Synergistic Regulation of Pigment Cell Precursors’ Differentiation and Migration by ednrb1a and ednrb2 in Nile Tilapia
Abstract
Highlights
- ednrb1a−/−; ednrb2−/− mutants exhibit complete iridophore loss, similarly to mpv17 mutants.
- ednrb mutants display no defects in the guanine synthesis pathway in tilapia.
- ednrb1a and ednrb2 synergistically regulate iridophore and erythrophore development.
- mitfa mRNA injection rescues the phenotype of ednrb1a−/−;ednrb2−/− mutants.
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Phylogenetic and Amino Acid Sequence Alignment Analyses
2.3. Quantitative Real-Time PCR (qPCR)
2.4. Mutation of ednrb1a,ednrb2 and Establishment of Homozygotes
- ednrb1a-gRNA-F: TAATACGACTCACTATAGGTCATCAGGGTGTAGAAAAGTTTTAGAGCTAGAAATAGC
- ednrb2-gRNA-F: TAATACGACTCACTATAGGTCGGCCTGGCTGCGAGGGGTTTTAGAGCTAGAAATAGC
- ednrb1a -gRNA-R: AGCACCGACTCGGTGCCAC
- ednrb2 -gRNA-R: AGCACCGACTCGGTGCCAC
2.5. Phenotype Analysis and Chromatophore Counting
2.6. H.E. Staining
2.7. Fluorescence In Situ Hybridization
2.8. Elisa (Enzyme-Linked Immunosorbent Assay)
2.9. mitfa and pnp4a mRNA Rescue
2.10. Statistic Analysis
3. Results
3.1. Comparative Analysis of Ednrb Amino Acid Sequences in Vertebrates
3.2. Phylogenetic Analysis of Ednrb in Vertebrates
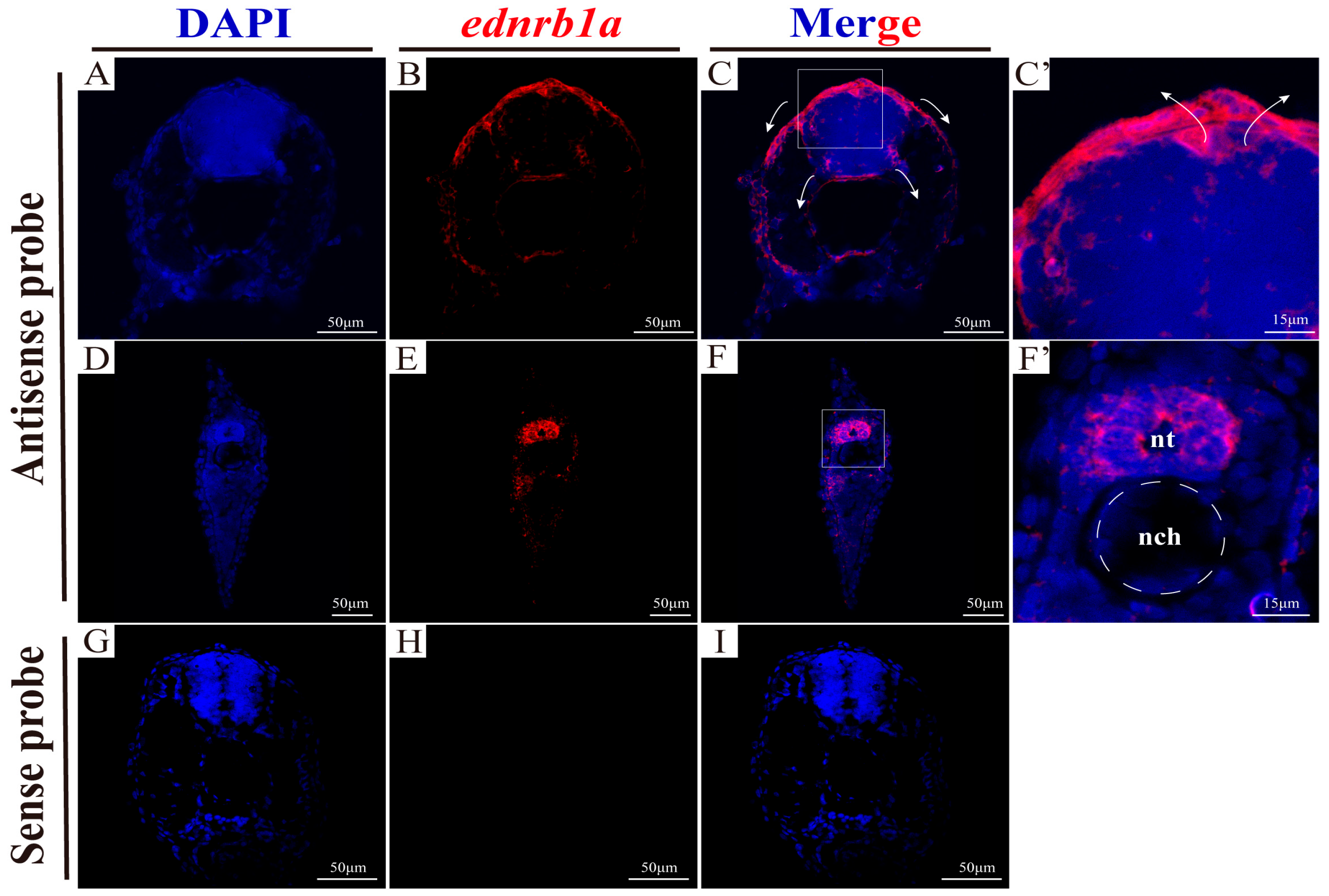
3.3. The Expression Patterns of ednrb1a and ednrb2
3.4. Establishment of Tilapia ednrb1a,ednrb2 and ednrb1a;ednrb2 Homozygous Mutants
3.5. Early-Stage Ednrb Mutants Exhibit Variable Iridophore Reduction in Tilapia
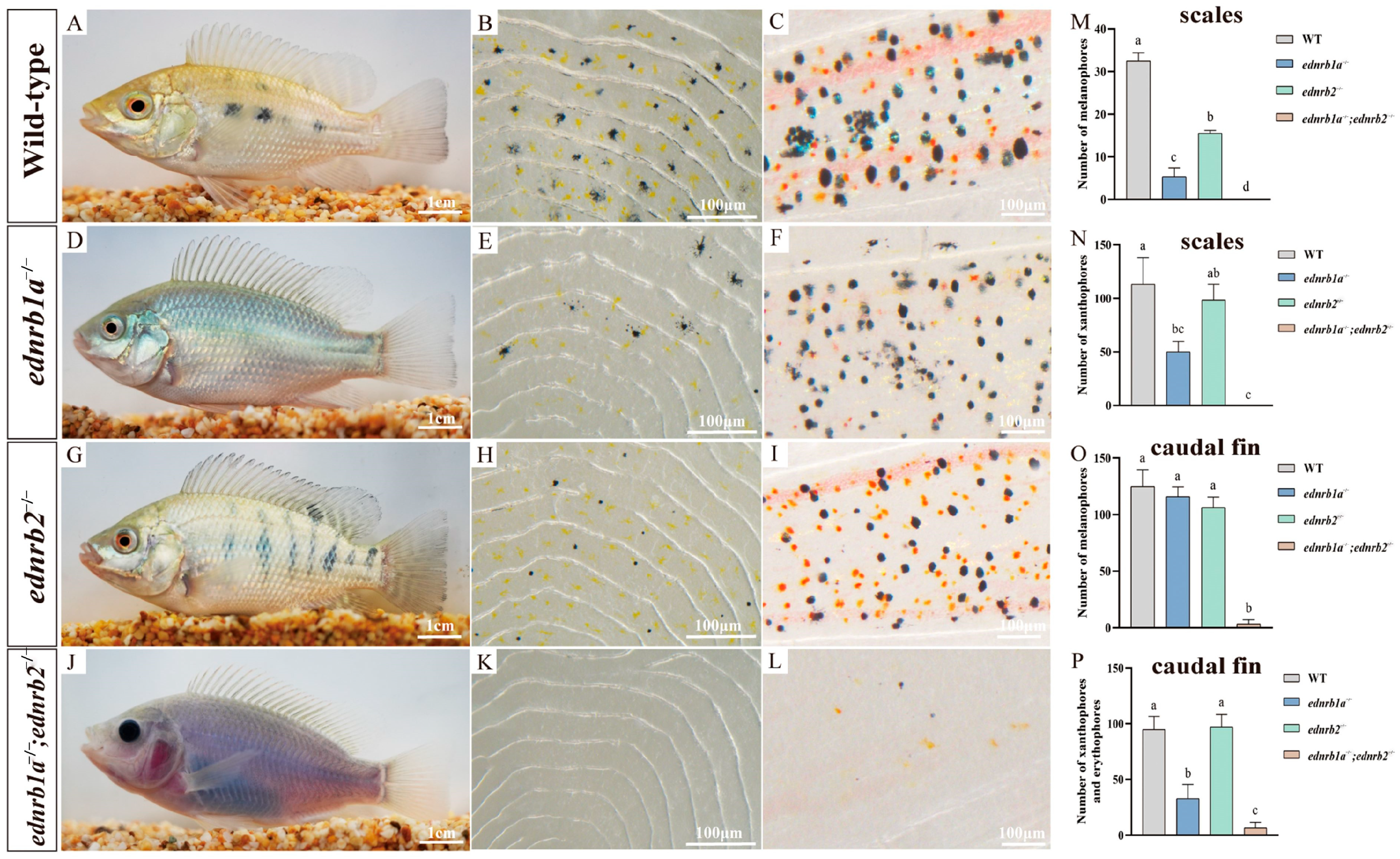
3.6. Impact of Ednrb Mutations on Pigment Cells in Tilapia at 90 dpfs
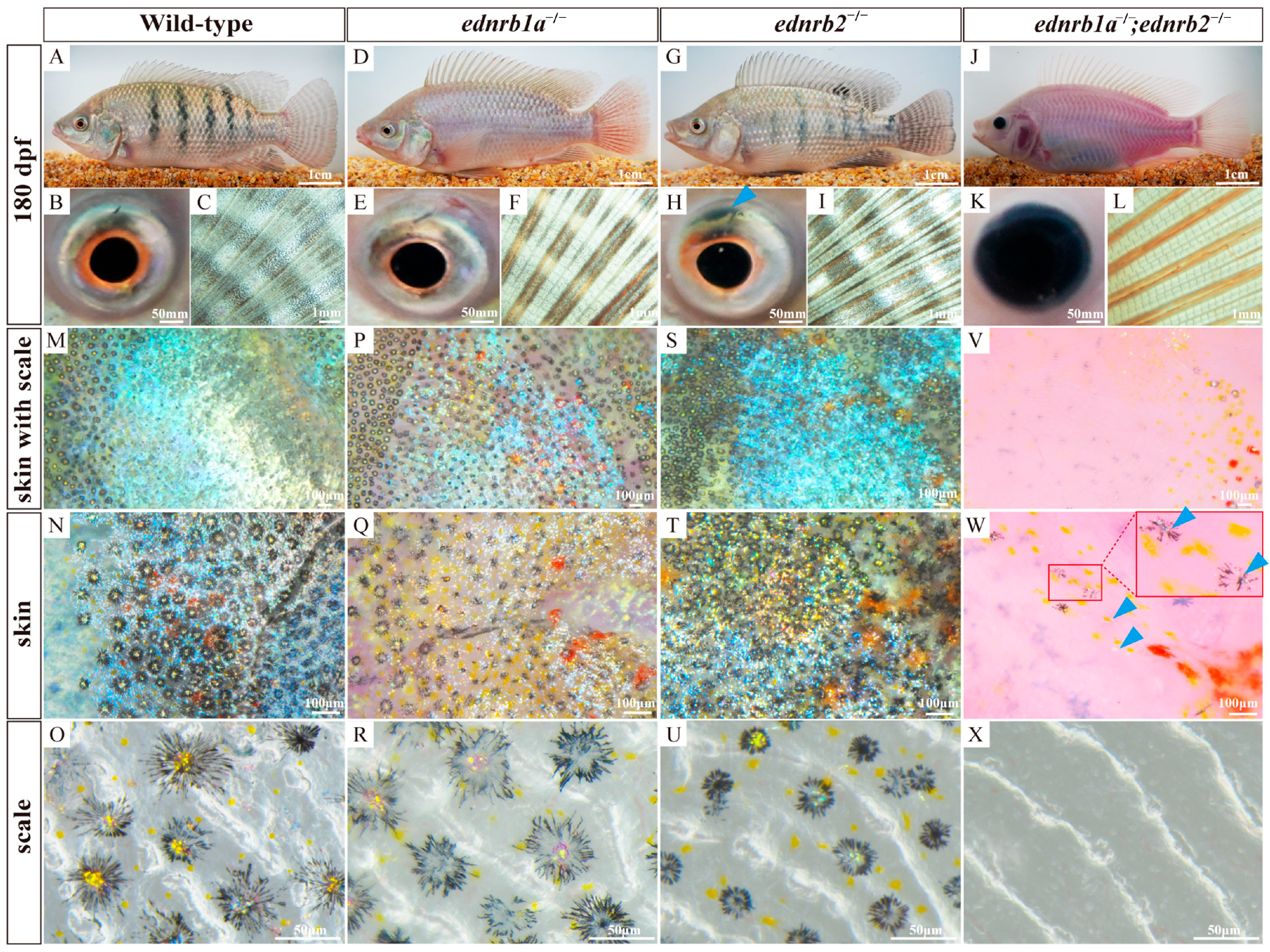
3.7. Ednrb Mutants Exhibit Ornamental Coloration in Tilapia at 180 dpfs
3.8. Ednrb Mutants Display No Impairment of Guanine Synthesis
3.9. Co-Expression of ednrb1a and ednrb2 in NCCs and Eyes
3.10. Microinjection of mitfa mRNA but Not pnp4a mRNA Rescues Embryonic Phenotypes in ednrb1a−/−;ednrb2−/− Mutants
4. Discussion
4.1. ednrb1a and ednrb2 Are Indispensable for Iridophore Development in Tilapia
4.2. Synergistic Roles of ednrb1a and ednrb2 in Tilapia Pigment Cell Development
4.3. ednrb1a and ednrb2 Regulate Erythrophore Development
4.4. Ednrb Mutants Do Not Affect the Guanine Synthesis Pathway in Tilapia
4.5. mitfa mRNA Injection Successfully Rescues the Pigment Cell Deficiency Phenotype in ednrb1a−/−;ednrb2−/− Mutants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Braasch, I.; Schartl, M. Evolution of endothelin receptors in vertebrates. Gen. Comp. Endocrinol. 2014, 209, 21–34. [Google Scholar] [CrossRef]
- Braasch, I.; Volff, J.-N.; Schartl, M. The Endothelin System: Evolution of Vertebrate-Specific Ligand-Receptor Interactions by Three Rounds of Genome Duplication. Mol. Biol. Evol. 2009, 26, 783–799. [Google Scholar] [CrossRef]
