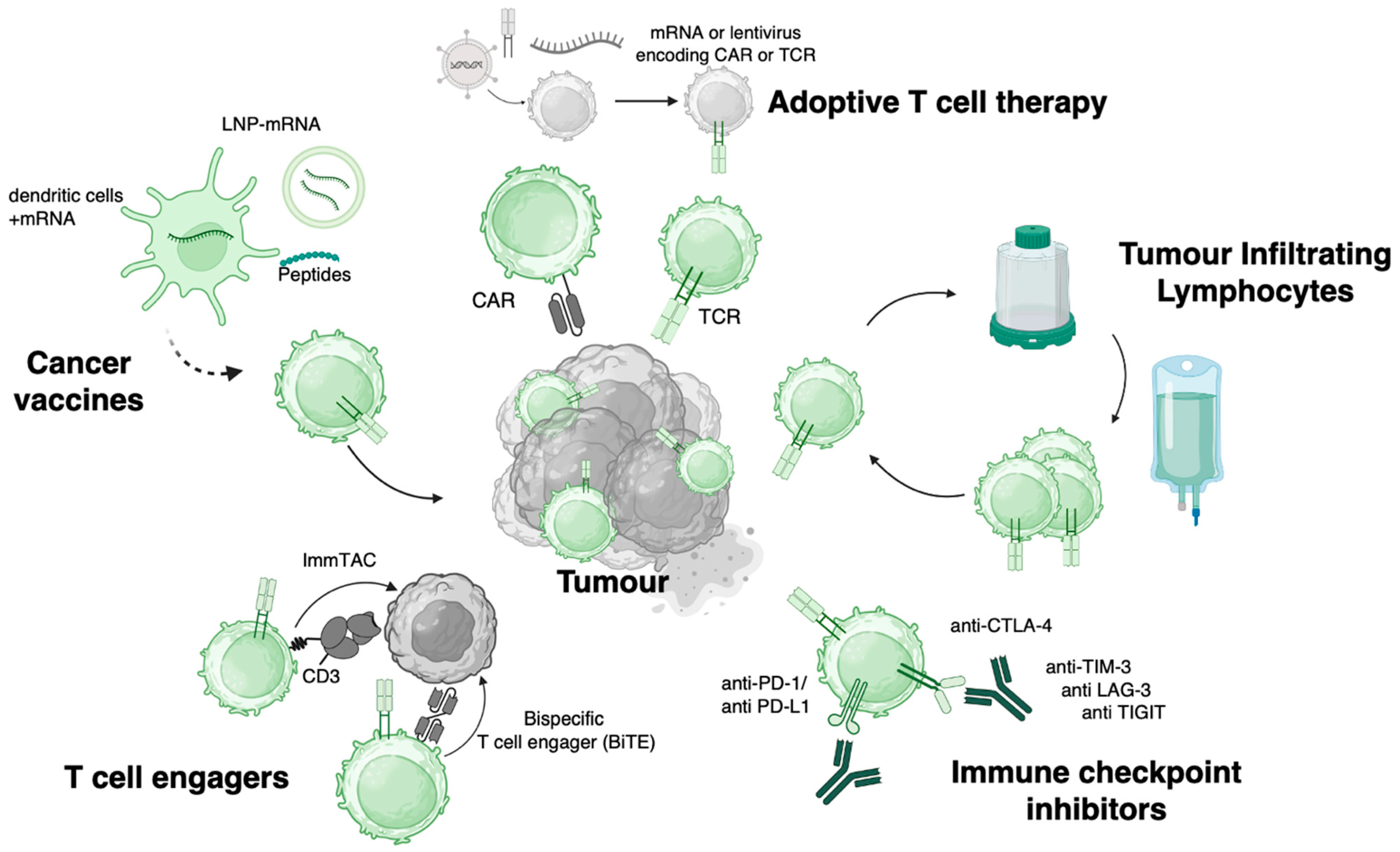
5. T Cell Engineering
5.1. Negatively Selecting for Cross-Reactivities Against Healthy Tissue
5.2. Genetic Engineering of TCRs to Manufacture Therapeutic Cell Products
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AAV | Adeno-associated virus |
| APC | Antigen-presenting cell |
| CAR | Chimeric antigen receptor |
| CDR | Complementarity determining region |
| CTA | Cancer testis antigens |
| EBV | Epstein–Barr virus |
| HBV | Hepatitis B virus |
| HCV | Hepatitis C virus |
| HTLV-1 | Human T cell leukaemia virus 1 |
| RCC | Renal cell carcinoma |
| TAA | Tumour-associated antigen |
| TCR | T cell receptor |
| TME | Tumour microenvironment |
| TRAC | T cell receptor alpha constant |
| TRBC | T cell receptor beta constant |
| Treg | T regulatory cell |
| TRM | T resident memory |
| TIL | Tumour-infiltrating lymphocyte |
| BCMA | B cell maturation antigen |
| FDA | Food and Drug Administration |
| IPSC | Induced pluripotent stem cell |
| K.I | Knock-in |
| K.O | Knock-out |
| PD-1 | Programmed cell death protein 1 |
| MAGE-A4 | Melanoma-associated antigen gene A4 |
| MHC | Major histocompatibility complex |
| NSCLC | Non-small cell lung cancer |
| NK | Natural killer |
| scFv | Single-chain variable fragment |
References
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Packard, B.S.; Aebersold, P.M.; Solomon, D.; Topalian, S.L.; Toy, S.T.; Simon, P.; Lotze, M.T.; Yang, J.C.; Seipp, C.A.; et al. Use of Tumor-Infiltrating Lymphocytes and Interleukin-2 in the Immunotherapy of Patients with Metastatic Melanoma. N. Engl. J. Med. 1988, 319, 1676–1680. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Lotze, M.T.; Muul, L.M.; Leitman, S.; Chang, A.E.; Ettinghausen, S.E.; Matory, Y.L.; Skibber, J.M.; Shiloni, E.; Vetto, J.T.; et al. Observations on the Systemic Administration of Autologous Lymphokine-Activated Killer Cells and Recombinant Interleukin-2 to Patients with Metastatic Cancer. N. Engl. J. Med. 1985, 313, 1485–1492. [Google Scholar] [CrossRef]
- Hamid, O.; Robert, C.; Daud, A.; Hodi, F.S.; Hwu, W.J.; Kefford, R.; Wolchok, J.D.; Hersey, P.; Joseph, R.W.; Weber, J.S.; et al. Safety and Tumor Responses with Lambrolizumab (Anti–PD-1) in Melanoma. N. Engl. J. Med. 2013, 369, 134–144. [Google Scholar] [CrossRef]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pagès, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, Density, and Location of Immune Cells Within Human Colorectal Tumors Predict Clinical Outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.M.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 Blockade Induces Responses by Inhibiting Adaptive Immune Resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef]
- Zhang, L.; Conejo-Garcia, J.R.; Katsaros, D.; Gimotty, P.A.; Massobrio, M.; Regnani, G.; Makrigiannakis, A.; Gray, H.; Schlienger, K.; Liebman, M.N.; et al. Intratumoral T Cells, Recurrence, and Survival in Epithelial Ovarian Cancer. N. Engl. J. Med. 2003, 348, 203–213. [Google Scholar] [CrossRef]
- D’Angelo, S.P.; Araujo, D.M.; Abdul Razak, A.R.; Agulnik, M.; Attia, S.; Blay, J.-Y.; Carrasco Garcia, I.; Charlson, J.A.; Choy, E.; Demetri, G.D.; et al. Afamitresgene autoleucel for advanced synovial sarcoma and myxoid round cell liposarcoma (SPEARHEAD-1): An international, open-label, phase 2 trial. Lancet 2024, 403, 1460–1471. [Google Scholar] [CrossRef]
- Schumacher, T.N.; Schreiber, R.D. Neoantigens in Cancer Immunotherapy. Science 2015, 348, 69–74. [Google Scholar] [CrossRef]
