B-Cell Lymphomas Secrete Novel Inhibitory Molecules That Disrupt HLA Class II-Mediated CD4+ T-Cell Recognition
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Peptide
2.3. Antigen Presentation Assays
2.4. Collection of Cell Eluates
2.5. Enzyme-Linked Immunosorbent Assay
2.6. Protein Extraction and Digestion
2.7. Mass Spectrometric Analysis Matrix-Assisted Laser Desorption/Ionization Time of Flight/Time of Flight (MALDI-TOF/TOF)
2.8. Statistical Analyses
3. Results
3.1. Deficiencies in CD4+ T-Cell Recognition of B-Cell Lymphoma Associated with HLA Class II Molecule Presentation
3.2. Disruption of CD4+ T-Cell Activation and MHC Specificity of BL-Associated Inhibitory Molecules
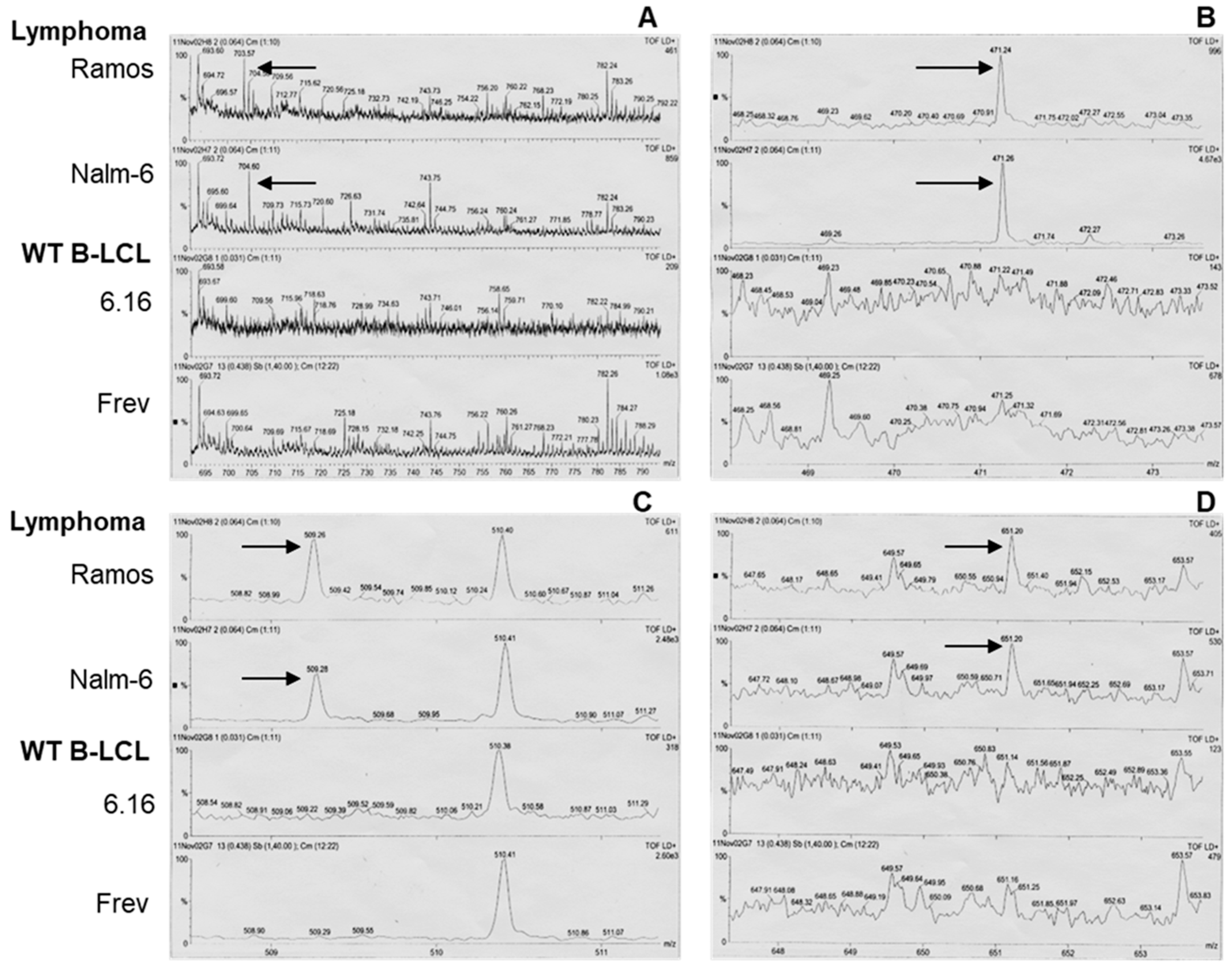
3.3. Mass Spectrometry of BL-Eluted Molecules Showed Different Spectra from BL Cells
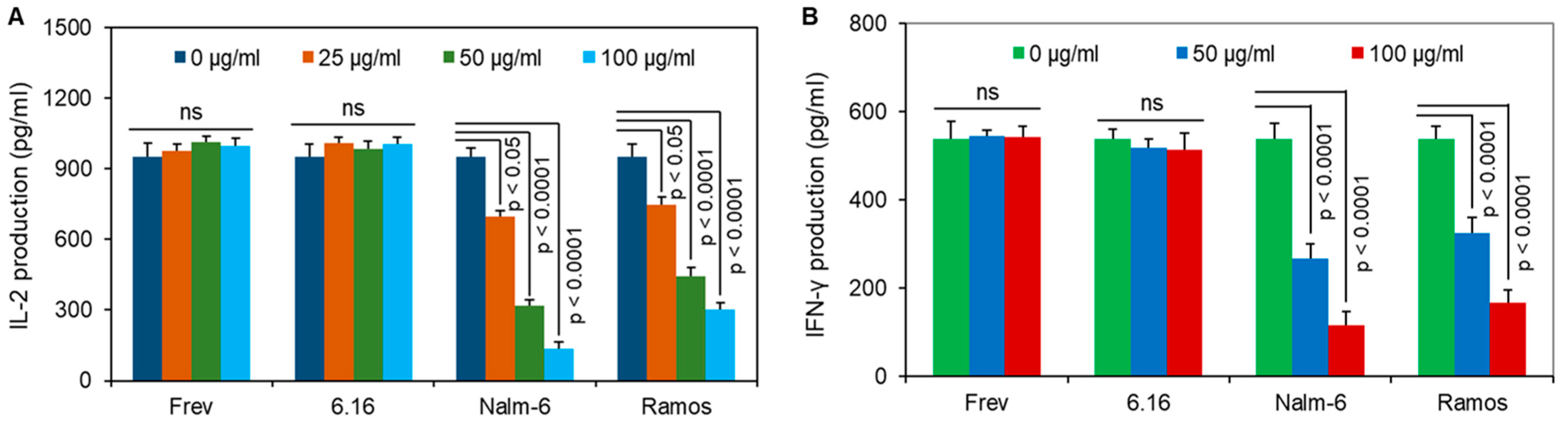
3.4. BL Shed Molecules Dose-Dependently Disrupt HLA Class II Ag Presentation
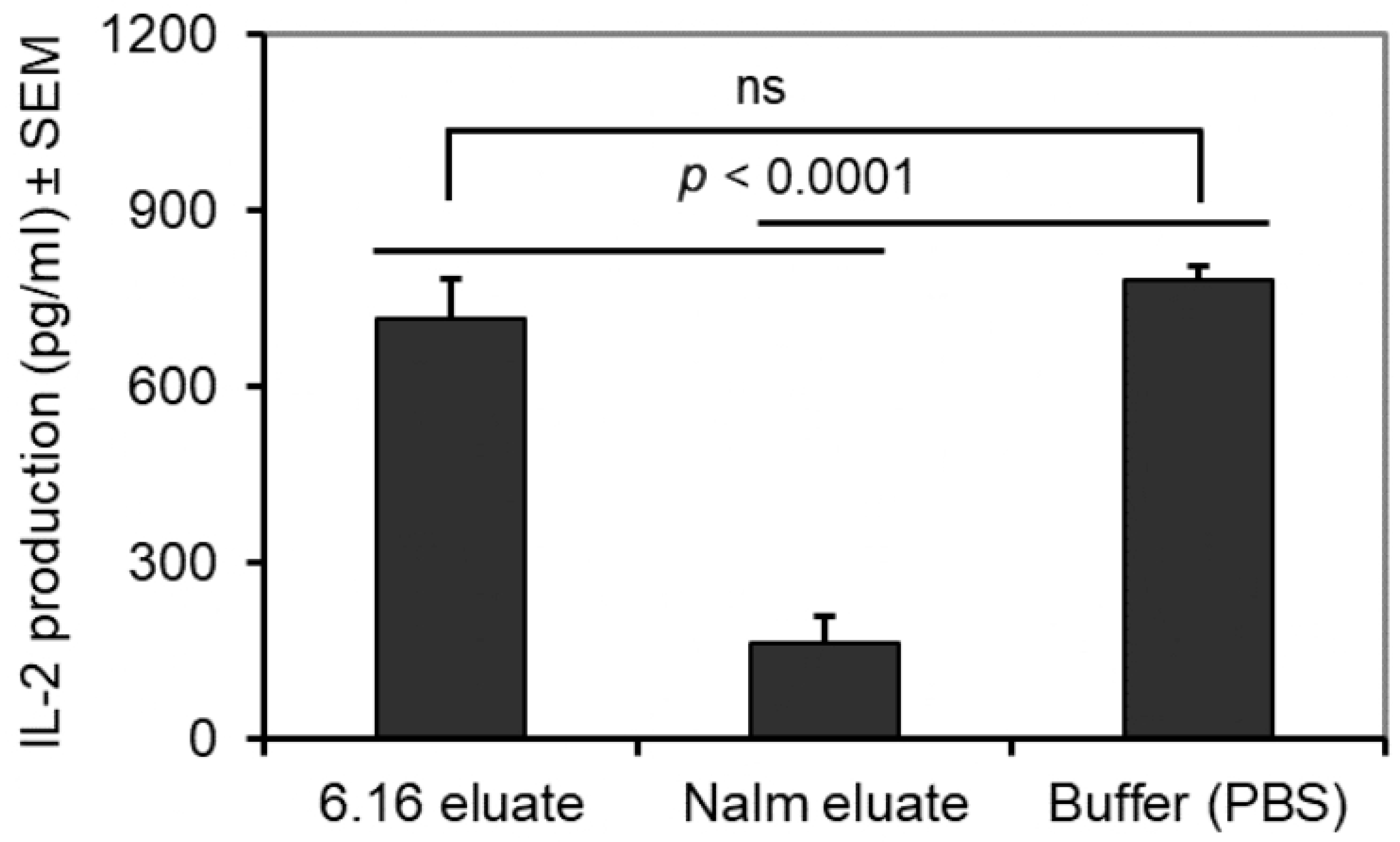
3.5. BL Shed Molecules Also Inhibit HLA Class II Ag Presentation by DC
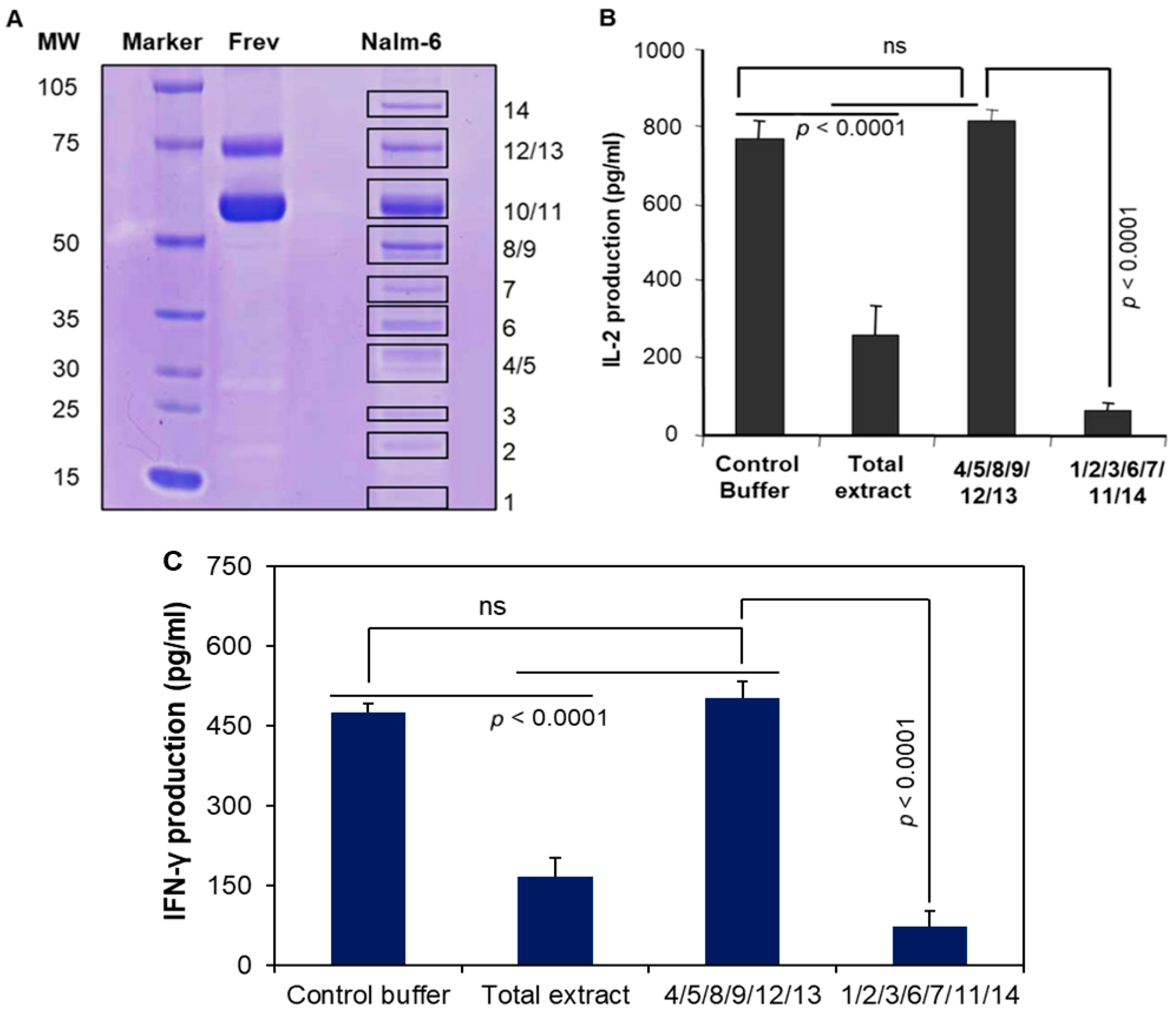
3.6. Pools of Protein Bands Exhibit Inhibitory Activity
3.7. MALDI Mass Spectroscopy and Amino Acid Sequencing Detected Protein Bands
4. Discussion
5. Conclusions
6. Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Patil, S.; Rajput, S.; Patil, S.; Mhaiskar, A. B-cell lymphoma: Advances in pathogenesis, diagnosis, and targeted therapies. Pathol. Res. Pract. 2025, 271, 156036. [Google Scholar] [CrossRef]
- Brauge, B.; Dessauge, E.; Creusat, F.; Tarte, K. Modeling the crosstalk between malignant B cells and their microenvironment in B-cell lymphomas: Challenges and opportunities. Front. Immunol. 2023, 14, 1288110. [Google Scholar] [CrossRef]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.; De, O.; Berti, E.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–7148. [Google Scholar] [CrossRef]
- Solares, S.; León, J.; García-Gutiérrez, L. The Functional Interaction Between Epstein–Barr Virus and MYC in the Pathogenesis of Burkitt Lymphoma. Cancers 2024, 16, 4212. [Google Scholar] [CrossRef]
- Rochford, R. Reframing Burkitt lymphoma: Virology not epidemiology defines clinical variants. Ann. Lymphoma 2021, 5, 22. [Google Scholar] [CrossRef]
