9-cis-Retinoic Acid Improves Disease Modelling in iPSC-Derived Liver Organoids
Abstract
1. Introduction
2. Materials and Methods
2.1. iPSC Culture Maintenance
2.2. Differentiation of iPSCs Towards Hepatic Stellate Cells (iHSCs)
2.3. Differentiation of iPSCs Towards Hepatocytes (iHepatocytes)
2.4. iPSC-Derived Liver Organoids
2.5. Isolation of pHSCs and Hepatocytes
2.6. pHSC/HepaRG Spheroids
2.7. Organoid Exposure to TGFβ and APAP
2.8. Cell Viability Assay
2.9. RNA Extraction and RT-qPCR
2.10. Immunofluorescence Stainings
2.11. Picrosirius Staining
2.12. Flow Cytometry Analysis
2.13. CYP3A4 Activity Assay
2.14. RNA Sequencing and Analysis
2.15. Statistical Analysis
3. Results
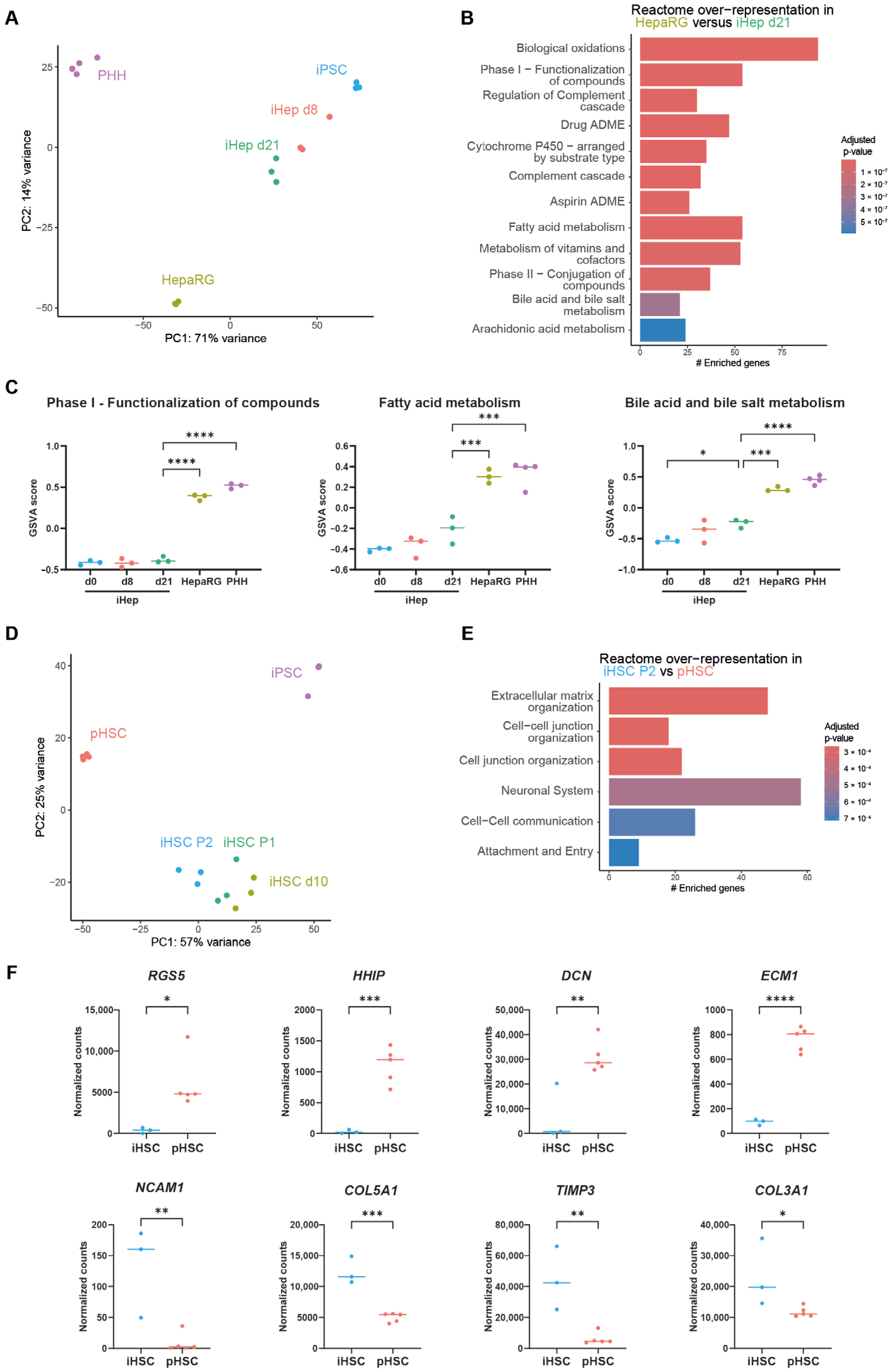
3.1. iPSC-Derived Liver Organoids Are More Fibrotic than HepaRG/pHSC Organoids
3.2. Immaturity of iPSC-Derived Liver Cells
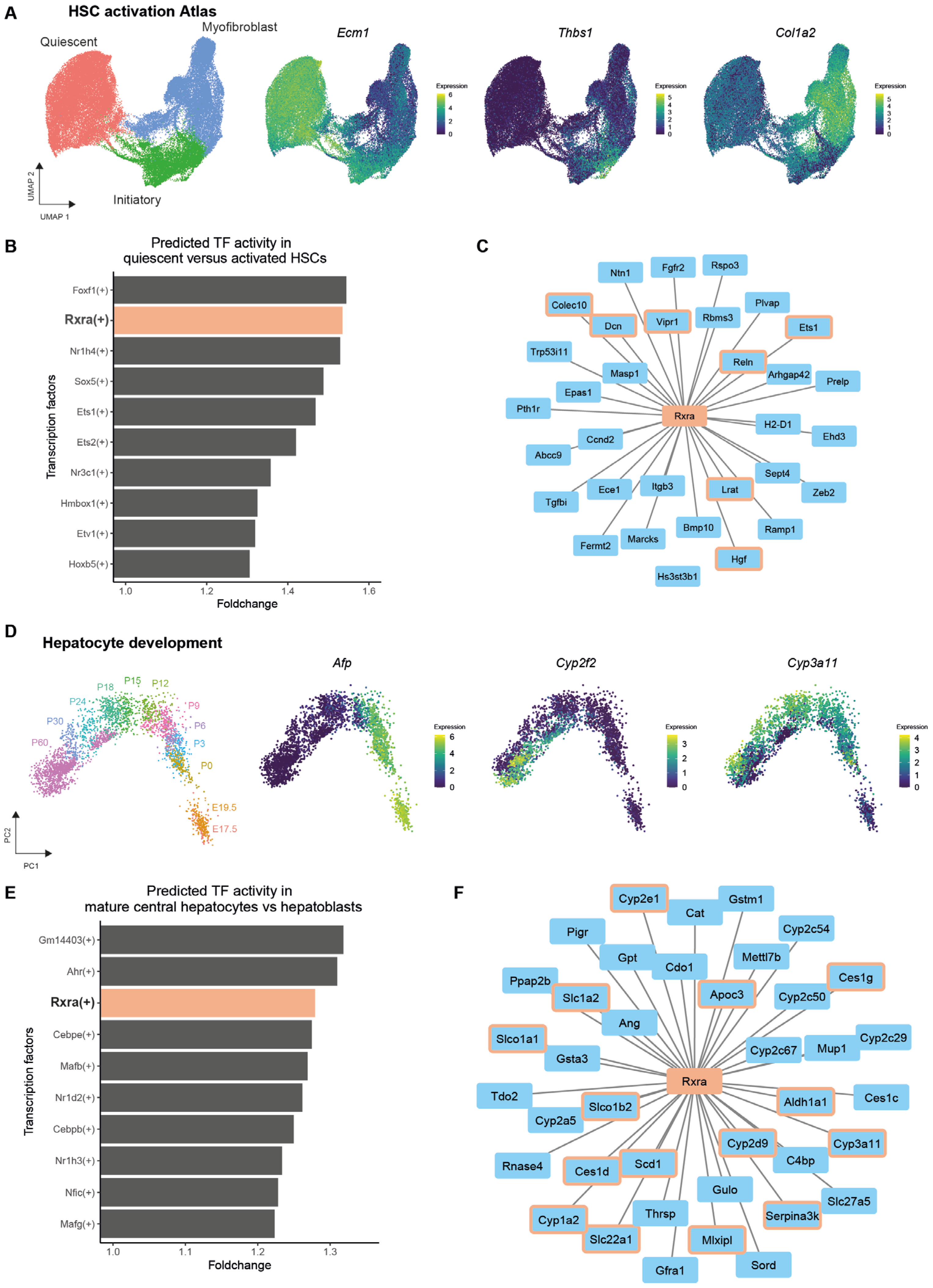
3.3. RXRA as a Target for Improving iPSC-Derived Liver Cells
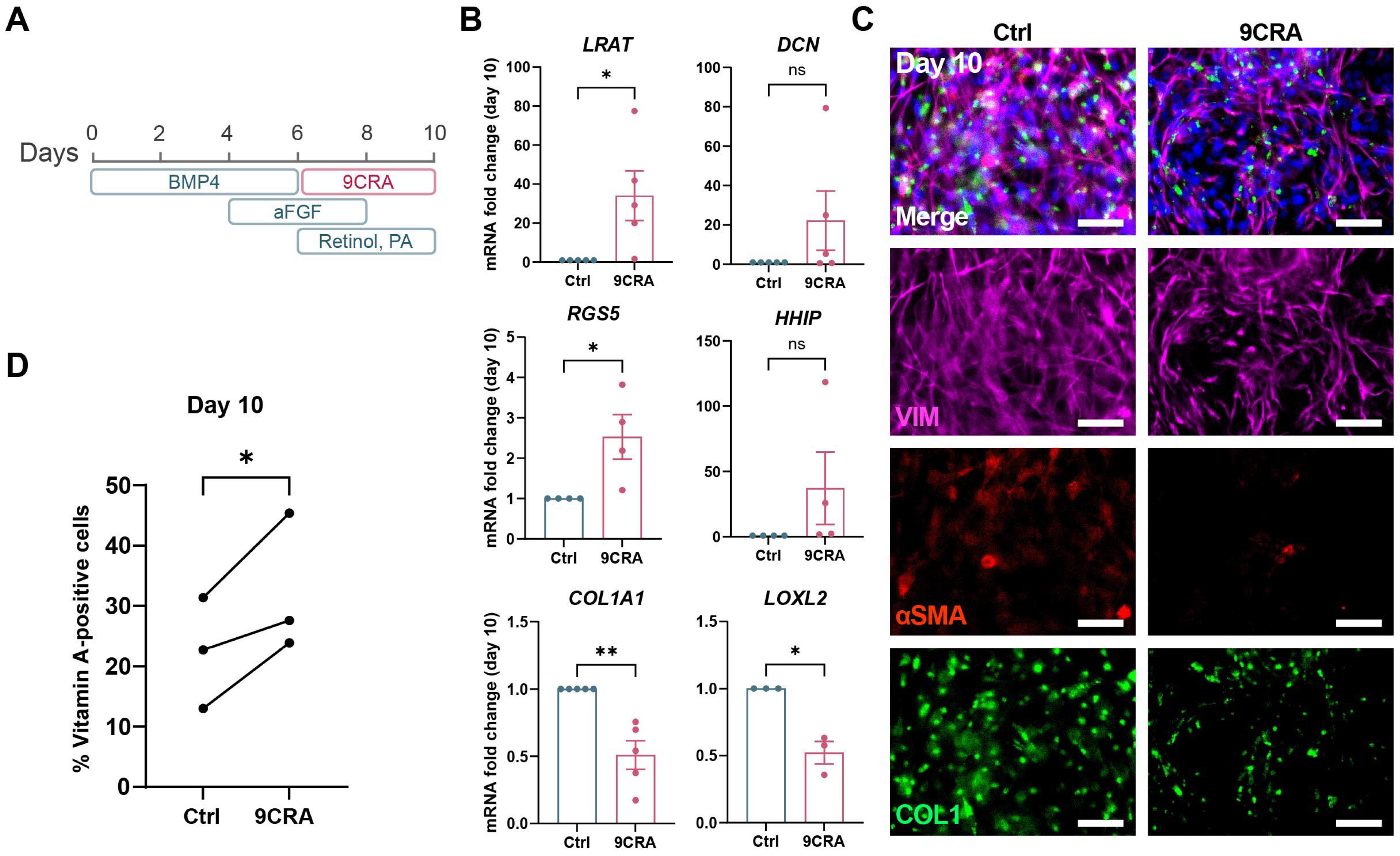
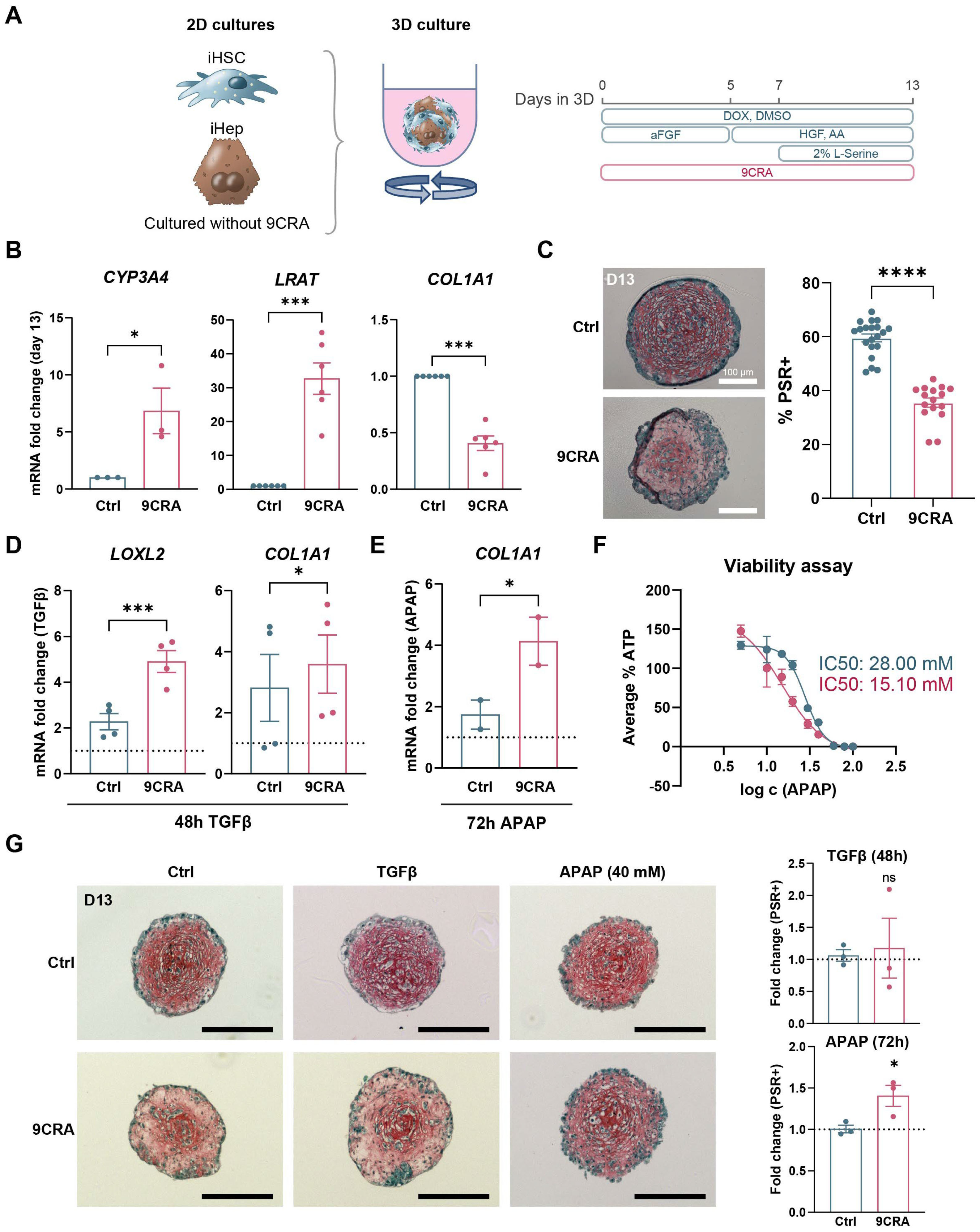
3.4. 9-cis-Retinoic Acid Treatment Improves HSC Differentiation from iPSCs
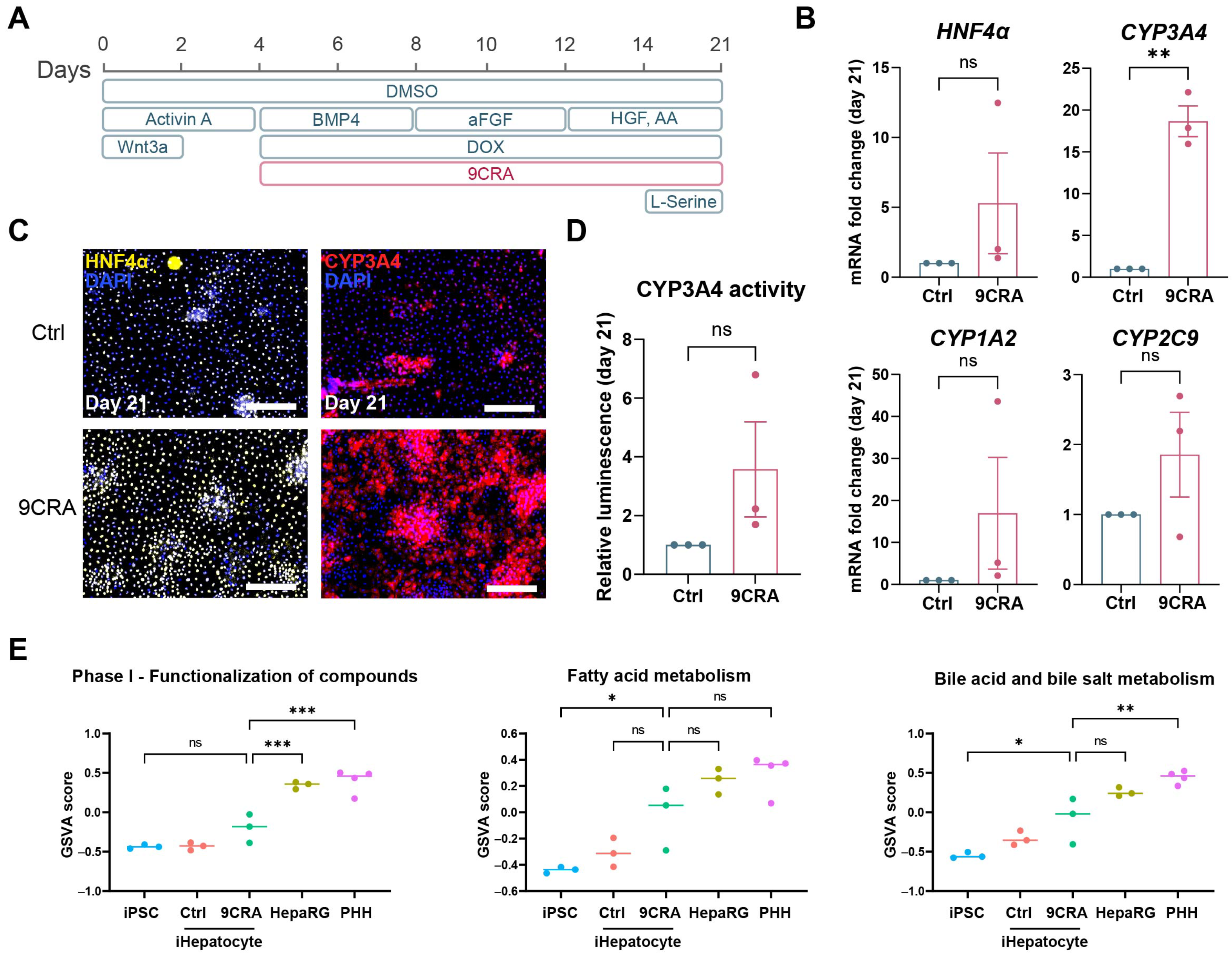
3.5. 9-cis-Retinoic Acid Makes iHepatocytes More Mature
3.6. 9-cis-Retinoic Acid Improves iPSC-Derived Liver Fibrosis Modelling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HSC | Hepatic stellate cell |
| iPSC | Induced pluripotent stem cell |
| iHep | Induced pluripotent stem cell-derived hepatoblast |
| pHSC | Primary human hepatic stellate cell |
| iHSC | Induced pluripotent stem cell-derived hepatic stellate cell |
| iHepatocyte | Induced pluripotent stem cell-derived hepatocyte |
| 9CRA | 9-cis-retinoic acid |
| LDM | Liver differentiation medium |
| DMSO | Dimethyl sulfoxide |
| DOX | Doxycycline hyclate |
| TGFβ | Transforming growth factor beta |
| APAP | Acetaminophen |
| BSA | Bovine serum albumin |
| FBS | Foetal bovine serum |
| GSVA | Gene set variation analysis |
| RXRA | Retinoid X receptor alpha |
References