- Shafer, P.; Kelly, L.M.; Hoyos, V. Cancer Therapy With TCR-Engineered T Cells: Current Strategies, Challenges, and Prospects. Front. Immunol. 2022, 13, 835762. [Google Scholar]
- Cheever, M.A.; Allison, J.P.; Ferris, A.S.; Finn, O.J.; Hastings, B.M.; Hecht, T.T.; Mellman, I.; Prindiville, S.A.; Viner, J.L.; Weiner, L.M.; et al. The Prioritization of Cancer Antigens: A National Cancer Institute Pilot Project for the Acceleration of Translational Research. Clin. Cancer Res. 2009, 15, 5323–5337. [Google Scholar] [CrossRef]
- Shah, N.M.; Jang, H.J.; Liang, Y.; Maeng, J.H.; Tzeng, S.-C.; Wu, A.; Basri, N.L.; Qu, X.; Fan, C.; Li, A.; et al. Pan-Cancer Analysis Identifies Tumor-Specific Antigens Derived from Transposable Elements. Nat. Genet. 2023, 55, 631–639. [Google Scholar] [CrossRef]
- Kwok, D.W.; Stevers, N.O.; Etxeberria, I.; Nejo, T.; Colton Cove, M.; Chen, L.H.; Jung, J.; Okada, K.; Lakshmanachetty, S.; Gallus, M.; et al. Tumour-wide RNA splicing aberrations generate actionable public neoantigens. Nature 2025, 639, 463–473. [Google Scholar] [CrossRef]
- Ferreira, H.J.; Stevenson, B.J.; Pak, H.; Yu, F.; Almeida Oliveira, J.; Huber, F.; Taillandier-Coindard, M.; Michaux, J.; Ricart-Altimiras, E.; Kraemer, A.I.; et al. Immunopeptidomics-Based Identification of Naturally Presented Non-Canonical circRNA-Derived Peptides. Nat. Commun. 2024, 15, 2357. [Google Scholar] [CrossRef]
- Kalaora, S.; Nagler, A.; Nejman, D.; Alon, M.; Barbolin, C.; Barnea, E.; Ketelaars, S.L.C.; Cheng, K.; Vervier, K.; Shental, N.; et al. Identification of Bacteria-Derived HLA-Bound Peptides in Melanoma. Nature 2021, 592, 138–143. [Google Scholar] [CrossRef]
- Naghavian, R.; Faigle, W.; Oldrati, P.; Wang, J.; Toussaint, N.C.; Qiu, Y.; Medici, G.; Wacker, M.; Freudenmann, L.K.; Bonté, P.-E.; et al. Microbial Peptides Activate Tumour-Infiltrating Lymphocytes in Glioblastoma. Nature 2023, 617, 807–817. [Google Scholar] [CrossRef]
- Shaban, M.; Khurram, S.A.; Fraz, M.M.; Alsubaie, N.; Masood, I.; Mushtaq, S.; Hassan, M.; Loya, A.; Rajpoot, N.M. A Novel Digital Score for Abundance of Tumour Infiltrating Lymphocytes Predicts Disease Free Survival in Oral Squamous Cell Carcinoma. Sci. Rep. 2019, 9, 13341. [Google Scholar] [CrossRef]
- Nakkireddy, S.R.; Jang, I.; Kim, M.; Yin, L.X.; Rivera, M.; Garcia, J.J.; Bartemes, K.R.; Routman, D.M.; Moore, E.J.; Abdel-Halim, C.N.; et al. Integrative Analysis of H&E and IHC Identifies Prognostic Immune Subtypes in HPV Related Oropharyngeal Cancer. Commun. Med. 2024, 4, 190. [Google Scholar] [CrossRef]
- Jost, L. Entropy and Diversity. Oikos 2006, 113, 363–375. [Google Scholar] [CrossRef]
- Chiffelle, J.; Genolet, R.; Perez, M.A.; Coukos, G.; Zoete, V.; Harari, A. T-cell Repertoire Analysis and Metrics of Diversity and Clonality. Curr. Opin. Biotechnol. 2020, 65, 284–295. [Google Scholar] [CrossRef]
- Laydon, D.J.; Bangham, C.R.M.; Asquith, B. Estimating T-cell Repertoire Diversity: Limitations of Classical Estimators and a New Approach. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140291. [Google Scholar] [CrossRef]
- Tiffeau-Mayer, A. Unbiased Estimation of Sampling Variance for Simpson’s Diversity Index. Phys. Rev. E 2024, 109, 064411. [Google Scholar] [CrossRef]
- Simpson, E.H. Measurement of Diversity. Nature 1949, 163, 688. [Google Scholar] [CrossRef]
- Mimura, K.; Ogata, T.; Nguyen, P.H.; Roy, S.; Kared, H.; Yuan, Y.C.; Fehlings, M.; Yoshimoto, Y.; Yoshida, D.; Nakajima, S.; et al. Combination of Oligo-Fractionated Irradiation with Nivolumab can Induce Immune Modulation in Gastric Cancer. J. Immunother. Cancer 2024, 12, e008385. [Google Scholar] [CrossRef]