- Hyndman, K.A.; Miyamoto, M.M.; Evans, D.H. Phylogeny, taxonomy, and evolution of the endothelin receptor gene family. Mol. Phylogenet. Evol. 2009, 52, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, K.; Hammer, R.E.; Richardson, J.A.; Baynash, A.G.; Cheung, J.C.; Giaid, A.; Yanagisawa, M. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell 1994, 79, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Lecoin, L.; Sakurai, T.; Ngo, M.T.; Abe, Y.; Yanagisawa, M.; Le Douarin, N.M. Cloning and characterization of a novel endothelin receptor subtype in the avian class. Proc. Natl. Acad. Sci. USA 1998, 95, 3024–3029. [Google Scholar] [CrossRef]
- Parichy, D.M.; Mellgren, E.M.; Rawls, J.F.; Lopes, S.S.; Kelsh, R.N.; Johnson, S.L. Mutational Analysis of Endothelin Receptor b1 (rose) during Neural Crest and Pigment Pattern Development in the Zebrafish Danio rerio. Dev. Biol. 2000, 227, 294–306. [Google Scholar] [CrossRef]
- Adameyko, I.; Lallemend, F.; Aquino, J.B.; Pereira, J.A.; Topilko, P.; Müller, T.; Fritz, N.; Beljajeva, A.; Mochii, M.; Liste, I.; et al. Schwann Cell Precursors from Nerve Innervation Are a Cellular Origin of Melanocytes in Skin. Cell 2009, 139, 366–379. [Google Scholar] [CrossRef]
- Adameyko, I.; Lallemend, F. Glial versus melanocyte cell fate choice: Schwann cell precursors as a cellular origin of melanocytes. Cell. Mol. Life Sci. 2010, 67, 3037–3055. [Google Scholar] [CrossRef] [PubMed]
- Ahi, E.P.; Lecaudey, L.A.; Ziegelbecker, A.; Steiner, O.; Glabonjat, R.; Goessler, W.; Hois, V.; Wagner, C.; Lass, A.; Sefc, K.M. Comparative transcriptomics reveals candidate carotenoid color genes in an East African cichlid fish. BMC Genom. 2020, 21, 54. [Google Scholar] [CrossRef]
- Alibardi, L. Observations on the ultrastructure and distribution of chromatophores in the skin of chelonians. Acta Zool. 2013, 94, 222–232. [Google Scholar] [CrossRef]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endo-nuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.J. Stem cells and transcription factors in the development of the mammalian neural crest. FASEB J. 1994, 8, 707–713. [Google Scholar] [CrossRef]
- Hirata, M.; Nakamura, K.; Kanemaru, T.; Shibata, Y.; Kondo, S. Pigment cell organization in the hypodermis of zebrafish. Dev. Dyn. 2003, 227, 497–503. [Google Scholar] [CrossRef] [PubMed]
- Andrade, P.; Carneiro, M. Pterin-based pigmentation in animals. Biol. Lett. 2021, 17, 20210221. [Google Scholar] [CrossRef]
- Bae, C.-J.; Saint-Jeannet, J.-P. Chapter 2—Induction and specification of neural crest cells: Extracellular signals and transcriptional switches. In Neural Crest Cells: Evolution, Development and Disease; Elsevier: Amsterdam, The Netherlands, 2014; pp. 27–49. [Google Scholar]
- Bagnara, J.T.; Taylor, J.D.; Hadley, M.E. The dermal chromatophore unit. J. Cell Biol. 1968, 38, 67–79. [Google Scholar] [CrossRef]
- Balani, K.; Brito, F.C.; Kos, L.; Agarwal, A. Melanocyte pigmentation stiffens murine cardiac tricuspid valve leaflet. J. R. Soc. Interface 2009, 6, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lu, B.; Li, T.; Liang, G.; Xu, M.; Liu, X.; Tao, W.; Zhou, L.; Kocher, T.D.; Wang, D.; et al. Nile Tilapia: A Model for Studying Teleost Color Patterns. J. Hered. 2021, 112, 469–484. [Google Scholar] [CrossRef]