- Asrani, S.K.; Devarbhavi, H.; Eaton, J.; Kamath, P.S. Burden of liver diseases in the world. J. Hepatol. 2019, 70, 151–171. [Google Scholar] [CrossRef]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Van Grunsven, L.A. 3D in vitro models of liver fibrosis. Adv. Drug Deliv. Rev. 2017, 121, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Boon, R.; Kumar, M.; Tricot, T.; Elia, I.; Ordovas, L.; Jacobs, F.; One, J.; De Smedt, J.; Eelen, G.; Bird, M.; et al. Amino acid levels determine metabolism and CYP450 function of hepatocytes and hepatoma cell lines. Nat. Commun. 2020, 11, 1393. [Google Scholar] [CrossRef] [PubMed]
- Coll, M.; Perea, L.; Boon, R.; Leite, S.B.; Vallverdu, J.; Mannaerts, I.; Smout, A.; El Taghdouini, A.; Blaya, D.; Rodrigo-Torres, D.; et al. Generation of Hepatic Stellate Cells from Human Pluripotent Stem Cells Enables In Vitro Modeling of Liver Fibrosis. Cell Stem Cell 2018, 23, 101–113.E7. [Google Scholar] [CrossRef]
- Wu, X.; Jiang, D.; Yang, Y.; Li, S.; Ding, Q. Modeling drug-induced liver injury and screening for anti-hepatofibrotic compounds using human PSC-derived organoids. Cell Regen. 2023, 12, 6. [Google Scholar] [CrossRef]
- Cools, L.; Dastjerd, M.K.; Smout, A.; Merens, V.; Yang, Y.; Reynaert, H.; Messaoudi, N.; Smet, V.; Kumar, M.; Verhulst, S.; et al. Human iPSC-derived liver co-culture spheroids to model liver fibrosis. Biofabrication 2024, 16, 3. [Google Scholar] [CrossRef]
- Ouchi, R.; Togo, S.; Kimura, M.; Shinozawa, T.; Koido, M.; Koike, H.; Thompson, W.; Karns, R.A.; Mayhew, C.N.; McGrath, P.S.; et al. Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metab. 2019, 30, 374–384.E6. [Google Scholar] [CrossRef]
- Leite, S.B.; Roosens, T.; El Taghdouini, A.; Mannaerts, I.; Smout, A.J.; Najimi, M.; Sokal, E.; Noor, F.; Chesne, C.; van Grunsven, L.A. Novel human hepatic organoid model enables testing of drug-induced liver fibrosis in vitro. Biomaterials 2016, 78, 1–10. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner; Lawrence Berkeley National Laboratory, LBNL Report #, LBNL-7065E; LBL Publications: Berkeley, CA, USA, 2014. [Google Scholar]
- Pouzat, C.; Chaffiol, A. Automatic spike train analysis and report generation. An implementation with R, R2HTML and STAR. J. Neurosci. Methods 2009, 181, 119–144. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef]