- Shingleton, J.; Wang, J.; Baloh, C.; Dave, T.; Davis, N.; Happ, L.; Jadi, O.; Kositsky, R.; Li, X.; Love, C.; et al. Non-Hodgkin Lymphomas: Malignancies Arising from Mature B Cells. Cold Spring Harb. Perspect. Med. 2020, 11, a034843. [Google Scholar] [CrossRef]
- Kolijn, P.M.; Langerak, A.W. Immune dysregulation as a leading principle for lymphoma development in diverse immunological backgrounds. Immunol. Lett. 2023, 263, 46–59. [Google Scholar] [CrossRef]
- Fowler, N.H.; Cheah, C.Y.; Gascoyne, R.D.; Gribben, J.; Neelapu, S.S.; Ghia, P.; Bollard, C.; Ansell, S.; Curran, M.; Wilson, W.H.; et al. Role of the tumor microenvironment in mature B-cell lymphoid malignancies. Haematologica 2016, 101, 531–540. [Google Scholar] [CrossRef]
- Masnikosa, R.; Cvetković, Z.; Pirić, D. Tumor Biology Hides Novel Therapeutic Approaches to Diffuse Large B-Cell Lymphoma: A Narrative Review. Int. J. Mol. Sci. 2024, 25, 11384. [Google Scholar] [CrossRef]
- Raskov, H.; Orhan, A.; Christensen, J.P.; Gögenur, I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br. J. Cancer 2021, 124, 359–367. [Google Scholar] [CrossRef]
- Kumar, S.; Singh, S.K.; Rana, B.; Rana, A. Tumor-infiltrating CD8+ T cell antitumor efficacy and exhaustion: Molecular insights. Drug Discov. Today 2021, 26, 951–967. [Google Scholar] [CrossRef]
- Kravtsov, D.S.; Erbe, A.K.; Sondel, P.M.; Rakhmilevich, A.L. Roles of CD4+ T cells as mediators of antitumor immunity. Front. Immunol. 2022, 13, 972021. [Google Scholar] [CrossRef]
- Ostroumov, D.; Fekete-Drimusz, N.; Saborowski, M.; Kühnel, F.; Woller, N. CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell. Mol. Life Sci. 2017, 75, 689–713. [Google Scholar] [CrossRef]
- van der Leun, A.M.; Thommen, D.S.; Schumacher, T.N. CD8+ T cell states in human cancer: Insights from single-cell analysis. Nat. Rev. Cancer 2020, 20, 218–232. [Google Scholar] [CrossRef]
- Chen, Y.; Yu, D.; Qian, H.; Shi, Y.; Tao, Z. CD8+ T cell-based cancer immunotherapy. J. Transl. Med. 2024, 22, 394. [Google Scholar] [CrossRef]
- Montauti, E.; Oh, D.Y.; Fong, L. CD4+ T cells in antitumor immunity. Trends Cancer 2024, 10, 969–985. [Google Scholar] [CrossRef]