- Lu, B.; Wang, C.; Liang, G.; Xu, M.; Kocher, T.D.; Sun, L.; Wang, D. Generation of ornamental Nile tilapia with distinct gray and black body color pattern by csf1ra mutation. Aquac. Rep. 2022, 23, 101077. [Google Scholar] [CrossRef]
- Johnston, C.E.; Eales, J.G. Purines in the Integument of the Atlantic Salmon (Salmo salar) During Parr–Smolt Transformation. J. Fish. Res. Board Can. 1967, 24, 955–964. [Google Scholar] [CrossRef]
- Kampmann, M. CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem. Biol. 2017, 13, 406–416. [Google Scholar] [CrossRef]
- Cuervo, J.J.; Belliure, J.; Negro, J.J. Coloration reflects skin pterin concentration in a red-tailed lizard. Comp. Biochem. Physiol. Part B: Biochem. Mol. Biol. 2016, 193, 17–24. [Google Scholar] [CrossRef]
- Curran, K.; Lister, J.A.; Kunkel, G.R.; Prendergast, A.; Parichy, D.M.; Raible, D.W. Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev. Biol. 2010, 344, 107–118. [Google Scholar] [CrossRef]
- Dawes, J.H.P.; Kelsh, R.N. Cell Fate Decisions in the Neural Crest, from Pigment Cell to Neural Development. Int. J. Mol. Sci. 2021, 22, 13531. [Google Scholar] [CrossRef]
- Day, J.A. Gene Targeting in Ciona Intestinalis via Zinc Finger Nuclease Mediated Homologous Recombination. Master’s Thesis, San Diego State University, San Diego, CA, USA, 2011. [Google Scholar]
- Cornean, A.; Gierten, J.; Welz, B.; Mateo, J.L.; Thumberger, T.; Wittbrodt, J. Precise in vivo functional analysis of DNA variants with base editing using ACEofBASEs target prediction. eLife 2022, 11, e72124. [Google Scholar] [CrossRef]
- Cropp, C.D.; Komori, T.; Shima, J.E.; Urban, T.J.; Yee, S.W.; More, S.S.; Giacomini, K.M. Organic Anion Transporter 2 (SLC22A7) Is a Facilitative Transporter of cGMP. Mol. Pharmacol. 2008, 73, 1151–1158. [Google Scholar] [CrossRef]
- D’aGati, G.; Beltre, R.; Sessa, A.; Burger, A.; Zhou, Y.; Mosimann, C.; White, R.M. A defect in the mitochondrial protein Mpv17 underlies the transparent casper zebrafish. Dev. Biol. 2017, 430, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Minoux, M.; Rijli, F.M. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development 2010, 137, 2605–2621. [Google Scholar] [CrossRef] [PubMed]
- Miwa, M.; Inoue-Murayama, M.; Aoki, H.; Kunisada, T.; Hiragaki, T.; Mizutani, M.; Ito, S. Endothelin receptor B2 (EDNRB2) is associated with the panda plumage colour mutation in Japanese quail. Anim. Genet. 2007, 38, 103–108. [Google Scholar] [CrossRef]
- Nagao, A. Absorption and metabolism of dietary carotenoids. BioFactors 2011, 37, 83–87. [Google Scholar] [CrossRef] [PubMed]
- Nagao, Y.; Takada, H.; Miyadai, M.; Adachi, T.; Seki, R.; Kamei, Y.; Hara, I.; Taniguchi, Y.; Naruse, K.; Hibi, M.; et al. Distinct interactions of Sox5 and Sox10 in fate specification of pigment cells in medaka and zebrafish. PLOS Genet. 2018, 14, e1007260. [Google Scholar] [CrossRef]