- Merens, V.; Knetemann, E.; Gurbuz, E.; De Smet, V.; Messaoudi, N.; Reynaert, H.; Verhulst, S.; van Grunsven, L.A. Hepatic stellate cell single cell atlas reveals a highly similar activation process across liver disease aetiologies. JHEP Rep. 2025, 7, 101223. [Google Scholar] [CrossRef]
- Yang, L.; Wang, X.; Zheng, J.X.; Xu, Z.R.; Li, L.C.; Xiong, Y.L.; Zhou, B.C.; Gao, J.; Xu, C.R. Determination of key events in mouse hepatocyte maturation at the single-cell level. Dev. Cell 2023, 58, 1996–2010.E6. [Google Scholar] [CrossRef]
- Van de Sande, B.; Flerin, C.; Davie, K.; De Waegeneer, M.; Hulselmans, G.; Aibar, S.; Seurinck, R.; Saelens, W.; Cannoodt, R.; Rouchon, Q.; et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 2020, 15, 2247–2276. [Google Scholar] [CrossRef]
- Sison-Young, R.L.; Lauschke, V.M.; Johann, E.; Alexandre, E.; Antherieu, S.; Aerts, H.; Gerets, H.H.J.; Labbe, G.; Hoet, D.; Dorau, M.; et al. A multicenter assessment of single-cell models aligned to standard measures of cell health for prediction of acute hepatotoxicity. Arch. Toxicol. 2017, 91, 1385–1400. [Google Scholar] [CrossRef]
- McGill, M.R.; Yan, H.M.; Ramachandran, A.; Murray, G.J.; Rollins, D.E.; Jaeschke, H. HepaRG cells: A human model to study mechanisms of acetaminophen hepatotoxicity. Hepatology 2011, 53, 974–982. [Google Scholar] [CrossRef]
- De Smet, V.; Gurbuz, E.; Eysackers, N.; Dewyse, L.; Smout, A.; Kazemzadeh Dastjerd, M.; Lefesvre, P.; Messaoudi, N.; Reynaert, H.; Verhulst, S.; et al. Orphan receptor GPR176 in hepatic stellate cells exerts a profibrotic role in chronic liver disease. JHEP Rep. 2024, 6, 101036. [Google Scholar] [CrossRef] [PubMed]
- Maepa, S.W.; Ndlovu, H. Advances in generating liver cells from pluripotent stem cells as a tool for modeling liver diseases. Stem Cells 2020, 38, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Schrooders, Y.; Hauser, D.; van Herwijnen, M.; Albrecht, W.; Ter Braak, B.; Brecklinghaus, T.; Castell, J.V.; Elenschneider, L.; Escher, S.; et al. Comparing in vitro human liver models to in vivo human liver using RNA-Seq. Arch. Toxicol. 