- Tay, R.E.; Richardson, E.K.; Toh, H.C. Revisiting the role of CD4+ T cells in cancer immunotherapy—New insights into old paradigms. Cancer Gene Ther. 2020, 28, 5–17. [Google Scholar] [CrossRef]
- God, J.M.; Haque, A. Burkitt Lymphoma: Pathogenesis and Immune Evasion. J. Oncol. 2010, 2010, 516047. [Google Scholar] [CrossRef]
- Staege, M.S.; Lee, S.P.; Frisan, T.; Mautner, J.; Scholz, S.; Pajic, A.; Rickinson, A.B.; Masucci, M.G.; Polack, A.; Bornkamm, G.W. MYC overexpression imposes a nonimmunogenic phenotype on Epstein–Barr virus-infected B cells. Proc. Natl. Acad. Sci. USA 2002, 99, 4550–4555. [Google Scholar] [CrossRef]
- God, J.M.; Zhao, D.; Cameron, C.A.; Amria, S.; Bethard, J.R.; Haque, A. Disruption of HLA class II antigen presentation in Burkitt lymphoma: Implication of a 47 000 MW acid labile protein in CD4+ T-cell recognition. Immunology 2014, 142, 492–505. [Google Scholar] [CrossRef]
- Hossain, A.; God, J.M.; Radwan, F.F.Y.; Amria, S.; Zhao, D.; Bethard, J.R.; Haque, A. HLA Class II Defects in Burkitt Lymphoma: Bryostatin-1-Induced 17 kDa Protein Restores CD4+ T-Cell Recognition. J. Immunol. Res. 2011, 2011, 780839. [Google Scholar] [CrossRef]
- Fu, T.; Voo, K.S.; Wang, R.-F. Critical role of EBNA1-specific CD4+ T cells in the control of mouse Burkitt lymphoma in vivo. J. Clin. Investig. 2004, 114, 542–550. [Google Scholar] [CrossRef]
- Yang, Y.; Ma, X.; Li, Y.; Jin, L.; Zhou, X. The evolving tumor-associated adipose tissue microenvironment in breast cancer: From cancer initiation to metastatic outgrowth. Clin. Transl. Oncol. 2024, 27, 2778–2788. [Google Scholar] [CrossRef]
- de Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef]
- Del Prete, A.; Salvi, V.; Soriani, A.; Laffranchi, M.; Sozio, F.; Bosisio, D.; Sozzani, S. Dendritic cell subsets in cancer immunity and tumor antigen sensing. Cell. Mol. Immunol. 2023, 20, 432–447. [Google Scholar] [CrossRef]
- Mazzoccoli, L.; Liu, B. Dendritic Cells in Shaping Anti-Tumor T Cell Response. Cancers 2024, 16, 2211. [Google Scholar] [CrossRef]
- MacNabb, B.W.; Chen, X.; Tumuluru, S.; Godfrey, J.; Kasal, D.N.; Yu, J.; Jongsma, M.L.; Spaapen, R.M.; Kline, D.E.; Kline, J. Dendritic cells can prime anti-tumor CD8+ T cell responses through major histocompatibility complex cross-dressing. Immunity 2022, 55, 982–997.e8. [Google Scholar] [CrossRef]
