miR-302a/b/d-3p Differentially Expressed During Frontonasal Development Is Sensitive to Retinoic Acid Exposure
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis
2.2. Cell Culture
2.3. Cell Proliferation and Cell Death Assay
2.4. Cell Migration Assay
2.5. Quantitative RT-PCR
2.6. Statistical Analysis
3. Results
3.1. miRNAs Expressed in the Frontonasal Process in a Temporal-Specific Manner During Frontonasal Development
3.2. Overexpression of miR-28a-5p, miR-302a-3p, miR-302b-3p, and miR-302d-3p Inhibits Cell Proliferation Through Downregulation of Genes Related to Frontonasal Malformations in MEFM and O9-1 Cells
3.3. Identification of miRNAs Altered by Excessive atRA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| atRA | all-trans retinoic acid |
| miRNA | microRNA |
| MEFM | Mouse embryonic frontonasal mesenchymal |
| CNC | Cranial neural crest |
| CL/P | Cleft lip with/without cleft palate |
| ES | Embryonic stem |
| SHH | Sonic hedgehog |
References
- Suzuki, A.; Sangani, D.R.; Ansari, A.; Iwata, J. Molecular mechanisms of midfacial developmental defects. Dev. Dyn. 2016, 245, 276–293. [Google Scholar] [CrossRef]
- Johnston, M.C.; Bronsky, P.T. Prenatal craniofacial development: New insights on normal and abnormal mechanisms. Crit. Rev. Oral Biol. Med. Off. Publ. Am. Assoc. Oral Biol. 1995, 6, 368–422. [Google Scholar] [CrossRef]
- Krapels, I.P.; van Rooij, I.A.; Ocke, M.C.; West, C.E.; van der Horst, C.M.; Steegers-Theunissen, R.P. Maternal nutritional status and the risk for orofacial cleft offspring in humans. J. Nutr. 2004, 134, 3106–3113. [Google Scholar] [CrossRef] [PubMed]
- Sulik, K.K. Genesis of alcohol-induced craniofacial dysmorphism. Exp. Biol. Med. 2005, 230, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Kietzman, H.W.; Everson, J.L.; Sulik, K.K.; Lipinski, R.J. The teratogenic effects of prenatal ethanol exposure are exacerbated by Sonic Hedgehog or GLI2 haploinsufficiency in the mouse. PLoS ONE 2014, 9, e89448. [Google Scholar] [CrossRef] [PubMed]