- Jin, Y.; Luo, W.; Zhang, G.; Lin, K.; Cui, J.; Chen, X.; Pan, Y.; Mao, X.; Tang, J.; Wang, Y. TCR Repertoire Profiling of Tumors, Adjacent Normal Tissues, and Peripheral Blood Predicts Survival in Nasopharyngeal Carcinoma. Cancer Immunol. Immunother. 2018, 67, 1719–1730. [Google Scholar] [CrossRef]
- Bai, X.; Zhang, Q.; Wu, S.; Zhang, X.; Wang, M.; He, F.; Wei, T.; Yang, J.; Lou, Y.; Cai, Z.; et al. Characteristics of Tumor Infiltrating Lymphocyte and Circulating Lymphocyte Repertoires in Pancreatic Cancer by the Sequencing of T Cell Receptors. Sci. Rep. 2015, 5, 13664. [Google Scholar] [CrossRef]
- Pruessmann, W.; Rytlewski, J.; Wilmott, J.; Mihm, M.C.; Attrill, G.H.; Dyring-Andersen, B.; Fields, P.; Zhan, Q.; Colebatch, A.J.; Ferguson, P.M.; et al. Molecular Analysis of Primary Melanoma T cells Identifies Patients at Risk for Metastatic Recurrence. Nat. Cancer 2020, 1, 197–209. [Google Scholar] [CrossRef]
- Chiffelle, J.; Barras, D.; Pétremand, R.; Orcurto, A.; Bobisse, S.; Arnaud, M.; Auger, A.; Rodrigo, B.N.; Ghisoni, E.; Sauvage, C.; et al. Tumor-reactive T Cell Clonotype Dynamics Underlying Clinical Response to TIL Therapy in Melanoma. Immunity 2024, 57, 2466–2482.e12. [Google Scholar] [CrossRef]
- Kamphorst, A.O.; Pillai, R.N.; Yang, S.; Nasti, T.H.; Akondy, R.S.; Wieland, A.; Sica, G.L.; Yu, K.; Koenig, L.; Patel, N.T.; et al. Proliferation of PD-1+ CD8 T Cells in Peripheral Blood After PD-1–Targeted Therapy in Lung Cancer Patients. Proc. Natl. Acad. Sci. USA 2017, 114, 4993–4998. [Google Scholar] [CrossRef]
- Yost, K.E.; Satpathy, A.T.; Wells, D.K.; Qi, Y.; Wang, C.; Kageyama, R.; McNamara, K.L.; Granja, J.M.; Sarin, K.Y.; Brown, R.A.; et al. Clonal Replacement of Tumor-Specific T Cells Following PD-1 Blockade. Nat. Med. 2019, 25, 1251–1259. [Google Scholar] [CrossRef]
- Wang, Z.; Zhong, Y.; Zhang, Z.; Zhou, K.; Huang, Z.; Yu, H.; Liu, L.; Liu, S.; Yang, H.; Zhou, J.; et al. Characteristics and Clinical Significance of T-Cell Receptor Repertoire in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 847263. [Google Scholar]
- Sherwood, A.M.; Emerson, R.O.; Scherer, D.; Habermann, N.; Buck, K.; Staffa, J.; Desmarais, C.; Halama, N.; Jaeger, D.; Schirmacher, P.; et al. Tumor-infiltrating lymphocytes in Colorectal Tumors Display a Diversity of T Cell Receptor Sequences that Differ from the T Cells in Adjacent Mucosal Tissue. Cancer Immunol. Immunother. 2013, 62, 1453–1461. [Google Scholar] [CrossRef]
- Beausang, J.F.; Wheeler, A.J.; Chan, N.H.; Hanft, V.R.; Dirbas, F.M.; Jeffrey, S.S.; Quake, S.R. T Cell Receptor Sequencing of Early-Stage Breast Cancer Tumors Identifies Altered Clonal Structure of the T cell Repertoire. Proc. Natl. Acad. Sci. USA 2017, 114, E10409–E10417. [Google Scholar] [CrossRef]
- Wang, T.; Wang, C.; Wu, J.; He, C.; Zhang, W.; Liu, J.; Zhang, R.; Lv, Y.; Li, Y.; Zeng, X.; et al. The Different T-cell Receptor Repertoires in Breast Cancer Tumors, Draining Lymph Nodes, and Adjacent Tissues. Cancer Immunol. Res. 2017, 5, 148–156. [Google Scholar] [CrossRef]
- Serana, F.; Sottini, A.; Caimi, L.; Palermo, B.; Natali, P.G.; Nisticò, P.; Imberti, L. Identification of a public CDR3 motif and a biased utilization of T-cell receptor V beta and J beta chains in HLA-A2/Melan-A-specific T-cell Clonotypes of Melanoma Patients. J. Transl. Med. 2009, 7, 21. [Google Scholar] [CrossRef]