- Jing, H.; Gao, Y.; Sun, Z.; Liu, S. Recent advances in novel tumor immunotherapy strategies based on regulating the tumor microenvironment and immune checkpoints. Front. Immunol. 2025, 16, 1529403. [Google Scholar] [CrossRef]
- Kuang, L.; Wu, L.; Li, Y. Extracellular vesicles in tumor immunity: Mechanisms and novel insights. Mol. Cancer 2025, 24, 45. [Google Scholar] [CrossRef]
- Garcia-Lacarte, M.; Grijalba, S.C.; Melchor, J.; Pascual, M.; Goñi, E.; Clemente-Larramendi, I.; Morales-Sánchez, S.; Burrell, M.A.; Blanco, O.; Arnaiz-Leché, A.; et al. IL-10 from tumoral B cells modulates the diffuse large B-cell lymphoma microenvironment and response to immunotherapy. Blood 2025, 145, 2746–2761. [Google Scholar] [CrossRef]
- Sinkarevs, S.; Strumfs, B.; Volkova, S.; Strumfa, I. Tumour Microenvironment: The General Principles of Pathogenesis and Implications in Diffuse Large B Cell Lymphoma. Cells 2024, 13, 1057. [Google Scholar] [CrossRef]
- Xiong, J.; Chi, H.; Yang, G.; Zhao, S.; Zhang, J.; Tran, L.J.; Xia, Z.; Yang, F.; Tian, G. Revolutionizing anti-tumor therapy: Unleashing the potential of B cell-derived exosomes. Front. Immunol. 2023, 14, 1188760. [Google Scholar] [CrossRef]
- Kempkes, B.; Spitkovsky, D.; Jansen-Dürr, P.; Ellwart, J.; Kremmer, E.; Delecluse, H.; Rottenberger, C.; Bornkamm, G.; Hammerschmidt, W. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 1995, 14, 88–96. [Google Scholar] [CrossRef]
- Haque, M.A.; Hawes, J.W.; Blum, J.S. Cysteinylation of MHC Class II Ligands: Peptide Endocytosis and Reduction Within APC Influences T Cell Recognition. J. Immunol. 2001, 166, 4543–4551. [Google Scholar] [CrossRef]
- Radwan, F.F.Y.; Zhang, L.; Hossain, A.; Doonan, B.P.; God, J.M.; Haque, A. Mechanisms regulating enhanced human leukocyte antigen class II-mediated CD4 + T cell recognition of human B-cell lymphoma by resveratrol. Leuk. Lymphoma 2011, 53, 305–314. [Google Scholar] [CrossRef]
- Amria, S.; Hajiaghamohseni, L.M.; Harbeson, C.; Zhao, D.; Goldstein, O.; Blum, J.S.; Haque, A. HLA-DM negatively regulates HLA-DR4-restricted collagen pathogenic peptide presentation and T cell recognition. Eur. J. Immunol. 2008, 38, 1961–1970. [Google Scholar] [CrossRef]