- Ahlgren, S.C.; Thakur, V.; Bronner-Fraser, M. Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proc. Natl. Acad. Sci. USA 2002, 99, 10476–10481. [Google Scholar] [CrossRef]
- Schneider, R.A.; Hu, D.; Rubenstein, J.L.; Maden, M.; Helms, J.A. Local retinoid signaling coordinates forebrain and facial morphogenesis by maintaining FGF8 and SHH. Development 2001, 128, 2755–2767. [Google Scholar] [CrossRef]
- Dupe, V.; Matt, N.; Garnier, J.M.; Chambon, P.; Mark, M.; Ghyselinck, N.B. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc. Natl. Acad. Sci. USA 2003, 100, 14036–14041. [Google Scholar] [CrossRef]
- Mark, M.; Lohnes, D.; Mendelsohn, C.; Dupe, V.; Vonesch, J.L.; Kastner, P.; Rijli, F.; Bloch-Zupan, A.; Chambon, P. Roles of retinoic acid receptors and of Hox genes in the patterning of the teeth and of the jaw skeleton. Int. J. Dev. Biol. 1995, 39, 111–121. [Google Scholar]
- Padmanabhan, R.; Shafiullah, M. Effect of maternal diabetes and ethanol interactions on embryo development in the mouse. Mol. Cell. Biochem. 2004, 261, 43–56. [Google Scholar] [CrossRef]
- Alade, A.; Ismail, W.; Nair, R.; Schweizer, M.; Awotoye, W.; Oladayo, A.; Ryckman, K.; Butali, A. Periconceptional use of vitamin A and the risk of giving birth to a child with nonsyndromic orofacial clefts-A meta-analysis. Birth Defects Res. 2022, 114, 467–477. [Google Scholar] [CrossRef]
- Lammer, E.J.; Chen, D.T.; Hoar, R.M.; Agnish, N.D.; Benke, P.J.; Braun, J.T.; Curry, C.J.; Fernhoff, P.M.; Grix, A.W., Jr.; Lott, I.T.; et al. Retinoic acid embryopathy. N. Engl. J. Med. 1985, 313, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Tsuchiya, S.; Meltzer, S.J.; Shimizu, K. MicroRNAs and epigenetics. FEBS J. 2011, 278, 1598–1609. [Google Scholar] [CrossRef]