2021, 95, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Liu, T.; Chen, W.; Hammad, S.; Longerich, T.; Hausser, I.; Fu, Y.; Li, N.; He, Y.; Liu, C.; et al. ECM1 Prevents Activation of Transforming Growth Factor beta, Hepatic Stellate Cells, and Fibrogenesis in Mice. Gastroenterology 2019, 157, 1352–1367.e13. [Google Scholar] [CrossRef]
- Lian, N.; Jiang, Y.; Zhang, F.; Jin, H.; Lu, C.; Wu, X.; Lu, Y.; Zheng, S. Curcumin regulates cell fate and metabolism by inhibiting hedgehog signaling in hepatic stellate cells. Lab. Investig. 2015, 95, 790–803. [Google Scholar] [CrossRef]
- Baghy, K.; Iozzo, R.V.; Kovalszky, I. Decorin-TGFbeta axis in hepatic fibrosis and cirrhosis. J. Histochem. Cytochem. 2012, 60, 262–268. [Google Scholar] [CrossRef]
- Bahrami, A.J.; Gunaje, J.J.; Hayes, B.J.; Riehle, K.J.; Kenerson, H.L.; Yeung, R.S.; Stempien-Otero, A.S.; Campbell, J.S.; Mahoney, W.M., Jr. Regulator of G-protein signaling-5 is a marker of hepatic stellate cells and expression mediates response to liver injury. PLoS ONE 2014, 9, e108505. [Google Scholar] [CrossRef]
- Haaker, M.W.; Vaandrager, A.B.; Helms, J.B. Retinoids in health and disease: A role for hepatic stellate cells in affecting retinoid levels. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158674. [Google Scholar] [CrossRef]
- Kotulkar, M.; Robarts, D.R.; Apte, U. HNF4alpha in Hepatocyte Health and Disease. Semin. Liver Dis. 2023, 43, 234–244. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Graffmann, N.; Scherer, B.; Adjaye, J. In vitro differentiation of pluripotent stem cells into hepatocyte like cells—Basic principles and current progress. Stem Cell Res. 2022, 61, 102763. [Google Scholar] [CrossRef] [PubMed]
- Tate, B.F.; Levin, A.A.; Grippo, J.F. The discovery of 9-cis retinoic acid: A hormone that binds the retinoid-X receptor. Trends Endocrinol. Metab. 1994, 5, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Fadel, L.; Reho, B.; Volko, J.; Bojcsuk, D.; Kolostyak, Z.; Nagy, G.; Muller, G.; Simandi, Z.; Hegedus, E.; Szabo, G.; et al. Agonist binding directs dynamic competition among nuclear receptors for heterodimerization with retinoid X receptor. J. Biol. Chem. 2020, 295, 10045–10061. [Google Scholar] [CrossRef]