- Haque, M.A.; Li, P.; Jackson, S.K.; Zarour, H.M.; Hawes, J.W.; Phan, U.T.; Maric, M.; Cresswell, P.; Blum, J.S. Absence of γ-Interferon–inducible Lysosomal Thiol Reductase in Melanomas Disrupts T Cell Recognition of Select Immunodominant Epitopes. J. Exp. Med. 2002, 195, 1267–1277. [Google Scholar] [CrossRef]
- Ma, C.; Whiteley, P.E.; Cameron, P.M.; Freed, D.C.; Pressey, A.; Chen, S.-L.; Garni-Wagner, B.; Fang, C.; Zaller, D.M.; Wicker, L.S.; et al. Role of APC in the Selection of Immunodominant T Cell Epitopes. J. Immunol. 1999, 163, 6413–6423. [Google Scholar] [CrossRef]
- Balkan, W.; Martinez, A.F.; Fernandez, I.; Rodriguez, M.A.; Pang, M.; Troen, B.R. Identification of NFAT binding sites that mediate stimulation of cathepsin K promoter activity by RANK ligand. Gene 2009, 446, 90–98. [Google Scholar] [CrossRef]
- Pathak, S.S.; Blum, J.S. Endocytic Recycling is Required for the Presentation of an Exogenous Peptide via MHC Class II Molecules. Traffic 2000, 1, 561–569. [Google Scholar] [CrossRef]
- Zhao, D.; Amria, S.; Hossain, A.; Sundaram, K.; Komlosi, P.; Nagarkatti, M.; Haque, A. Enhancement of HLA class II-restricted CD4+ T cell recognition of human melanoma cells following treatment with bryostatin-1. Cell. Immunol. 2011, 271, 392–400. [Google Scholar] [CrossRef]
- Hossain, A.; Radwan, F.F.Y.; Doonan, B.P.; God, J.M.; Zhang, L.; Bell, P.D.; Haque, A. A possible cross-talk between autophagy and apoptosis in generating an immune response in melanoma. Apoptosis 2012, 17, 1066–1078. [Google Scholar] [CrossRef]
- God, J.M.; Cameron, C.; Figueroa, J.; Amria, S.; Hossain, A.; Kempkes, B.; Bornkamm, G.W.; Stuart, R.K.; Blum, J.S.; Haque, A. Elevation of c-MYC Disrupts HLA Class II–Mediated Immune Recognition of Human B Cell Tumors. J. Immunol. 2015, 194, 1434–1445. [Google Scholar] [CrossRef]
- Mentlein, R. Cell-surface peptidases. Int Rev Cytol. 2004, 235, 165–213. [Google Scholar] [CrossRef]
- Nelson, J.B.; Carducci, M.A. Small Bioactive Peptides and Cell Surface Peptidases in Androgen-Independent Prostate Cancer. Cancer Investig. 2000, 18, 87–96. [Google Scholar] [CrossRef]
- A Villadangos, J.; Ploegh, H.L. Proteolysis in MHC Class II Antigen Presentation. Immunity 2000, 12, 233–239. [Google Scholar] [CrossRef]