- Li, M.; Huo, X.; Davuljigari, C.B.; Dai, Q.; Xu, X. MicroRNAs and their role in environmental chemical carcinogenesis. Environ. Geochem. Health 2019, 41, 225–247. [Google Scholar] [CrossRef] [PubMed]
- Warner, D.R.; Mukhopadhyay, P.; Brock, G.; Webb, C.L.; Michele Pisano, M.; Greene, R.M. MicroRNA expression profiling of the developing murine upper lip. Dev. Growth Differ. 2014, 56, 434–447. [Google Scholar] [CrossRef]
- Shin, J.O.; Lee, J.M.; Cho, K.W.; Kwak, S.; Kwon, H.J.; Lee, M.J.; Cho, S.W.; Kim, K.S.; Jung, H.S. MiR-200b is involved in Tgf-beta signaling to regulate mammalian palate development. Histochem. Cell Biol. 2012, 137, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhyay, P.; Smolenkova, I.; Warner, D.; Pisano, M.M.; Greene, R.M. Spatio-Temporal Expression and Functional Analysis of miR-206 in Developing Orofacial Tissue. MicroRNA 2019, 8, 43–60. [Google Scholar] [CrossRef]
- Yoshioka, H.; Mikami, Y.; Ramakrishnan, S.S.; Suzuki, A.; Iwata, J. MicroRNA-124-3p Plays a Crucial Role in Cleft Palate Induced by Retinoic Acid. Front. Cell Dev. Biol. 2021, 9, 621045. [Google Scholar] [CrossRef]
- Rueda, A.; Barturen, G.; Lebrón, R.; Gómez-Martín, C.; Alganza, Á.; Oliver, J.L.; Hackenberg, M. sRNAtoolbox: An integrated collection of small RNA research tools. Nucleic Acids Res. 2015, 43, W467–W473. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Chivero, E.T.; Dagur, R.S.; Peeples, E.S.; Sil, S.; Liao, K.; Ma, R.; Chen, L.; Gurumurthy, C.B.; Buch, S.; Hu, G. Biogenesis, physiological functions and potential applications of extracellular vesicles in substance use disorders. Cell. Mol. Life Sci. 2021, 78, 4849–4865. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Salazar, E.G.; Gayosso-Gomez, L.V.; Baez-Saldana, R.; Falfan-Valencia, R.; Perez-Padilla, R.; Higuera-Iglesias, A.L.; Vazquez-Manriquez, M.E.; Ortiz-Quintero, B. Cigarette Smoking Alters the Expression of Circulating microRNAs and Its Potential Diagnostic Value in Female Lung Cancer Patients. Biology 2021, 10, 793. [Google Scholar] [CrossRef] [PubMed]
- Francavilla, A.; Gagliardi, A.; Piaggeschi, G.; Tarallo, S.; Cordero, F.; Pensa, R.G.; Impeduglia, A.; Caviglia, G.P.; Ribaldone, D.G.; Gallo, G.; et al. Faecal miRNA profiles associated with age, sex, BMI, and lifestyle habits in healthy individuals. Sci. Rep. 2021, 11, 20645. [Google Scholar] [CrossRef]
- Clark, J.; Eaves, L.A.; Gaona, A.R.; Santos, H.P., Jr.; Smeester, L.; Bangma, J.T.; Rager, J.E.; O’Shea, T.M.; Fry, R.C. Pre-pregnancy BMI-associated miRNA and mRNA expression signatures in the placenta highlight a sexually-dimorphic response to maternal underweight status. Sci. Rep. 2021, 11, 15743. [Google Scholar] [CrossRef]
- Iwata, J.; Hosokawa, R.; Sanchez-Lara, P.A.; Urata, M.; Slavkin, H.; Chai, Y. Transforming growth factor-beta regulates basal transcriptional regulatory machinery to control cell proliferation and differentiation in cranial neural crest-derived osteoprogenitor cells. J. Biol. Chem. 2010, 285, 4975–4982. [Google Scholar] [CrossRef]
- Yoshioka, H.; Suzuki, A.; Iwaya, C.; Iwata, J. Suppression of microRNA 124-3p and microRNA 340-5p ameliorates retinoic acid-induced cleft palate in mice. Development 2022, 149, dev200476. [Google Scholar] [CrossRef]
- Yoshioka, H.; Ramakrishnan, S.S.; Shim, J.; Suzuki, A.; Iwata, J. Excessive All-Trans Retinoic Acid Inhibits Cell Proliferation Through Upregulated MicroRNA-4680-3p in Cultured Human Palate Cells. Front. Cell Dev. Biol. 2021, 9, 618876. [Google Scholar] [CrossRef]