- Köhidai, L.; Tóth, K.; Ruskoaho, H.; Csaba, G. Effect of vasoactive peptides on Tetrahymena. Chemotactic properties of endothelins (ET-1, ET-2, ET-3, fragment 11-21 of ET-1 and big endothelin-1): A short-term inducible signalling mechanism of chemotaxis. Cell Biol. 2001, 25, 1173–1177. [Google Scholar] [CrossRef][Green Version]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Kyriakoudi, S.A.; Chatzi, D.; Dermitzakis, I.; Gargani, S.; Manthou, M.E.; Meditskou, S.; Theotokis, P. Genetic Identity of Neural Crest Cell Differentiation in Tissue and Organ Development. Front. Biosci. 2024, 29, 261. [Google Scholar] [CrossRef]
- La, M.; Reid, J.J. Endothelin-1 and the regulation of vascular tone. Clin. Exp. Pharmacol. Physiol. 1995, 22, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Laghari, M.Y.; Lashari, P.; Zhang, Y.; Sun, X. Identification of Quantitative Trait Loci (QTLs) in Aquaculture Species. Rev. Fish. Sci. Aquacult. 2014, 22, 221–238. [Google Scholar] [CrossRef]
- Li, L.; Li, D.; Liu, L.; Li, S.; Feng, Y.; Peng, X.; Gong, Y.; Shellman, Y.G. Endothelin Receptor B2 (EDNRB2) Gene Is Associated with Spot Plumage Pattern in Domestic Ducks (Anas platyrhynchos). PLoS ONE 2015, 10, e0125883. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Zhang, B.; Wang, D.; Jiang, D.; Zhu, C.; Deng, S.; Chen, H.; Li, G.; Shi, H. Metabolism, Function, Molecular Mechanism, and Application of Carotenoids in Coloration of Aquatic Animals. Rev. Aquac. 2025, 17, e70016. [Google Scholar] [CrossRef]
- Gudjohnsen, S.A.H.; Atacho, D.A.M.; Gesbert, F.; Raposo, G.; Hurbain, I.; Larue, L.; Steingrimsson, E.; Petersen, P.H. Meningeal Melanocytes in the Mouse: Distribution and Dependence on Mitf. Front. Neuroanat. 2015, 9, 149. [Google Scholar] [CrossRef]
- Guo, X.; Wu, L.; Lyu, Y.; Chowanadisai, W.; Clarke, S.L.; Lucas, E.A.; Smith, B.J.; He, H.; Wang, W.; Medeiros, D.M.; et al. Ablation of β, β-carotene-9′,10′-oxygenase 2 remodels the hypothalamic metabolome leading to metabolic disorders in mice. J. Nutr. Biochem. 2017, 46, 74–82. [Google Scholar] [CrossRef]
- Le Guyader, S.; Maier, J.; Jesuthasan, S. Esrom, an ortholog of PAM (protein associated with c-myc), regulates pteridine synthesis in the zebrafish. Dev. Biol. 2005, 277, 378–386. [Google Scholar] [CrossRef]
- Harris, J.; Hunt, S. The fine structure of iridophores in the skin of the Atlantic Salmon (Salmo Salar L.). Tissue Cell 1973, 5, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, H.; Goda, M.; Futahashi, R.; Kelsh, R.; Akiyama, T. Pigments, Pigment Cells and Pigment Patterns; Springer: Singapore, 2021. [Google Scholar]
- Huang, D.; Lewis, V.M.; Foster, T.N.; Toomey, M.B.; Corbo, J.C.; Parichy, D.M. Development and genetics of red coloration in the zebrafish relative Danio albolineatus. eLife 2021, 10, e70253. [Google Scholar] [CrossRef]
- Davenport, A.P.; Hyndman, K.A.; Dhaun, N.; Southan, C.; Kohan, D.E.; Pollock, J.S.; Pollock, D.M.; Webb, D.J.; Maguire, J.J. Endothelin. Pharmacol. Rev. 2016, 68, 357–418. [Google Scholar] [CrossRef] [PubMed]
- David, L.; Rothbard, S.; Rubinstein, I.; Katzman, H.; Hulata, G.; Hillel, J.; Lavi, U. Aspects of red and black color inheritance in the Japanese ornamental (Koi) carp (Cyprinus carpio L.). Aquaculture 2003, 233, 129–147. [Google Scholar] [CrossRef]