- Campoli, M.; Ferrone, S. HLA antigen changes in malignant cells: Epigenetic mechanisms and biologic significance. Oncogene 2008, 27, 5869–5885. [Google Scholar] [CrossRef]
- Hazini, A.; Fisher, K.; Seymour, L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J. Immunother. Cancer 2021, 9, e002899. [Google Scholar] [CrossRef]
- Krishnakumar, S.; Abhyankar, D.; Lakshmi, S.A.; Shanmugam, M.P.; Pushparaj, V.; Biswas, J. HLA class II antigen expression in uveal melanoma: Correlation with clinicopathological features. Exp. Eye Res. 2003, 77, 175–180. [Google Scholar] [CrossRef]
- Vigneron, N.; Eynde, B.J.V.D. Proteasome Subtypes and Regulators in the Processing of Antigenic Peptides Presented by Class I Molecules of the Major Histocompatibility Complex. Biomolecules 2014, 4, 994–1025. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef]
- A Wu, A.; Drake, V.; Huang, H.-S.; Chiu, S.; Zheng, L. Reprogramming the tumor microenvironment: Tumor-induced immunosuppressive factors paralyze T cells. OncoImmunology 2015, 4, e1016700. [Google Scholar] [CrossRef]
- Wang, D.; Cui, Q.; Yang, Y.J.; Liu, A.; Zhang, G.; Yu, J.C. Application of dendritic cells in tumor immunotherapy and progress in the mechanism of anti-tumor effect of Astragalus polysaccharide (APS) modulating dendritic cells: A review. Biomed. Pharmacother. 2022, 155, 113541. [Google Scholar] [CrossRef]
- Tominaga, M.; Uto, T.; Fukaya, T.; Mitoma, S.; Riethmacher, D.; Umekita, K.; Yamashita, Y.; Sato, K. Crucial role of dendritic cells in the generation of anti-tumor T-cell responses and immunogenic tumor microenvironment to suppress tumor development. Front. Immunol. 2024, 15, 1200461. [Google Scholar] [CrossRef]
- Alfei, F.; Ho, P.-C.; Lo, W.-L. DCision-making in tumors governs T cell anti-tumor immunity. Oncogene 2021, 40, 5253–5261. [Google Scholar] [CrossRef]
- Kallingal, A.; Olszewski, M.; Maciejewska, N.; Brankiewicz, W.; Baginski, M. Cancer immune escape: The role of antigen presentation machinery. J. Cancer Res. Clin. Oncol. 2023, 149, 8131–8141. [Google Scholar] [CrossRef]
- Murshid, A.; Gong, J.; Calderwood, S.K. Hsp90–peptide complexes stimulate antigen presentation through the class II pathway after binding scavenger receptor SREC-I. Immunobiology 2014, 219, 924–931. [Google Scholar] [CrossRef]