- Yoshioka, H.; Ramakrishnan, S.S.; Suzuki, A.; Iwata, J. Phenytoin Inhibits Cell Proliferation through microRNA-196a-5p in Mouse Lip Mesenchymal Cells. Int. J. Mol. Sci. 2021, 22, 1746. [Google Scholar] [CrossRef]
- Webster, W.S.; Howe, A.M.; Abela, D.; Oakes, D.J. The relationship between cleft lip, maxillary hypoplasia, hypoxia and phenytoin. Curr. Pharm. Des. 2006, 12, 1431–1448. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.Y.; Chen, J.S.; Ying, S.Y. The microRNA and the perspectives of miR-302. Heliyon 2019, 5, e01167. [Google Scholar] [CrossRef]
- Parchem, R.J.; Moore, N.; Fish, J.L.; Parchem, J.G.; Braga, T.T.; Shenoy, A.; Oldham, M.C.; Rubenstein, J.L.; Schneider, R.A.; Blelloch, R. miR-302 Is Required for Timing of Neural Differentiation, Neural Tube Closure, and Embryonic Viability. Cell Rep. 2015, 12, 760–773. [Google Scholar] [CrossRef]
- Li, X.; Zhang, L.; Yin, X.; Gao, Z.; Zhang, H.; Liu, X.; Pan, X.; Li, N.; Yu, Z. Retinoic acid remodels extracellular matrix (ECM) of cultured human fetal palate mesenchymal cells (hFPMCs) through down-regulation of TGF-beta/Smad signaling. Toxicol. Lett. 2014, 225, 208–215. [Google Scholar] [CrossRef]
- Yao, Z.; Chen, D.; Wang, A.; Ding, X.; Liu, Z.; Ling, L.; He, Q.; Zhao, T. Folic acid rescue of ATRA-induced cleft palate by restoring the TGF-beta signal and inhibiting apoptosis. J. Oral Pathol. Med. 2011, 40, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gao, Y.; Yu, M.; Wu, H.; Ai, Z.; Wu, Y.; Liu, H.; Du, J.; Guo, Z.; Zhang, Y. Retinoic Acid Induces Embryonic Stem Cell Differentiation by Altering Both Encoding RNA and microRNA Expression. PLoS ONE 2015, 10, e0132566. [Google Scholar] [CrossRef]
- Keuls, R.A.; Kojima, K.; Lozzi, B.; Steele, J.W.; Chen, Q.; Gross, S.S.; Finnell, R.H.; Parchem, R.J. MiR-302 Regulates Glycolysis to Control Cell-Cycle during Neural Tube Closure. Int. J. Mol. Sci. 2020, 21, 7534. [Google Scholar] [CrossRef] [PubMed]