- Ruggiero, E.; Nicolay, J.P.; Fronza, R.; Arens, A.; Paruzynski, A.; Nowrouzi, A.; Ürenden, G.; Lulay, C.; Schneider, S.; Goerdt, S.; et al. High-resolution Analysis of the Human T-cell Receptor Repertoire. Nat. Commun. 2015, 6, 8081. [Google Scholar] [CrossRef]
- Chung, Y.-L.; Wu, M.-L. Spatiotemporal Homogeneity and Distinctness of the T-Cell Receptor β-Chain Repertoires in Epstein–Barr Virus-Associated Primary and Metastatic Nasopharyngeal Carcinomas. Int. J. Cancer 2018, 143, 610–620. [Google Scholar] [CrossRef]
- Mayer-Blackwell, K.; Schattgen, S.; Cohen-Lavi, L.; Crawford, J.C.; Souquette, A.; Gaevert, J.A.; Hertz, T.; Thomas, P.G.; Bradley, P.; Fiore-Gartland, A. TCR Meta-Clonotypes for Biomarker Discovery with Tcrdist3 Enabled Identification of Public, HLA-Restricted Clusters of SARS-CoV-2 TCRs. eLife 2021, 10, e68605. [Google Scholar] [CrossRef]
- Lagattuta, K.A.; Kang, J.B.; Nathan, A.; Pauken, K.E.; Jonsson, A.H.; Rao, D.A.; Sharpe, A.H.; Ishigaki, K.; Raychaudhuri, S. Repertoire Analyses Reveal T cell Antigen Receptor Sequence Features that Influence T cell fate. Nat. Immunol. 2022, 23, 446–457. [Google Scholar] [CrossRef]
- Kajihara, R.; Long, M.D.; Hoki, T.; Chen, H.; Yamauchi, T.; Kanemaru, H.; Segal, B.H.; Dy, G.K.; Ito, F. Divergent Transcriptional States and Kinetics of Circulating Tumor-Infiltrating Lymphocyte Repertoires with Highly Homologous T-cell Receptor Sequences in a Patient During Immunotherapy. J. Immunother. Cancer 2025, 13, e010092. [Google Scholar] [CrossRef]
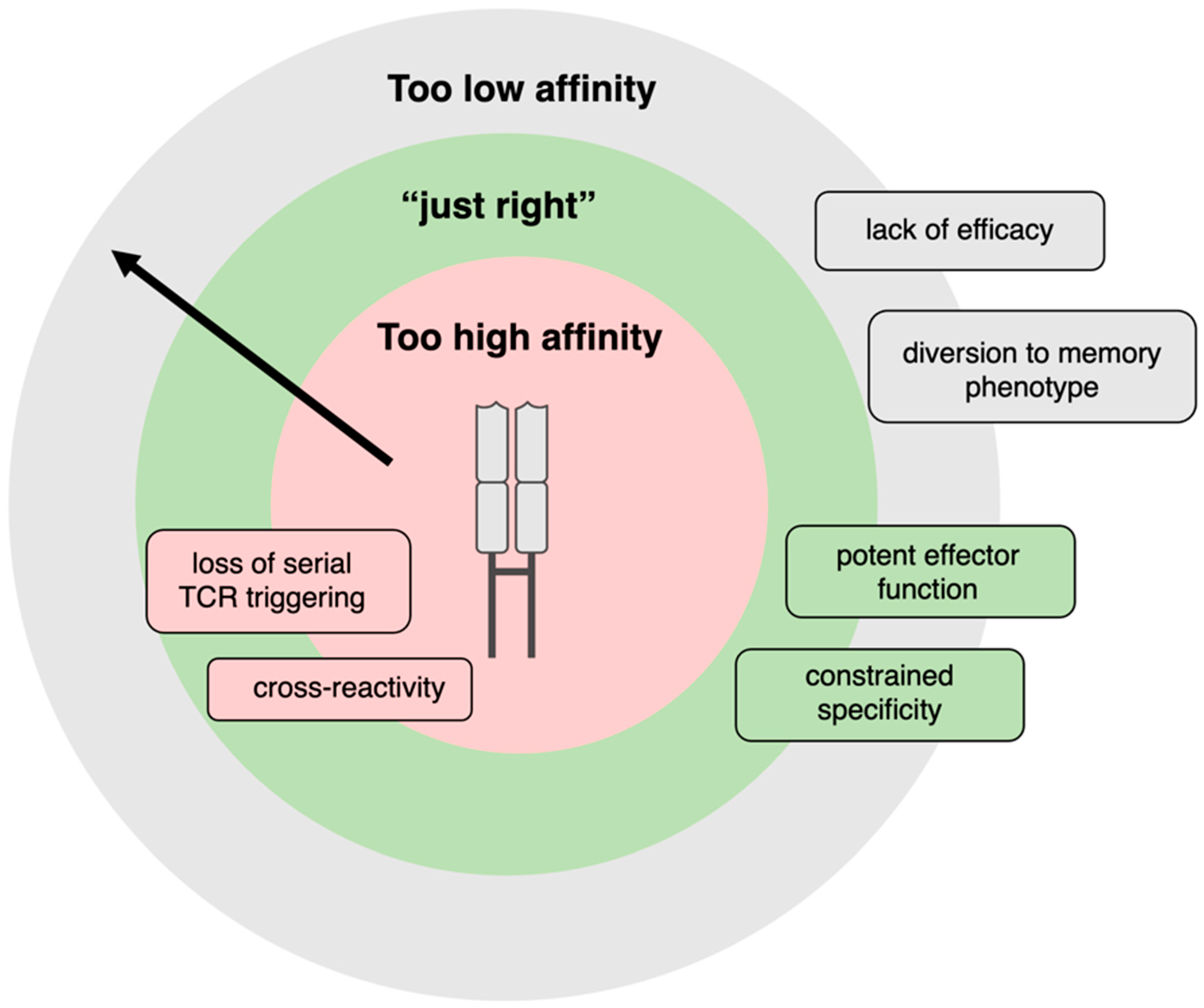
- Thomas, S.; Xue, S.-A.; Bangham, C.R.M.; Jakobsen, B.K.; Morris, E.C.; Stauss, H.J. Human T cells Expressing Affinity-Matured TCR Display Accelerated Responses but Fail to Recognize Low Density of MHC-Peptide Antigen. Blood 2011, 118, 319–329. [Google Scholar] [CrossRef]
- Tan, M.P.; Gerry, A.B.; Brewer, J.E.; Melchiori, L.; Bridgeman, J.S.; Bennett, A.D.; Pumphrey, N.J.; Jakobsen, B.K.; Price, D.A.; Ladell, K.; et al. T Cell Receptor Binding Affinity Governs THE Functional Profile of Cancer-Specific CD8+ T Cells. Clin. Exp. Immunol. 2015, 180, 255–270. [Google Scholar] [CrossRef]
- Lowery, F.J.; Krishna, S.; Yossef, R.; Parikh, N.B.; Chatani, P.D.; Zacharakis, N.; Parkhurst, M.R.; Levin, N.; Sindiri, S.; Sachs, A.; et al. Molecular Signatures of Antitumor Neoantigen-Reactive T cells from Metastatic Human Cancers. Science 2022, 375, 877–884. [Google Scholar] [CrossRef]
- Spindler, M.J.; Nelson, A.L.; Wagner, E.K.; Oppermans, N.; Bridgeman, J.S.; Heather, J.M.; Adler, A.S.; Asensio, M.A.; Edgar, R.C.; Lim, Y.W.; et al. Massively Parallel Interrogation and Mining Of Natively Paired Human TCRαβ Repertoires. Nat. Biotechnol. 2020, 38, 609–619. [Google Scholar] [CrossRef]
- Vazquez-Lombardi, R.; Jung, J.S.; Schlatter, F.S.; Mei, A.; Mantuano, N.R.; Bieberich, F.; Hong, K.-L.; Kucharczyk, J.; Kapetanovic, E.; Aznauryan, E.; et al. High-throughput T Cell Receptor Engineering by Functional Screening Identifies Candidates with Enhanced Potency and Specificity. Immunity 2022, 55, 1953–1966.e10. [Google Scholar] [CrossRef]
- Kidani, Y.; Nogami, W.; Yasumizu, Y.; Kawashima, A.; Tanaka, A.; Sonoda, Y.; Tona, Y.; Nashiki, K.; Matsumoto, R.; Hagiwara, M.; et al. CCR8-targeted specific depletion of clonally expanded Treg cells in tumor tissues evokes potent tumor immunity with long-lasting memory. Proc. Natl. Acad. Sci. USA 2022, 119, e2114282119. [Google Scholar] [CrossRef]
- Gambardella, V.; Ong, M.; Rodriguez-Ruiz, M.E.; Machiels, J.-P.; Sanmamed, M.F.; Galvao, V.; Spreafico, A.; Renouf, D.J.; Luen, S.J.; Galot, R.; et al. Safety and Antitumor Activity of a Novel aCD25 Treg Depleter RG6292 as a Single Agent and in Combination with Atezolizumab in Patients with Solid Tumors. Cancer Res. Commun. 2025, 5, 422–432. [Google Scholar] [CrossRef]