- Nagy, L.; Schwabe, J.W. Mechanism of the nuclear receptor molecular switch. Trends Biochem. Sci. 2004, 29, 317–324. [Google Scholar] [CrossRef]
- Gantier, M.; Rispal, R.; Fourrier, A.; Menoret, S.; Delbos, F.; Anegon, I.; Nguyen, T.H. Cryopreserved cGMP-compliant human pluripotent stem cell-derived hepatic progenitors rescue mice from acute liver failure through rapid paracrine effects on liver cells. Stem Cell Res. Ther. 2024, 15, 71. [Google Scholar] [CrossRef]
- Fourrier, A.; Delbos, F.; Menoret, S.; Collet, C.; Thi Thuy, L.T.; Myara, A.; Petit, F.; Tolosa, L.; Laplanche, S.; Gomez-Lechon, M.J.; et al. Regenerative cell therapy for the treatment of hyperbilirubinemic Gunn rats with fresh and frozen human induced pluripotent stem cells-derived hepatic stem cells. Xenotransplantation 2020, 27, e12544. [Google Scholar] [CrossRef]
- Shapiro, A.M.J.; Thompson, D.; Donner, T.W.; Bellin, M.D.; Hsueh, W.; Pettus, J.; Wilensky, J.; Daniels, M.; Wang, R.M.; Brandon, E.P.; et al. Insulin expression and C-peptide in type 1 diabetes subjects implanted with stem cell-derived pancreatic endoderm cells in an encapsulation device. Cell Rep. Med. 2021, 2, 100466. [Google Scholar] [CrossRef]
- Heyman, R.A.; Mangelsdorf, D.J.; Dyck, J.A.; Stein, R.B.; Eichele, G.; Evans, R.M.; Thaller, C. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell 1992, 68, 397–406. [Google Scholar] [CrossRef]
- Kane, M.A. Analysis, occurrence, and function of 9-cis-retinoic acid. Biochim. Biophys. Acta 2012, 1821, 10–20. [Google Scholar] [CrossRef]
- Yoo, H.S.; Moss, K.O.; Cockrum, M.A.; Woo, W.; Napoli, J.L. Energy status regulates levels of the RAR/RXR ligand 9-cis-retinoic acid in mammalian tissues: Glucose reduces its synthesis in beta-cells. J. Biol. Chem. 2023, 299, 105255. [Google Scholar] [CrossRef]
- Govaere, O.; Cockell, S.; Tiniakos, D.; Queen, R.; Younes, R.; Vacca, M.; Alexander, L.; Ravaioli, F.; Palmer, J.; Petta, S.; et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci. Transl. Med. 2020, 12, eaba4448. [Google Scholar] [CrossRef] [PubMed]