- Li, H.L.; Wei, J.F.; Fan, L.Y.; Wang, S.H.; Zhu, L.; Li, T.P.; Lin, G.; Sun, Y.; Sun, Z.J.; Ding, J.; et al. miR-302 regulates pluripotency, teratoma formation and differentiation in stem cells via an AKT1/OCT4-dependent manner. Cell Death Dis. 2016, 7, e2078. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, T.; Miura, T.; Kawasaki, T.; Umezawa, A.; Akutsu, H. The hsa-miR-302 cluster controls ectodermal differentiation of human pluripotent stem cell via repression of DAZAP2. Regen. Ther. 2020, 15, 1–9. [Google Scholar] [CrossRef]
- Pajanoja, C.; Hsin, J.; Olinger, B.; Schiffmacher, A.; Yazejian, R.; Abrams, S.; Dapkunas, A.; Zainul, Z.; Doyle, A.D.; Martin, D.; et al. Maintenance of pluripotency-like signature in the entire ectoderm leads to neural crest stem cell potential. Nat. Commun. 2023, 14, 5941. [Google Scholar] [CrossRef]
- Keuls, R.A.; Oh, Y.S.; Patel, I.; Parchem, R.J. Post-transcriptional regulation in cranial neural crest cells expands developmental potential. Proc. Natl. Acad. Sci. USA 2023, 120, e2212578120. [Google Scholar] [CrossRef] [PubMed]
- Patel, I.; Parchem, R.J. Regulation of Oct4 in stem cells and neural crest cells. Birth Defects Res. 2022, 114, 983–1002. [Google Scholar] [CrossRef] [PubMed]
- Lignell, A.; Kerosuo, L.; Streichan, S.J.; Cai, L.; Bronner, M.E. Identification of a neural crest stem cell niche by Spatial Genomic Analysis. Nat. Commun. 2017, 8, 1830. [Google Scholar] [CrossRef] [PubMed]
- Kaltschmidt, B.; Kaltschmidt, C.; Widera, D. Adult craniofacial stem cells: Sources and relation to the neural crest. Stem Cell Rev. 2012, 8, 658–671. [Google Scholar] [CrossRef]
- Card, D.A.; Hebbar, P.B.; Li, L.; Trotter, K.W.; Komatsu, Y.; Mishina, Y.; Archer, T.K. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol. Cell. Biol. 2008, 28, 6426–6438. [Google Scholar] [CrossRef]