- Núñez, N.G.; Tosello Boari, J.; Ramos, R.N.; Richer, W.; Cagnard, N.; Anderfuhren, C.D.; Niborski, L.L.; Bigot, J.; Meseure, D.; De La Rochere, P.; et al. Tumor Invasion in Draining Lymph Nodes is Associated with Treg Accumulation in Breast Cancer Patients. Nat. Commun. 2020, 11, 3272. [Google Scholar] [CrossRef]
- Bredel, D.; Tihic, E.; Mouraud, S.; Danlos, F.-X.; Susini, S.; Aglave, M.; Alfaro, A.; Mohamed-Djalim, C.; Rouanne, M.; Halse, H.; et al. Immune checkpoints are predominantly co-expressed by clonally expanded CD4+FoxP3+ intratumoral T-cells in primary human cancers. J. Exp. Clin. Cancer Res. 2023, 42, 333. [Google Scholar] [CrossRef]
- Sainz-Perez, A.; Lim, A.; Lemercier, B.; Leclerc, C. The T-cell Receptor Repertoire of Tumor-Infiltrating Regulatory T Lymphocytes Is Skewed Toward Public Sequences. Cancer Res. 2012, 72, 3557–3569. [Google Scholar] [CrossRef]
- Oh, D.Y.; Kwek, S.S.; Raju, S.S.; Li, T.; McCarthy, E.; Chow, E.; Aran, D.; Ilano, A.; Pai, C.-C.S.; Rancan, C.; et al. Intratumoral CD4+ T Cells Mediate Anti-tumor Cytotoxicity in Human Bladder Cancer. Cell 2020, 181, 1612–1625.e13. [Google Scholar] [CrossRef]
- Liotta, F.; Gacci, M.; Frosali, F.; Querci, V.; Vittori, G.; Lapini, A.; Santarlasci, V.; Serni, S.; Cosmi, L.; Maggi, L.; et al. Frequency of Regulatory T Cells in Peripheral Blood and in Tumour-Infiltrating Lymphocytes Correlates with Poor Prognosis in Renal Cell Carcinoma. BJU Int. 2011, 107, 1500–1506. [Google Scholar] [CrossRef]
- Bergsland, C.H.; Jeanmougin, M.; Moosavi, S.H.; Svindland, A.; Bruun, J.; Nesbakken, A.; Sveen, A.; Lothe, R.A. Spatial Analysis and CD25-Expression Identify Regulatory T Cells As Predictors of a Poor Prognosis in Colorectal Cancer. Mod. Pathol. 2022, 35, 1236–1246. [Google Scholar] [CrossRef]
- Maeda, N.; Yoshimura, K.; Yamamoto, S.; Kuramasu, A.; Inoue, M.; Suzuki, N.; Watanabe, Y.; Maeda, Y.; Kamei, R.; Tsunedomi, R.; et al. Expression of B7-H3, a Potential Factor of Tumor Immune Evasion in Combination with the Number of Regulatory T Cells, Affects Against Recurrence-Free Survival in Breast Cancer Patients. Ann. Surg. Oncol. 2014, 21, 546–554. [Google Scholar] [CrossRef]
- Saillard, M.; Cenerenti, M.; Reichenbach, P.; Guillaume, P.; Su, Z.; Hafezi, M.; Schmidt, J.; Cesbron, J.; Genolet, R.; Queiroz, L.; et al. Engineered CD4 TCR T Cells with Conserved High-Affinity TCRs Targeting NY-ESO-1 for Advanced Cellular Therapies in Cancer. Sci. Adv. 2025, 11, eadu5754. [Google Scholar] [CrossRef]
- Brightman, S.E.; Becker, A.; Thota, R.R.; Naradikian, M.S.; Chihab, L.; Zavala, K.S.; Ramamoorthy Premlal, A.L.; Griswold, R.Q.; Dolina, J.S.; Cohen, E.E.W.; et al. Neoantigen-Specific Stem Cell Memory-Like CD4+ T Cells Mediate CD8+ T Cell-Dependent Immunotherapy of MHC class II-negative Solid Tumors. Nat. Immunol. 2023, 24, 1345–1357. [Google Scholar] [CrossRef]
- Tran, E.; Turcotte, S.; Gros, A.; Robbins, P.F.; Lu, Y.-C.; Dudley, M.E.; Wunderlich, J.R.; Somerville, R.P.; Hogan, K.; Hinrichs, C.S.; et al. Cancer Immunotherapy Based on Mutation-Specific CD4+ T Cells in a Patient with Epithelial Cancer. Science 2014, 344, 641–645. [Google Scholar] [CrossRef]
- Veatch, J.R.; Lee, S.M.; Fitzgibbon, M.; Chow, I.-T.; Jesernig, B.; Schmitt, T.; Kong, Y.Y.; Kargl, J.; Houghton, A.M.; Thompson, J.A.; et al. Tumor-Infiltrating BRAFV600E-specific CD4+ T cells Correlated with Complete Clinical Response in Melanoma. J. Clin. Investig. 2018, 128, 1563–1568. [Google Scholar] [CrossRef]