- Philpott, J.; Kazimierczyk, S.; Korgaonkar, P.; Bordt, E.; Zois, J.; Vasudevan, C.; Meng, D.; Bhatia, I.; Lu, N.; Jimena, B.; et al. RXRalpha Regulates the Development of Resident Tissue Macrophages. Immunohorizons 2022, 6, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Den Braver-Sewradj, S.P.; den Braver, M.W.; Vermeulen, N.P.; Commandeur, J.N.; Richert, L.; Vos, J.C. Inter-donor variability of phase I/phase II metabolism of three reference drugs in cryopreserved primary human hepatocytes in suspension and monolayer. Toxicol. In Vitro 2016, 33, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Le, T.V.; Diep, T.T.N.; Nhan, V.T.; Uyen, N.L.T.; Thanh, D.M.; Truong, N.H. A simple and effective scaffold for mouse hepatic stellate cell primary culture. Am. J. Physiol. Cell Physiol. 2023, 324, C1213–C1222. [Google Scholar] [CrossRef]
- Li, H.; Zhang, J.; Huang, G.; Zhang, N.; Chen, Q.; Zhang, X. Effect of retinoid kappa receptor alpha (RXRalpha) transfection on the proliferation and phenotype of rat hepatic stellate cells in vitro. Chin. Med. J. 2002, 115, 928–932. [Google Scholar]
- Sharvit, E.; Abramovitch, S.; Reif, S.; Bruck, R. Amplified inhibition of stellate cell activation pathways by PPAR-gamma, RAR and RXR agonists. PLoS ONE 2013, 8, e76541. [Google Scholar] [CrossRef]
- Panebianco, C.; Oben, J.A.; Vinciguerra, M.; Pazienza, V. Senescence in hepatic stellate cells as a mechanism of liver fibrosis reversal: A putative synergy between retinoic acid and PPAR-gamma signalings. Clin. Exp. Med. 2017, 17, 269–280. [Google Scholar] [CrossRef]
- Guan, Y.; Enejder, A.; Wang, M.; Fang, Z.; Cui, L.; Chen, S.Y.; Wang, J.; Tan, Y.; Wu, M.; Chen, X.; et al. A human multi-lineage hepatic organoid model for liver fibrosis. Nat. Commun. 2021, 12, 6138. [Google Scholar] [CrossRef]
- Tsang, H.Y.; Yi Lo, P.H.; Ho Lee, K.K. Generation of Liver Organoids from Human Induced Pluripotent Stem Cells as Liver Fibrosis and Steatosis Models. BioRxiv. 2021. Available online: https://www.researchgate.net/publication/352869786_Generation_of_liver_organoids_from_human_induced_pluripotent_stem_cells_as_liver_fibrosis_and_steatosis_models (accessed on 22 June 2025).