- Kurz, K.S.; Ott, M.; Kalmbach, S.; Steinlein, S.; Kalla, C.; Horn, H.; Ott, G.; Staiger, A.M. Large B-Cell Lymphomas in the 5th Edition of the WHO-Classification of Haematolymphoid Neoplasms—Updated Classification and New Concepts. Cancers 2023, 15, 2285. [Google Scholar] [CrossRef]
- D’alò, F.; Bellesi, S.; Maiolo, E.; Alma, E.; Bellisario, F.; Malafronte, R.; Viscovo, M.; Campana, F.; Hohaus, S. Novel Targets and Advanced Therapies in Diffuse Large B Cell Lymphomas. Cancers 2024, 16, 2243. [Google Scholar] [CrossRef]
- de Charette, M.; Houot, R. Hide or defend, the two strategies of lymphoma immune evasion: Potential implications for immunotherapy. Haematologica 2018, 103, 1256–1268. [Google Scholar] [CrossRef]
- Tie, Y.; Tang, F.; Wei, Y.-Q.; Wei, X.-W. Immunosuppressive cells in cancer: Mechanisms and potential therapeutic targets. J. Hematol. Oncol. 2022, 15, 61. [Google Scholar] [CrossRef]
- Petty, A.J.; Yang, Y. Tumor-Associated Macrophages in Hematologic Malignancies: New Insights and Targeted Therapies. Cells 2019, 8, 1526. [Google Scholar] [CrossRef]
- Alibrahim, M.N.; Gloghini, A.; Carbone, A. Pathobiological Features and Therapeutic Opportunities Linked to TNF Family Member Expression in Classic Hodgkin Lymphoma. Cancers 2024, 16, 4070. [Google Scholar] [CrossRef]
- Melaccio, A.; Reale, A.; Saltarella, I.; Desantis, V.; Lamanuzzi, A.; Cicco, S.; Frassanito, M.A.; Vacca, A.; Ria, R. Pathways of Angiogenic and Inflammatory Cytokines in Multiple Myeloma: Role in Plasma Cell Clonal Expansion and Drug Resistance. J. Clin. Med. 2022, 11, 6491. [Google Scholar] [CrossRef]
- Lam, H.P.J.; Amin, F.; Arulogun, S.O.; Gleeson, M. Nodal Peripheral T-Cell Lymphoma: Therapeutic Challenges and Future Perspectives. Cancers 2025, 17, 1134. [Google Scholar] [CrossRef]
- Luan, Y.; Li, X.; Luan, Y.; Luo, J.; Dong, Q.; Ye, S.; Li, Y.; Li, Y.; Jia, L.; Yang, J.; et al. Therapeutic challenges in peripheral T-cell lymphoma. Mol. Cancer 2024, 23, 2. [Google Scholar] [CrossRef]
- Stein, A.F.V.; Hallek, M.; Nguyen, P.-H. Role of the tumor microenvironment in CLL pathogenesis. Semin. Hematol. 2023, 61, 142–154. [Google Scholar] [CrossRef]

| HLA-Restriction and Antigenic Peptide Presentation | Inhibition of T Cell Responses |
|---|---|