- Lu, Y.-C.; Parker, L.L.; Lu, T.; Zheng, Z.; Toomey, M.A.; White, D.E.; Yao, X.; Li, Y.F.; Robbins, P.F.; Feldman, S.A.; et al. Treatment of Patients With Metastatic Cancer Using a Major Histocompatibility Complex Class II–Restricted T-Cell Receptor Targeting the Cancer Germline Antigen MAGE-A3. J. Clin. Oncol. 2017, 35, 3322–3329. [Google Scholar] [CrossRef]
- Johnson, L.A.; Heemskerk, B.; Powell, D.J., Jr.; Cohen, C.J.; Morgan, R.A.; Dudley, M.E.; Robbins, P.F.; Rosenberg, S.A. Gene Transfer of Tumor-Reactive TCR Confers Both High Avidity and Tumor Reactivity to Nonreactive Peripheral Blood Mononuclear Cells and Tumor-Infiltrating Lymphocytes1. J. Immunol. 2006, 177, 6548–6559. [Google Scholar] [CrossRef]
- Afeyan, A.B.; Wu, C.J.; Oliveira, G. Rapid Parallel Reconstruction and Specificity Screening of Hundreds of T Cell Receptors. Nat. Protoc. 2025, 20, 539–586. [Google Scholar] [CrossRef]
- Manfredi, F.; Stasi, L.; Buonanno, S.; Marzuttini, F.; Noviello, M.; Mastaglio, S.; Abbati, D.; Potenza, A.; Balestrieri, C.; Cianciotti, B.C.; et al. Harnessing T Cell Exhaustion and Trogocytosis to Isolate Patient-Derived Tumor-Specific TCR. Sci. Adv. 2023, 9, eadg8014. [Google Scholar] [CrossRef]
- Neeser, A.; Ramasubramanian, R.; Wang, C.; Ma, L. Engineering Enhanced Chimeric Antigen Receptor-T Cell Therapy for Solid Tumors. Immuno-Oncol. Technol. 2023, 19, 100385. [Google Scholar] [CrossRef]
- Labanieh, L.; Majzner, R.G.; Mackall, C.L. Programming CAR-T Cells to Kill Cancer. Nat. Biomed. Eng. 2018, 2, 377–391. [Google Scholar] [CrossRef]
- Kim, G.B.; Riley, J.L.; Levine, B.L. Engineering T Cells To Survive and Thrive In The Hostile Tumor Microenvironment. Curr. Opin. Biomed. Eng. 2022, 21, 100360. [Google Scholar] [CrossRef]
- Chapuis, A.; Greenberg, P.D. T Cell Receptor Gene Therapy Targeting WT1 Prevents Acute Myeloid Leukemia Relapse Post Transplant. NatMed 2019, 25, 1064–1072. [Google Scholar]
- Ng Tang, D.; Shen, Y.; Sun, J.; Wen, S.; Wolchok, J.D.; Yuan, J.; Allison, J.P.; Sharma, P. Increased Frequency of ICOS+ CD4 T Cells as a Pharmacodynamic Biomarker for Anti-CTLA-4 Therapy. Cancer Immunol. Res. 2013, 1, 229–234. [Google Scholar] [CrossRef]
- Sharma, N.; Fan, X.; Atolagbe, O.T.; Ge, Z.; Dao, K.N.; Sharma, P.; Allison, J.P. ICOS Costimulation in Combination with CTLA-4 Blockade Remodels Tumor-Associated Macrophages Toward an Antitumor Phenotype. J. Exp. Med. 2024, 221, e20231263. [Google Scholar] [CrossRef]
- Bai, Z.; Feng, B.; McClory, S.E.; de Oliveira, B.C.; Diorio, C.; Gregoire, C.; Tao, B.; Yang, L.; Zhao, Z.; Peng, L.; et al. Single-Cell CAR T Atlas Reveals Type 2 Function in 8-Year Leukaemia Remission. Nature 2024, 634, 702–711. [Google Scholar] [CrossRef]
- Ott, P.A.; Hu, Z.; Keskin, D.B.; Shukla, S.A.; Sun, J.; Bozym, D.J.; Zhang, W.; Luoma, A.; Giobbie-Hurder, A.; Peter, L.; et al. An Immunogenic Personal Neoantigen Vaccine for Patients with Melanoma. Nature 2017, 547, 217–221. [Google Scholar] [CrossRef]
- Braun, D.A.; Moranzoni, G.; Chea, V.; McGregor, B.A.; Blass, E.; Tu, C.R.; Vanasse, A.P.; Forman, C.; Forman, J.; Afeyan, A.B.; et al. A Neoantigen Vaccine Generates Antitumour Immunity in Renal Cell Carcinoma. Nature 2025, 639, 474–482. [Google Scholar] [CrossRef]
- Schreiber, S.; Honz, M.; Mamozai, W.; Kurktschiev, P.; Schiemann, M.; Witter, K.; Moore, E.; Zielinski, C.; Sette, A.; Protzer, U.; et al. Characterization of a Library of 20 HBV-Specific MHC Class II-Restricted T Cell Receptors. Mol. Ther. Methods Clin. Dev. 2021, 23, 476–489. [Google Scholar] [CrossRef]