- Buechler, M.B.; Pradhan, R.N.; Krishnamurty, A.T.; Cox, C.; Calviello, A.K.; Wang, A.W.; Yang, Y.A.; Tam, L.; Caothien, R.; Roose-Girma, M.; et al. Cross-tissue organization of the fibroblast lineage. Nature 2021, 593, 575–579. [Google Scholar] [CrossRef]

| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HNF4α (1/200) | Santa Cruz Biotechnology (Dallas, TX, USA) | Cat# Sc-8987 |
| CYP3A4 (1/200) | Cypex limited UK (Dundee, UK) | Cat# MA5-17064 |
| αSMA-Cy3 (1/100) | Merck (Darmstadt, Germany) | Cat# C6198 |
| VIM (1/100) | Dako (Glostrup, Denmark) | Cat# M0725 |
| COL1 (1/100) | SouthernBiotech (Birmingham, AL, USA) | Cat# 1310-01 |
| PDGFRβ (1/100) | Abcam (Cambridge, UK) | Cat# ab32570 |
| Donkey anti-Rabbit IgG (H + L) Alexa Fluor 647 (1/200) | Invitrogen (Waltham, MA, USA) | Cat# A31573 |
| Goat anti-Mouse IgG (H + L) Alexa Fluor 488 (1/200) | Invitrogen | Cat# A11029 |
| Goat anti-Rabbit IgG (H + L) Alexa Fluor 488 (1/200) | Invitrogen | Cat# A11008 |
| Donkey anti-Mouse IgG (H + L) Alexa Fluor 647 (1/200) | Thermo Fisher Scientific (Waltham, MA, USA) | Cat# A31571 |
| Donkey anti-Goat IgG (H + L) Alexa Fluor 488 (1/200) | Thermo Fisher Scientific | Cat# A11055 |
| Chemicals, peptides, and recombinant proteins | ||
| Biolaminin | Biolamina (Sundbyberg, Sweden) | Cat# LN521-05 |
| Essential 8 flex medium | Thermo Fisher Scientific | Cat# A2858501 |
| DPBS with calcium and magnesium | Thermo Fisher Scientific | Cat# 14040117 |
| StemPro Accutase Cell Dissociation Reagent | Thermo Fisher Scientific | Cat# A11105-01 |
| RevitaCell Supplement 100X | Gibco (Waltham, MA, USA) | Cat# A2644501 |
| mTeSR1 complete medium | Stemcell Technologies (Vancouver, BC, Canada) | Cat# 85850 |
| BMP | PeproTech (Cranbury, NJ, USA) | Cat# 120-05 |
| aFGF | PeproTech | Cat# 100-17A |
| Retinol | Merck Group | Cat# R7632 |
| Palmitic acid | Sigma-Aldrich (St. Louis, MO, USA) | Cat# P5585 |
| 9-cis-retinoic acid | Sigma-Aldrich | Cat# R4643 |
| Dimethyl sulfoxide | Merck Group | Cat# D2650 |
| Trypsin-EDTA 0.05% | Invitrogen | Cat# 25300-054 |
| Dulbecco’s modified medium | Capricorn (Ebsdorfergrund, Germany) | Cat# CA DMEM-HPA |
| HEPES Buffer solution | Capricorn | Cat# CA HEP-B |
| Penicillin-Streptomycin | Gibco | Cat# 15140122 |
| Foetal Bovine Serum | TICO Europe (Amstelveen, Netherlands) | Cat# FBSEU500 |
| Doxycycline hyclate | Merck Group | Cat# D9891 |
| Activin A | PeproTech | Cat# AF-120-14E |
| Wnt3a | PeproTech | Cat# 315-20 |
| HGF | PeproTech | Cat# 100-39 |
| Essential amino acids | Thermo Fisher Scientific | Cat# 11130036 |
| Non-essential amino acids | Thermo Fisher Scientific | Cat# 11140035 |
| L-Serine | Thermo Fisher Scientific | Cat# A11179.22 |
| Transforming growth factor beta | Peprotech | Cat# 100-21C |
| Acetaminophen | Merck Group | Cat# A7085 |
| IGL-1 organ preservation solution | Institut Georges Lopez (Lissieu, France) | Cat# IGU.IGL-1 REV07 |
| Pronase E | Merck | Cat# 1074330005 |
| Collagenase P | Roche (Basel, Switzerland) | Cat# 11 213873001 |
| Nycodenz gradient | Axis Shield Diagnostics (Dundee, UK) | Cat# 1002424 |
| DNase I | Roche | Cat# 10104159001 |
| Percoll gradient | GE Healthcare (Chicago, IL, USA) | Cat# GE17-0891-02 |
| Williams E medium | Gibco | Cat# A12176-01 |
| Glutamax supplement (200mM) | Thermo Fisher Scientific | Cat# 35050038 |
| EGF | PeproTech | Cat# 315-09 |
| Formaldehyde solution | Merck Group | Cat# 1004965000 |
| Triton-X100 | Merck Group | Cat# T8787 |
| Bovine serum albumin | Sigma-Aldrich | Cat# A2153 |
| Fluorescence Mounting Medium | Dako | Cat# S302380-2 |
| DAPI dilactate | Merck Group | Cat# D9564 |
| Xylene | VWR International (Radnor, PA, USA) | Cat# 28973.363 |
| 2-Propanol | Merck Group | Cat# 59300 |
| Methanol | VWR International | Cat# 20903.368 |
| Sirius Red | Sigma-Aldrich | Cat# 365548 |
| Fast Green FCF | Merck Group | Cat# F7258 |
| Picric acid solution | VWR International | Cat# 84512.260 |
| DPX mounting medium for histology | Merck Group | Cat# 06522 |
| Fixable viability stain 780 | BD Biosciences (Franklin Lakes, NJ, USA) | Cat# 565388 |
| Critical commercial assays | ||
| CellTiter-Glo Luminescent Cell Viability assay | Promega (Madison, WI, USA) | Cat# G7571 |
| ReliaPrep RNA Cell Miniprep System | Promega | Cat# Z6012 |
| M-MLV Reverse Transcriptase, RNase H Minus and Point Mutant | Promega | Cat# M5301 |
| GoTaq qPCR Master Mix with BRYT Green Dye | Promega | Cat# A6002 |