| HLA-DR4-restricted presentation of Igκ188–203 and Igκ145–2159 epitopes from whole IgGκ | Yes |
| HLA-DR4-restricted presentation of κ188–203 and κ145–159 synthetic peptides | Yes |
| HLA-DR4-restricted presentation of HSA64–76K epitope from whole HSA | Yes |
| HLA-DR4-restricted presentation of HSA64–76K synthetic peptide | Yes |
| HLA-DR4-restricted presentation of HA-flu306–319 peptide | Yes |
| HLA-DR7-restricted presentation of EBNA1482–496 peptide | Yes |
| HLA-DR7-restricted presentation of Survivin20–34 peptide | Yes |
| HLA-DR1-restricted presentation of Igκ188–203 epitope from whole IgGκ | Yes |
| HLA-DR1-restricted presentation of κ188–203 synthetic peptides | Yes |
| HLA class I (HLA-A2)-restricted Ag presentation | No |
| Bands | Known/Unknown Protein Products |
|---|---|
| 1 | Profilin 1 |
| 2 | Unknown/modified protein |
| 3 | Peroxiredoxin 1 |
| 4/5 | NA |
| 6 | Unknown/modified protein |
| 7 | Aldolase A |
| 8/9 | NA |
| 10 | Heat shock protein 60 |
| 11 | Muscle pyruvate kinase, chain A + unknown/modified protein product |
| 12/13 | NA |
| 14 | Heat shock protein 90 + unknown/modified protein product |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
God, J.M.; Amria, S.; Cameron, C.A.; Zhang, L.; Bethard, J.R.; Haque, A. B-Cell Lymphomas Secrete Novel Inhibitory Molecules That Disrupt HLA Class II-Mediated CD4+ T-Cell Recognition. Cells 2025, 14, 1220. https://doi.org/10.3390/cells14151220
God JM, Amria S, Cameron CA, Zhang L, Bethard JR, Haque A. B-Cell Lymphomas Secrete Novel Inhibitory Molecules That Disrupt HLA Class II-Mediated CD4+ T-Cell Recognition. Cells. 2025; 14(15):1220. https://doi.org/10.3390/cells14151220
Chicago/Turabian StyleGod, Jason M., Shereen Amria, Christine A. Cameron, Lixia Zhang, Jennifer R. Bethard, and Azizul Haque. 2025. "B-Cell Lymphomas Secrete Novel Inhibitory Molecules That Disrupt HLA Class II-Mediated CD4+ T-Cell Recognition" Cells 14, no. 15: 1220. https://doi.org/10.3390/cells14151220
APA StyleGod, J. M., Amria, S., Cameron, C. A., Zhang, L., Bethard, J. R., & Haque, A. (2025). B-Cell Lymphomas Secrete Novel Inhibitory Molecules That Disrupt HLA Class II-Mediated CD4+ T-Cell Recognition. Cells, 14(15), 1220. https://doi.org/10.3390/cells14151220