- Diepeveen, E.T.; Salzburger, W. Molecular Characterization of Two Endothelin Pathways in East African Cichlid Fishes. J. Mol. Evol. 2011, 73, 355–368. [Google Scholar] [CrossRef][Green Version]
- Gasque, P.; Jaffar-Bandjee, M.C. The immunology and inflammatory responses of human melanocytes in infectious diseases. J. Infect. 2015, 71, 413–421. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Goda, M.; Fujii, R. Blue Chromatophores in Two Species of Callionymid Fish. Zool. Sci. 1995, 12, 811–813. [Google Scholar] [CrossRef]
- Goda, M.; Fujiyoshi, Y.; Sugimoto, M.; Fujii, R. Novel Dichromatic Chromatophores in the Integument of the Mandarin Fish Synchiropus splendidus. Biol. Bull. 2013, 224, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef]
- Kirkby, N.S.; Hadoke, P.W.F.; Bagnall, A.J.; Webb, D.J. The endothelin system as a therapeutic target in cardiovascular disease: Great expectations or bleak house? Br. J. Pharmacol. 2008, 153, 1105–1119. [Google Scholar] [CrossRef]
- Matsushima, Y.; Shinkai, Y.; Kobayashi, Y.; Sakamoto, M.; Kunieda, T.; Tachibana, M. A mouse model of Waardenburg syndrome type 4 with a new spontaneous mutation of the endothelin-B receptor gene. Mamm. Genome 2002, 13, 30–35. [Google Scholar] [CrossRef]
- Menter, D.G.; Obika, M.; Tchen, T.T.; Taylor, J.D. Leucophores and iridophores of Fundulus heteroclitus: Biophysical and ultrastructural properties. J. Morphol. 1979, 160, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Djurdjevič, I.; Kreft, M.E.; Bajec, S.S. Comparison of pigment cell ultrastructure and organisation in the dermis of marble trout and brown trout, and first description of erythrophore ultrastructure in salmonids. Am. J. Anat. 2015, 227, 583–595. [Google Scholar] [CrossRef]
- Dooley, C.M.; Mongera, A.; Walderich, B.; Nüsslein-Volhard, C. On the embryonic origin of adult melanophores: The role of ErbB and Kit signalling in establishing melanophore stem cells in zebrafish. Development 2013, 140, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Futter, C.E. The molecular regulation of organelle transport in mammalian retinal pigment epithelial cells. Pigment. Cell Res. 2006, 19, 104–111. [Google Scholar] [CrossRef]
- Garciacastro, M.; Bronnerfraser, M. Induction and differentiation of the neural crest. Curr. Opin. Cell Biol. 1999, 11, 695–698. [Google Scholar] [CrossRef]
- Goda, M.; Ohata, M.; Ikoma, H.; Fujiyoshi, Y.; Sugimoto, M.; Fujii, R. Integumental reddish-violet coloration owing to novel dichromatic chromatophores in the teleost fish, Pseudochromis diadema. Pigment. Cell Melanoma Res. 2011, 24, 614–617. [Google Scholar] [CrossRef] [PubMed]
- Granneman, J.G.; A Kimler, V.; Zhang, H.; Ye, X.; Luo, X.; Postlethwait, J.H.; Thummel, R. Lipid droplet biology and evolution illuminated by the characterization of a novel perilipin in teleost fish. eLife 2017, 6, e21771. [Google Scholar] [CrossRef]
- Huang, J.; Fang, W.; Li, J.; Cai, W.; Lu, J. Full-length transcriptome reveals alternative splicing regulation pattern of skin color variant in red tilapia (Oreochromis spp.). Aquaculture 2024, 598, 741963. [Google Scholar] [CrossRef]