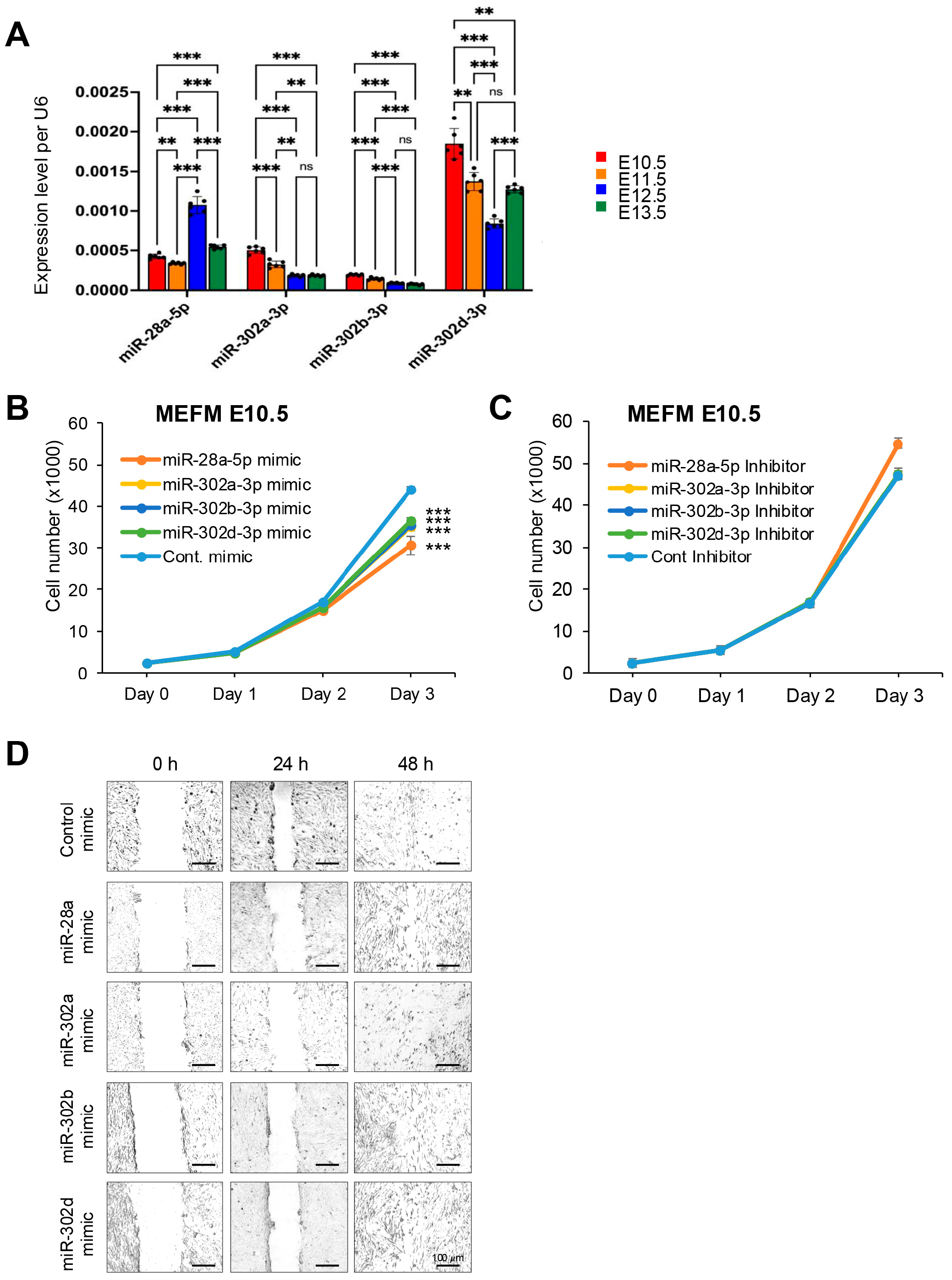
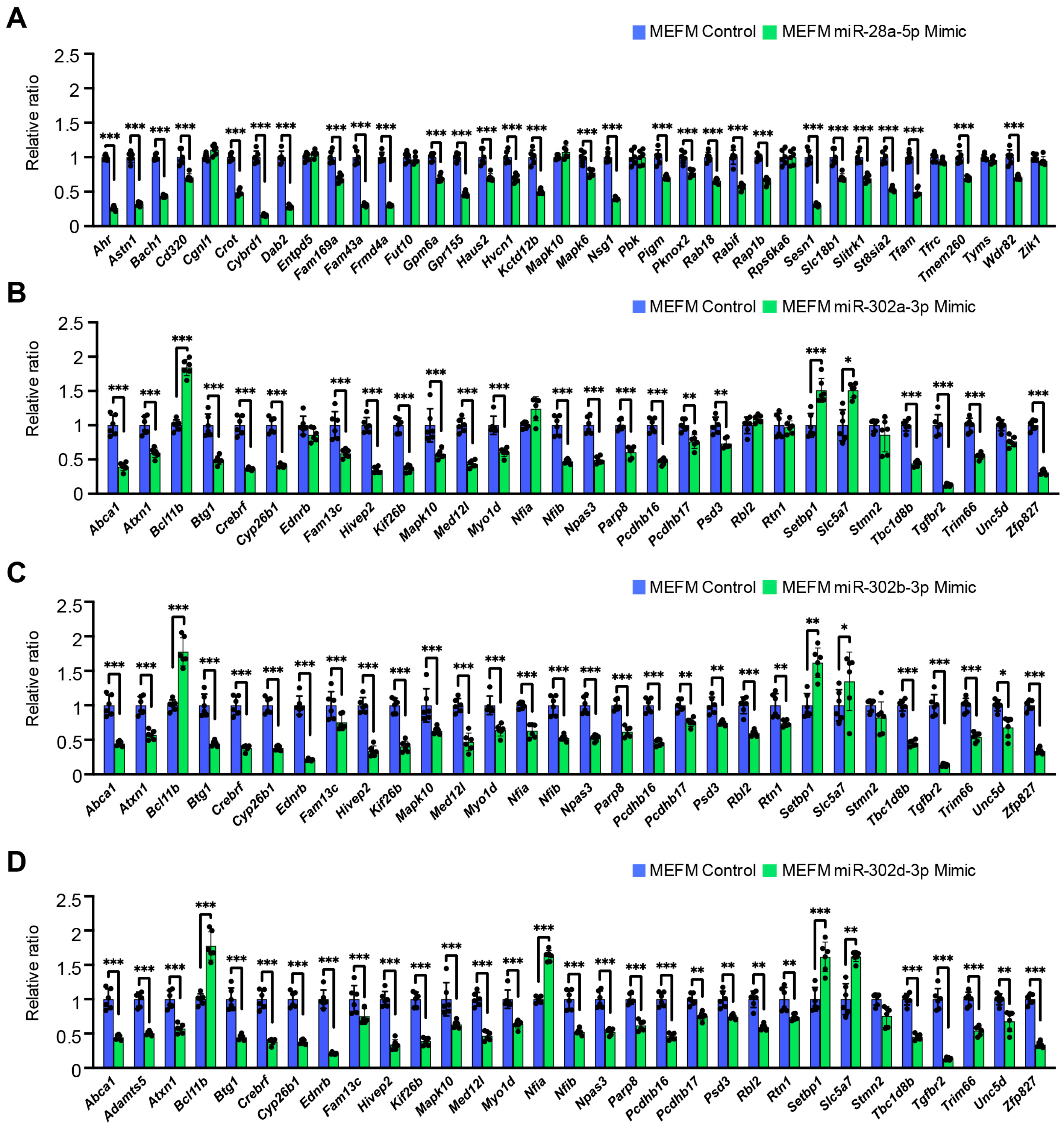
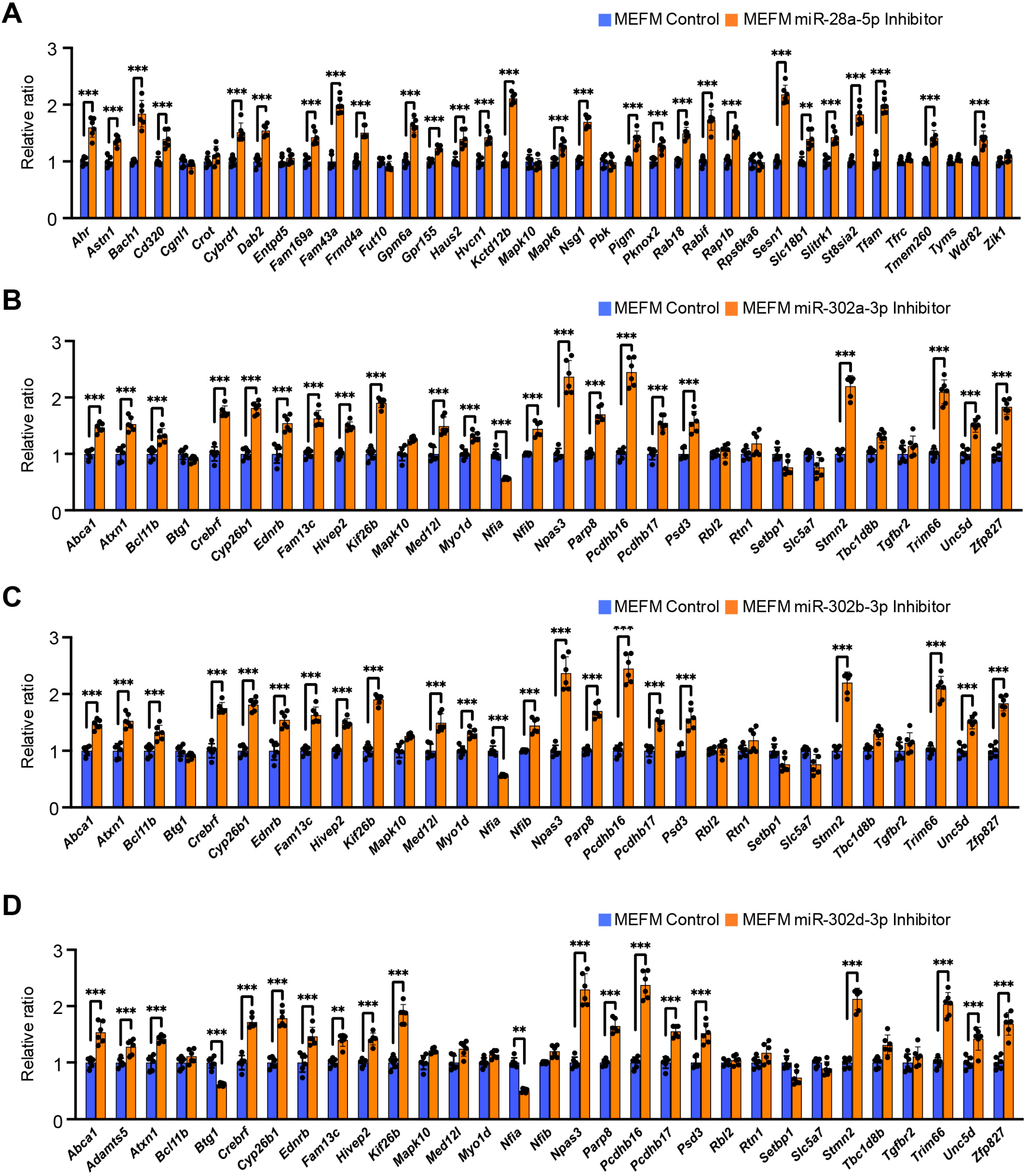
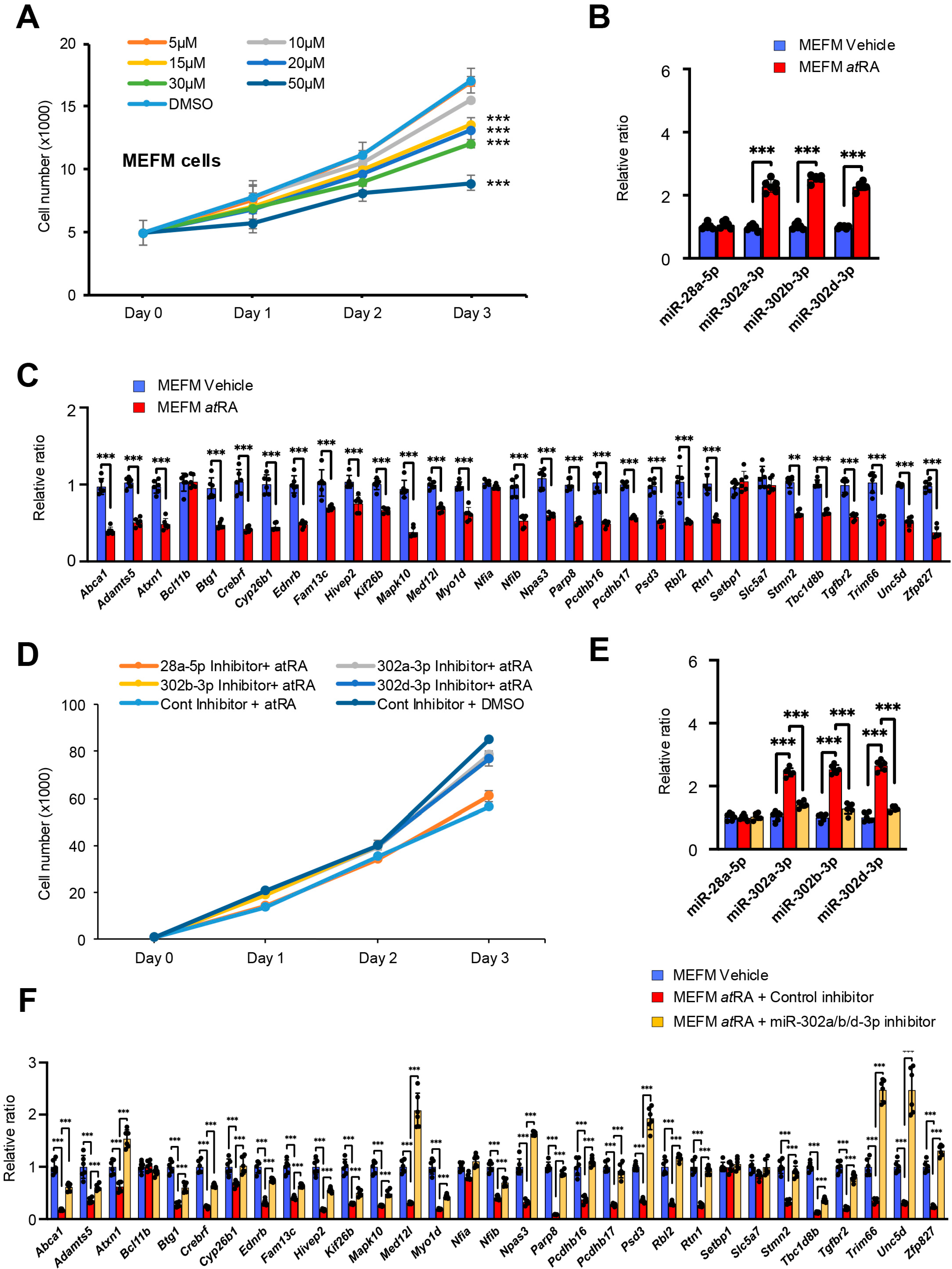
| A. E10.5 vs. E11.5 | ||
| miRNAs | logFC | FDR-Adjusted p-Value |
| mmu-miR-28a-5p | −4.37 | 1.86 × 10−4 |
| mmu-miR-302b-3p | −2.79 | 3.13 × 10−2 |
| mmu-miR-302a-3p | −2.76 | 3.13 × 10−2 |
| mmu-miR-302d-3p | −2.65 | 3.42 × 10−2 |
| mmu-miR-199a-5p | 1.44 | 3.13 × 10−2 |
| mmu-miR-125b-5p | 1.79 | 3.14 × 10−3 |
| mmu-miR-143-3p | 2.14 | 1.86 × 10−4 |
| mmu-miR-143-5p | 2.68 | 3.75 × 10−2 |
| mmu-miR-24-3p | 2.88 | 3.14 × 10−3 |
| B. E11.5 vs. E12.5 | ||
| miRNA | logFC | FDR-adjusted p-value |
| mmu-miR-300-5p | −5.39 | 1.74 × 10−2 |
| mmu-miR-302d-3p | −3.88 | 1.30 × 10−3 |
| mmu-miR-302a-3p | −3.71 | 3.03 × 10−3 |
| mmu-miR-302b-3p | −3.71 | 3.03 × 10−3 |
| mmu-miR-3063-3p | −3.56 | 1.30 × 10−3 |
| mmu-miR-341-3p | −2.37 | 6.18 × 10−4 |
| mmu-miR-20b-5p | −1.99 | 4.59 × 10−2 |
| mmu-miR-335-3p | −1.73 | 4.22 × 10−3 |
| mmu-miR-300-3p | −1.48 | 2.70 × 10−2 |
| mmu-miR-451a | −1.39 | 3.62 × 10−2 |
| mmu-miR-92b-3p | 1.58 | 3.10 × 10−2 |
| mmu-let-7g-5p | 1.71 | 1.01 × 10−2 |
| mmu-let-7i-5p | 1.82 | 1.43 × 10−2 |
| mmu-miR-224-5p | 2.97 | 1.74 × 10−2 |
| mmu-miR-28a-5p | 4.34 | 3.66 × 10−7 |
| C. E12.5 vs. E13.5 | ||
| miRNA | logFC | FDR-adjusted p-value |
| mmu-miR-300-5p | −5.39 | 1.74 × 10−2 |
| mmu-miR-302d-3p | −3.88 | 1.30 × 10−3 |
| mmu-miR-302a-3p | −3.71 | 3.03 × 10−3 |
| mmu-miR-302b-3p | −3.71 | 3.03 × 10−3 |
| mmu-miR-3063-3p | −3.56 | 1.30 × 10−3 |
| mmu-miR-341-3p | −2.37 | 6.18 × 10−4 |
| mmu-miR-20b-5p | −1.99 | 4.59 × 10−2 |
| mmu-miR-335-3p | −1.73 | 4.22 × 10−3 |
| mmu-miR-300-3p | −1.48 | 2.70 × 10−2 |
| mmu-miR-451a | −1.39 | 3.62 × 10−2 |
| mmu-miR-92b-3p | 1.58 | 3.10 × 10−2 |