- Jokinen, E.; Huuhtanen, J.; Mustjoki, S.; Heinonen, M.; Lähdesmäki, H. Predicting Recognition Between T Cell Receptors and Epitopes with TCRGP. PLoS Comput. Biol. 2021, 17, e1008814. [Google Scholar] [CrossRef]
- Tan, C.L.; Lindner, K.; Boschert, T.; Meng, Z.; Rodriguez Ehrenfried, A.; De Roia, A.; Haltenhof, G.; Faenza, A.; Imperatore, F.; Bunse, L.; et al. Prediction of Tumor-Reactive T Cell Receptors from scRNA-Seq Data for Personalized T cell therapy. Nat. Biotechnol. 2024, 43, 134–142. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef]
- Aoki, H.; Ueha, S.; Shichino, S.; Ogiwara, H.; Shitara, K.; Shimomura, M.; Suzuki, T.; Nakatsura, T.; Yamashita, M.; Kitano, S.; et al. Transient Depletion of CD4+ Cells Induces Remodeling of the TCR Repertoire in Gastrointestinal Cancer. Cancer Immunol. Res. 2021, 9, 624–636. [Google Scholar] [CrossRef]
- Stoeckius, M.; Hafemeister, C.; Stephenson, W.; Houck-Loomis, B.; Chattopadhyay, P.K.; Swerdlow, H.; Satija, R.; Smibert, P. Simultaneous Epitope and Transcriptome Measurement in Single Cells. Nat. Methods 2017, 14, 865–868. [Google Scholar] [CrossRef]
- Duhen, T.; Duhen, R.; Montler, R.; Moses, J.; Moudgil, T.; de Miranda, N.F.; Goodall, C.P.; Blair, T.C.; Fox, B.A.; McDermott, J.E.; et al. Co-Expression of CD39 and CD103 Identifies Tumor-Reactive CD8 T Cells in Human Solid Tumors. Nat. Commun. 2018, 9, 2724. [Google Scholar] [CrossRef]
- Li, Y.; Moysey, R.; Molloy, P.E.; Vuidepot, A.-L.; Mahon, T.; Baston, E.; Dunn, S.; Liddy, N.; Jacob, J.; Jakobsen, B.K.; et al. Directed Evolution of Human T-Cell Receptors with Picomolar Affinities by Phage Display. Nat. Biotechnol. 2005, 23, 349–354. [Google Scholar] [CrossRef]
- Schmid, D.A.; Irving, M.B.; Posevitz, V.; Hebeisen, M.; Posevitz-Fejfar, A.; Sarria, J.-C.F.; Gomez-Eerland, R.; Thome, M.; Schumacher, T.N.M.; Romero, P.; et al. Evidence for a TCR Affinity Threshold Delimiting Maximal CD8 T Cell Function. J. Immunol. 2010, 184, 4936–4946. [Google Scholar] [CrossRef]
- Valitutti, S.; Müller, S.; Cella, M.; Padovan, E.; Lanzavecchia, A. Serial Triggering of Many T-cell Receptors by a Few Peptide–MHC Complexes. Nature 1995, 375, 148–151. [Google Scholar] [CrossRef]
- Ishizuka, J.; Grebe, K.; Shenderov, E.; Peters, B.; Chen, Q.; Peng, Y.; Wang, L.; Dong, T.; Pasquetto, V.; Oseroff, C.; et al. Quantitating T Cell Cross-Reactivity for Unrelated Peptide Antigens. J. Immunol. 2009, 183, 4337–4345. [Google Scholar] [CrossRef]
- Johnson, L.A.; Morgan, R.A.; Dudley, M.E.; Cassard, L.; Yang, J.C.; Hughes, M.S.; Kammula, U.S.; Royal, R.E.; Sherry, R.M.; Wunderlich, J.R.; et al. Gene Therapy with Human and Mouse T-Cell Receptors Mediates Cancer Regression and Targets Normal Tissues Expressing Cognate Antigen. Blood 2009, 114, 535–546. [Google Scholar] [CrossRef]
- Parkhurst, M.R.; Yang, J.C.; Langan, R.C.; Dudley, M.E.; Nathan, D.-A.N.; Feldman, S.A.; Davis, J.L.; Morgan, R.A.; Merino, M.J.; Sherry, R.M.; et al. T Cells Targeting Carcinoembryonic Antigen Can Mediate Regression of Metastatic Colorectal Cancer but Induce Severe Transient Colitis. Mol. Ther. 2011, 19, 620–626. [Google Scholar] [CrossRef]
- Linette, G.P.; Stadtmauer, E.A.; Maus, M.V.; Rapoport, A.P.; Levine, B.L.; Emery, L.; Litzky, L.; Bagg, A.; Carreno, B.M.; Cimino, P.J.; et al. Cardiovascular Toxicity and Titin Cross-Reactivity of Affinity-Enhanced T Cells in Myeloma and Melanoma. Blood 2013, 122, 863–871. [Google Scholar] [CrossRef]