- Hubbard, J.K.; Uy, J.A.C.; Hauber, M.E.; Hoekstra, H.E.; Safran, R.J. Vertebrate pigmentation: From underlying genes to adaptive function. Trends Genet. 2010, 26, 231–239. [Google Scholar] [CrossRef]
- Ito, S.; Wakamatsu, K. Diversity of human hair pigmentation as studied by chemical analysis of eumelanin and pheomelanin. J. Eur. Acad. Dermatol. Venereol. 2011, 25, 1369–1380. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Duan, J.; Yuan, Q.; He, X.; Yang, G.; Zhu, S.; Wu, K.; Hu, W.; Gao, T.; Cheng, X.; et al. Structural basis of peptide recognition and activation of endothelin receptors. Nat. Commun. 2023, 14, 1268. [Google Scholar] [CrossRef]
- Karne, S.; Jayawickreme, C.; Lerner, M. Cloning and characterization of an endothelin-3 specific receptor (ETC receptor) from Xenopus laevis dermal melanophores. J. Biol. Chem. 1993, 268, 19126–19133. [Google Scholar] [CrossRef]
- Kedzierski, R.M.; Yanagisawa, M. Endothelin System: The Double-Edged Sword in Health and Disease. Annu. Rev. Pharmacol. Toxicol. 2001, 41, 851–876. [Google Scholar] [CrossRef]
- Kelsh, R.N.; Sosa, K.C.; Farjami, S.; Makeev, V.; Dawes, J.H.P.; Rocco, A. Cyclical fate restriction: A new view of neural crest cell fate specification. Development 2021, 148, dev176057. [Google Scholar] [CrossRef]
- Kikuchi, D.W.; Seymoure, B.M.; Pfennig, D.W. Mimicry’s palette: Widespread use of conserved pigments in the aposematic signals of snakes. Evol. Dev. 2014, 16, 61–67. [Google Scholar] [CrossRef]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156–1160. [Google Scholar] [CrossRef] [PubMed]
- Meulemans, D.; Bronner-Fraser, M. Gene-Regulatory Interactions in Neural Crest Evolution and Development. Dev. Cell 2004, 7, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Nannan, M.; Wenjun, W.; Ran, Z.; Yongsheng, S.; Rongyan, Z.; Hui, C.; Sumin, Z.; Hui, X. Population genomics reveals that a missense mutation in EDNRB2 contributes to white plumage color in pigeons. Poult. Sci. 2024, 103, 103225. [Google Scholar] [CrossRef]
- Ng, A.; Uribe, R.A.; Yieh, L.; Nuckels, R.; Gross, J.M. Zebrafish mutations in gart and paics identify crucial roles for de novo purine synthesis in vertebrate pigmentation and ocular development. Development 2009, 136, 2601–2611. [Google Scholar] [CrossRef]
- Nitzan, E.; Pfaltzgraff, E.R.; Labosky, P.A.; Kalcheim, C. Neural crest and Schwann cell progenitor-derived melanocytes are two spatially segregated populations similarly regulated by Foxd3. Proc. Natl. Acad. Sci. 2013, 110, 12709–12714. [Google Scholar] [CrossRef]
- Parichy, D.M. Evolution of pigment cells and patterns: Recent insights from teleost fishes. Curr. Opin. Genet. Dev. 2021, 69, 88–96. [Google Scholar] [CrossRef]
- Patterson, L.B.; Parichy, D.M.; Barsh, G.S. Interactions with Iridophores and the Tissue Environment Required for Patterning Melanophores and Xanthophores during Zebrafish Adult Pigment Stripe Formation. PLoS Genet. 2013, 9, e1003561. [Google Scholar] [CrossRef]
- Patterson, L.B.; Bain, E.J.; Parichy, D.M. Pigment cell interactions and differential xanthophore recruitment underlying zebrafish stripe reiteration and Danio pattern evolution. Nat. Commun. 2014, 5, 5299. [Google Scholar] [CrossRef]