| mmu-let-7g-5p | 1.71 | 1.01 × 10−2 |
| mmu-let-7i-5p | 1.82 | 1.43 × 10−2 |
| mmu-miR-224-5p | 2.97 | 1.74 × 10−2 |
| mmu-miR-28a-5p | 4.34 | 3.66 × 10−7 |
| Anti-Correlated and Predicted Target Genes | |
|---|---|
| miR-28a-5p E10.5 vs. E11.5 | Ahr, Astn1, Bach1, Cgnl1, Crot, Cybrd1, Dab2, Entpd5, Frmd4a, Fut10, Gpm6a, Gpr155, Kctd12b, Mapk10, Nsg1, Rab18, Rabif, Rap1b, Sesn1, Slc18b1, Slitrk1, St8sia2, Tmem260, Wdr82 |
| miR-28a-5p E11.5 vs. E12.5 | Cd320, Fam169a, Fam43a, Haus2, Hvcn1, Mapk6, Pbk, Pigm, Pknox2, Rps6ka6, Tfam, Tfrc, Tyms, Zik1 |
| miR-302a/b/d-3p E10.5 vs. E11.5 vs. E12.5 *miR-302d-3p only | Abca1, Adamts5*, Atxn1, Bcl11b, Btg1, Crebrf, Cyp26b1, Ednrb, Fam13c, Hivep2, Kif26b, Mapk10, Med12l, Myo1d, Nfia, Nfib, Npas3, Parp8, Pcdhb16, Pcdhb17, Psd3, Rbl2, Rtn1, Setbp1, Slc5a7, Stmn2, Tbc1d8b, Tgfbr2, Trim66, Unc5d, Zfp827 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iwaya, C.; Suzuki, A.; Jun, G.; Iwata, J. miR-302a/b/d-3p Differentially Expressed During Frontonasal Development Is Sensitive to Retinoic Acid Exposure. Cells 2025, 14, 1068. https://doi.org/10.3390/cells14141068
Iwaya C, Suzuki A, Jun G, Iwata J. miR-302a/b/d-3p Differentially Expressed During Frontonasal Development Is Sensitive to Retinoic Acid Exposure. Cells. 2025; 14(14):1068. https://doi.org/10.3390/cells14141068
Chicago/Turabian StyleIwaya, Chihiro, Akiko Suzuki, Goo Jun, and Junichi Iwata. 2025. "miR-302a/b/d-3p Differentially Expressed During Frontonasal Development Is Sensitive to Retinoic Acid Exposure" Cells 14, no. 14: 1068. https://doi.org/10.3390/cells14141068
APA StyleIwaya, C., Suzuki, A., Jun, G., & Iwata, J. (2025). miR-302a/b/d-3p Differentially Expressed During Frontonasal Development Is Sensitive to Retinoic Acid Exposure. Cells, 14(14), 1068. https://doi.org/10.3390/cells14141068