| P450-Glo CYP3A4 activity assay | Promega | Cat# V9001 |
| QuantSeq 3’ mRNA-Seq V2 Library Prep Kit FWD with UDI 12 nt set B1 | Lexogen (Vienna, Austria) | Cat# 192.96 |
| Deposited data | ||
| All RNAseq SRA files are available at NCBI under Password for reviewers onolugasdncbnyn This includes Normalized_counts and Raw_counts files | GEO ID: GSE292959 | |
| All other data files are deposited at Zenodo | 10.5281/zenodo.15095698 | |
| Experimental models: cell lines | ||
| iPSCs | Sigma-Aldrich | IPSC0028-1VL |
| HepaRG | Biopredic (Saint-Grégoire, France) | Cat# HPR116 |
| Oligonucleotides | ||
| DCN forward (CCAATATCACCAGCATTCCTC) | Integrated DNA Technologies (IDT, Coralville, IA, USA) | NA |
| DCN reverse (CTGCTGATTTTGTTGCCATC) | IDT | NA |
| LRAT forward (GCTGGGCTTTACCCCCTA) | IDT | NA |
| LRAT reverse (CGAATAATTATCTTCACAGTCTCACAA) | IDT | NA |
| COL3A1 forward (GGAGCTGGCTACTTCTCGC) | IDT | NA |
| COL3A1 reverse (GGGAACATCCTCCTTCAACAG) | IDT | NA |
| COL1A1 forward (CCGGCTCCTGCTCCTCTTAGCG) | IDT | NA |
| COL1A1 reverse (CGTTCTGTACGCAGGTGATTGGTGG) | IDT | NA |
| LOXL2 forward (GGAGAGGACATACAATACCAAAGTG) | IDT | NA |
| LOXL2 reverse (CCATGGAGAATGGCCAGTAG) | IDT | NA |
| ALB forward (TGGCACAATGAAGTGGGTAA) | IDT | NA |
| ALB reverse (CTGAGCAAAGGCAATCAACA) | IDT | NA |
| CYP3A4 forward (TTCCTCCCTGAAAGATTCAGC) | IDT | NA |
| CY3A4 reverse (GTTGAAGAAGTCCTCCTAAGCT) | IDT | NA |
| CYP1A2 forward (CTGGGCACTTCGACCCTTAC) | IDT | NA |
| CYP1A2 reverse (TCTCATCGCTACTCTCAGGGA) | IDT | NA |
| CYP2C9 forward (CAGTCCCTGCAGCTCTCTTT) | IDT | NA |
| CYP2C9 reverse (TGCACAGTGAAACATAGGAAACTC) | IDT | NA |
| HNF4α forward (CACGGGCAAACACTACGGT) | IDT | NA |
| HNF4α reverse (TTGACCTTCGAGTGCTGATCC) | IDT | NA |
| GAPDH forward (AGCCACATCGCTCAGACAC) | IDT | NA |
| GAPDH reverse (GCCCAATACGACCAAATCC) | IDT | NA |
| RPL19 forward (ATTGGTCTCATTGGGGTCTAAC) | IDT | NA |
| RPL19 reverse (AGTATGCTCAGGCTTCAGAAGA) | IDT | NA |
| RGS5 forward (TTGCATGTGCCAGAAAGCAG) | IDT | NA |
| RGS5 reverse (TACCCCTTGGGTTTCGATGC) | IDT | NA |
| HHIP forward (CCGAGGCCATATTCCAGGTTT) | IDT | NA |
| HHIP reverse (TGGAAAGCACAACCCACCAT) | IDT | NA |
| Software and algorithms | ||
| Graphpad | 10.3.1 | https://www.graphpad.com/ |
| Flowjo | 10 | https://www.flowjo.com/ |
| Qupath | 0.4.4 | https://qupath.github.io/ |
| R | 4.3.1 | www.r-project.org |
| DESeq2 | 1.40.2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| clusterProfiler | 4.8.3 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| GSVA | 1.48.3 | https://bioconductor.org/packages/release/bioc/html/GSVA.html |
| Seurat | 4.1.0 | https://satijalab.org/seurat/ |
| Python | 3.7.7 | https://www.python.org/ |
| pySCENIC | 0.11.0 | https://pyscenic.readthedocs.io/en/latest/ |
| Other | ||
| 96-well ultra-low attachment U-bottom plates | Greiner Bio-One (Kremsmünster, Austria) | Cat# 650970 |
| ROTILABO round sieve, 75, 0.5 mm | Roth (Karlsruhe, Germany) | Cat# A624.1 |
| Glomax | Promega | Cat# GM3000 |
| Nanodrop 2000 Spectrophotometer | Thermo Fisher Scientific | Cat# G870 |
| QuantStudio 3 Real-Time PCR System | Applied Biosystems (Foster City, CA, USA) | Cat# 272310385 |
| EVOS | Thermo Fisher Scientific | Cat# AMF7000 |
| 40 µm cell strainer | Fisher Scientific (Geel, Belgium) | Cat# 11587522 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazemzadeh Dastjerd, M.; Merens, V.; Smout, A.; De Wolf, R.; Chesné, C.; Verfaillie, C.; Verhulst, S.; van Grunsven, L.A. 9-cis-Retinoic Acid Improves Disease Modelling in iPSC-Derived Liver Organoids. Cells 2025, 14, 983. https://doi.org/10.3390/cells14130983
Kazemzadeh Dastjerd M, Merens V, Smout A, De Wolf R, Chesné C, Verfaillie C, Verhulst S, van Grunsven LA. 9-cis-Retinoic Acid Improves Disease Modelling in iPSC-Derived Liver Organoids. Cells. 2025; 14(13):983. https://doi.org/10.3390/cells14130983
Chicago/Turabian StyleKazemzadeh Dastjerd, Mina, Vincent Merens, Ayla Smout, Rebeca De Wolf, Christophe Chesné, Catherine Verfaillie, Stefaan Verhulst, and Leo A. van Grunsven. 2025. "9-cis-Retinoic Acid Improves Disease Modelling in iPSC-Derived Liver Organoids" Cells 14, no. 13: 983. https://doi.org/10.3390/cells14130983
APA StyleKazemzadeh Dastjerd, M., Merens, V., Smout, A., De Wolf, R., Chesné, C., Verfaillie, C., Verhulst, S., & van Grunsven, L. A. (2025). 9-cis-Retinoic Acid Improves Disease Modelling in iPSC-Derived Liver Organoids. Cells, 14(13), 983. https://doi.org/10.3390/cells14130983