- Patterson, L.B.; Parichy, D.M. Zebrafish Pigment Pattern Formation: Insights into the Development and Evolution of Adult Form. Annu. Rev. Genet. 2019, 53, 505–530. [Google Scholar] [CrossRef]
- Greenhill, E.R.; Rocco, A.; Vibert, L.; Nikaido, M.; Kelsh, R.N.; Mullins, M.C. An Iterative Genetic and Dynamical Modelling Approach Identifies Novel Features of the Gene Regulatory Network Underlying Melanocyte Development. PLOS Genet. 2011, 7, e1002265. [Google Scholar] [CrossRef] [PubMed]
- Petratou, K.; Spencer, S.A.; Kelsh, R.N.; Lister, J.A.; Klymkowsky, M. The MITF paralog tfec is required in neural crest development for fate specification of the iridophore lineage from a multipotent pigment cell progenitor. PLoS ONE 2021, 16, e0244794. [Google Scholar] [CrossRef]
- Jang, H.S.; Chen, Y.; Ge, J.; Wilkening, A.N.; Hou, Y.; Lee, H.J.; Choi, Y.R.; Lowdon, R.F.; Xing, X.; Li, D.; et al. Epigenetic dynamics shaping melanophore and iridophore cell fate in zebrafish. Genome Biol. 2021, 22, 282. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Pavan, W.J. Transcriptional and signaling regulation in neural crest stem cell-derived melanocyte development: Do all roads lead to Mitf? Cell Res. 2008, 18, 1163–1176. [Google Scholar] [CrossRef] [PubMed]
- Miyadai, M.; Takada, H.; Shiraishi, A.; Kimura, T.; Watakabe, I.; Kobayashi, H.; Nagao, Y.; Naruse, K.; Higashijima, S.-I.; Shimizu, T.; et al. A gene regulatory network combining Pax3/7, Sox10 and Mitf generates diverse pigment cell types in medaka and zebrafish. Development 2023, 150, dev202114. [Google Scholar] [CrossRef]
- Petratou, K.; Subkhankulova, T.; Lister, J.A.; Rocco, A.; Schwetlick, H.; Kelsh, R.N.; Parichy, D.M. A systems biology approach uncovers the core gene regulatory network governing iridophore fate choice from the neural crest. PLoS Genet. 2018, 14, e1007402. [Google Scholar] [CrossRef]
- Pla, P.; Alberti, C.; Solov’EVa, O.; Pasdar, M.; Kunisada, T.; Larue, L. Ednrb2 orients cell migration towards the dorsolateral neural crest pathway and promotes melanocyte differentiation. Pigment. Cell Res. 2005, 18, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Sakata, R.C.; Ishiguro, S.; Mori, H.; Tanaka, M.; Tatsuno, K.; Ueda, H.; Yamamoto, S.; Seki, M.; Masuyama, N.; Nishida, K.; et al. Publisher Correction: Base editors for simultaneous introduction of C-to-T and A-to-G mutations. Nat. Biotechnol. 2020, 38, 901. [Google Scholar] [CrossRef]
- Krauss, J.; Astrinides, P.; Frohnhöfer, H.G.; Walderich, B.; Nüsslein-Volhard, C. transparent, a gene affecting stripe formation in Zebrafish, encodes the mitochondrial protein Mpv17 that is required for iridophore survival. Biol. Open 2013, 2, 703–710. [Google Scholar] [CrossRef]
- Samuel, R.M.; Navickas, A.; Maynard, A.; Gaylord, E.A.; Garcia, K.; Bhat, S.; Majd, H.; Richter, M.N.; Elder, N.; Le, D.; et al. Generation of Schwann cell derived melanocytes from hPSCs identifies pro-metastatic factors in melanoma. BioRxiv 2023, 531220, Preprint. [Google Scholar]