- Karapetyan, A.R.; Chaipan, C.; Winkelbach, K.; Wimberger, S.; Jeong, J.S.; Joshi, B.; Stein, R.B.; Underwood, D.; Castle, J.C.; van Dijk, M.; et al. TCR Fingerprinting and Off-Target Peptide Identification. Front. Immunol. 2019, 10, 2501. [Google Scholar] [CrossRef]
- Verdun, N.; Marks, P. Secondary Cancers after Chimeric Antigen Receptor T-Cell Therapy. N. Engl. J. Med. 2024, 390, 584–586. [Google Scholar] [CrossRef]
- Eyquem, J.; Mansilla-Soto, J.; Giavridis, T.; van der Stegen, S.J.C.; Hamieh, M.; Cunanan, K.M.; Odak, A.; Gönen, M.; Sadelain, M. Targeting a CAR to the TRAC Locus with CRISPR/Cas9 Enhances Tumour Rejection. Nature 2017, 543, 113–117. [Google Scholar] [CrossRef]
- Foy, S.P.; Jacoby, K.; Bota, D.A.; Hunter, T.; Pan, Z.; Stawiski, E.; Ma, Y.; Lu, W.; Peng, S.; Wang, C.L.; et al. Non-Viral Precision T Cell Receptor Replacement for Personalized Cell Therapy. Nature 2023, 615, 687–696. [Google Scholar] [CrossRef]
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-Engineered T Cells in Patients with Refractory Cancer. Science 2020, 367, eaba7365. [Google Scholar] [CrossRef]
- Nahmad, A.D.; Reuveni, E.; Goldschmidt, E.; Tenne, T.; Liberman, M.; Horovitz-Fried, M.; Khosravi, R.; Kobo, H.; Reinstein, E.; Madi, A.; et al. Frequent Aneuploidy in Primary Human T cells After CRISPR–Cas9 cleavage. Nat. Biotechnol. 2022, 40, 1807–1813. [Google Scholar] [CrossRef]
- Shy, B.R.; Vykunta, V.S.; Ha, A.; Talbot, A.; Roth, T.L.; Nguyen, D.N.; Pfeifer, W.G.; Chen, Y.Y.; Blaeschke, F.; Shifrut, E.; et al. High-Yield Genome Engineering in Primary Cells Using a Hybrid ssDNA repair template and small-molecule cocktails. Nat. Biotechnol. 2023, 41, 521–531. [Google Scholar] [CrossRef]
- Prommersberger, S.; Reiser, M.; Beckmann, J.; Danhof, S.; Amberger, M.; Quade-Lyssy, P.; Einsele, H.; Hudecek, M.; Bonig, H.; Ivics, Z. CARAMBA: A First-in-Human Clinical Trial with SLAMF7 CAR-T Cells Prepared by Virus-Free Sleeping Beauty Gene Transfer to Treat Multiple Myeloma. Gene Ther. 2021, 28, 560–571. [Google Scholar] [CrossRef]
- Janssens, I.; Campillo Davó, D.; Van den Bos, J.; De Reu, H.; Berneman, Z.N.; Wens, I.; Cools, N. Engineering of Regulatory T Cells by Means of mRNA Electroporation in a GMP-Compliant Manner. Cytotherapy 2022, 24, 659–672. [Google Scholar] [CrossRef]
- Mensali, N.; Myhre, M.R.; Dillard, P.; Pollmann, S.; Gaudernack, G.; Kvalheim, G.; Wälchli, S.; Inderberg, E.M. Preclinical Assessment of Transiently TCR Redirected T cells for Solid Tumour Immunotherapy. Cancer Immunol. Immunother. 2019, 68, 1235–1243. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demaël, U.M.; Rirkkrai, T.; Okus, F.Z.; Tiffeau-Mayer, A.; Stauss, H.J. Exploiting TCR Repertoire Analysis to Select Therapeutic TCRs for Cancer Immunotherapy. Cells 2025, 14, 1223. https://doi.org/10.3390/cells14151223
Demaël UM, Rirkkrai T, Okus FZ, Tiffeau-Mayer A, Stauss HJ. Exploiting TCR Repertoire Analysis to Select Therapeutic TCRs for Cancer Immunotherapy. Cells. 2025; 14(15):1223. https://doi.org/10.3390/cells14151223
Chicago/Turabian StyleDemaël, Ursule M., Thunchanok Rirkkrai, Fatma Zehra Okus, Andreas Tiffeau-Mayer, and Hans J. Stauss. 2025. "Exploiting TCR Repertoire Analysis to Select Therapeutic TCRs for Cancer Immunotherapy" Cells 14, no. 15: 1223. https://doi.org/10.3390/cells14151223
APA StyleDemaël, U. M., Rirkkrai, T., Okus, F. Z., Tiffeau-Mayer, A., & Stauss, H. J. (2025). Exploiting TCR Repertoire Analysis to Select Therapeutic TCRs for Cancer Immunotherapy. Cells, 14(15), 1223. https://doi.org/10.3390/cells14151223