- Sato-Jin, K.; Nishimura, E.K.; Akasaka, E.; Huber, W.; Nakano, H.; Miller, A.; Du, J.; Wu, M.; Hanada, K.; Sawamura, D.; et al. Epistatic connections between microphthalmia-associated transcription factor and endothelin signaling in Waardenburg syndrome and other pigmentary disorders. FASEB J. 2007, 22, 1155–1168. [Google Scholar] [CrossRef]
- Toews, D.P.; Hofmeister, N.R.; Taylor, S.A. The Evolution and Genetics of Carotenoid Processing in Animals. Trends Genet. 2017, 33, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Torrissen, O.J.; Hardy, R.W.; Shearer, K.D. Pigmentation of Salmonids-Carotenoid Deposition and Metabolism. Rev. Aquat. Sci. 1989, 1, 209–225. [Google Scholar]
- Bossche, K.V.D.; Naeyaert, J.; Lambert, J. The Quest for the Mechanism of Melanin Transfer. Traffic 2006, 7, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Hearing, V.J. Physiological factors that regulate skin pigmentation. BioFactors 2009, 35, 193–199. [Google Scholar] [CrossRef]
- Ytrestøyl, T.; Bjerkeng, B. Dose Response in Uptake and Deposition of Intraperitoneally Administered Astaxanthin in Atlantic Salmon (Salmo salar L.) and Atlantic Cod (Gadus morhua L.). Aquaculture 2007, 263, 179–191. [Google Scholar] [CrossRef]
- Kimura, T.; Takehana, Y.; Naruse, K. pnp4a is the causal gene of the medaka iridophore mutant guanineless. G3: Genes Genomes Genet. 2017, 7, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Leontovich, A.; Sarras, M.P. Molecular and functional evidence for early divergence of an endothelin-like system during metazoan evolution: Analysis of the Cnidarian, hydra. Development 2001, 128, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Nagao, Y.; Hashimoto, H.; Yamamoto-Shiraishi, Y.-I.; Yamamoto, S.; Yabe, T.; Takada, S.; Kinoshita, M.; Kuroiwa, A.; Naruse, K. Leucophores are similar to xanthophores in their specification and differentiation processes in medaka. Proc. Natl. Acad. Sci. USA 2014, 111, 7343–7348. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wen, Z.; Wu, J.; Yao, J.; Fang, F.; Ju, S.; Wang, C.; Liu, X.; Wang, D. Synergistic Regulation of Pigment Cell Precursors’ Differentiation and Migration by ednrb1a and ednrb2 in Nile Tilapia. Cells 2025, 14, 1213. https://doi.org/10.3390/cells14151213
Wen Z, Wu J, Yao J, Fang F, Ju S, Wang C, Liu X, Wang D. Synergistic Regulation of Pigment Cell Precursors’ Differentiation and Migration by ednrb1a and ednrb2 in Nile Tilapia. Cells. 2025; 14(15):1213. https://doi.org/10.3390/cells14151213
Chicago/Turabian StyleWen, Zilong, Jinzhi Wu, Jiawen Yao, Fugui Fang, Siyu Ju, Chenxu Wang, Xingyong Liu, and Deshou Wang. 2025. "Synergistic Regulation of Pigment Cell Precursors’ Differentiation and Migration by ednrb1a and ednrb2 in Nile Tilapia" Cells 14, no. 15: 1213. https://doi.org/10.3390/cells14151213
APA StyleWen, Z., Wu, J., Yao, J., Fang, F., Ju, S., Wang, C., Liu, X., & Wang, D. (2025). Synergistic Regulation of Pigment Cell Precursors’ Differentiation and Migration by ednrb1a and ednrb2 in Nile Tilapia. Cells, 14(15), 1213. https://doi.org/10.3390/cells14151213







