Abstract
Brain organoids are self-organized, three-dimensional (3D) aggregates derived from human embryonic stem cells, induced pluripotent stem cells, or primary organs with cell types and cellular architectures resembling those of the developing human brain. Recent studies have shown the use of region-specific brain organoids for modeling various diseases ranging from neurodevelopmental and neurodegenerative diseases to different brain cancers, which have numerous applications in fundamental research and the development of new drugs, personalized treatment, and regenerative medicine. Consequently, the use of brain organoids in drug discovery is complex and challenging and still an emerging area in this field. This review article summarizes the primary stem cells used in brain organoid generation, region-specific brain organoids, and the functional assays used in their characterization. In addition, we discuss the use of brain organoids in modeling neurodevelopmental and neurodegenerative diseases and pediatric brain cancers, as well as the application of organoids, assembloids, and tumoroids in cancer neuroscience. We further explore the recent advances in using brain organoids in high-throughput screening to improve their use for drug discovery.
1. Introduction
Organoids refer to cells cultivated in a specific three-dimensional (3D) environment in vitro, forming mini clusters that autonomously organize and differentiate into functional cell types, mirroring the structure and function of an organ in vivo [1]. Brain organoids are generated from human pluripotent stem cells (hPSCs) that self-assemble into an organized structure featuring progenitor, neuronal, and glial cells, resembling the human brain [2,3,4]. Additionally, certain brain organoids may display structural characteristics that emulate specific brain regions, known as organoids of a particular region [5]. In contrast to traditional two-dimensional (2D) cell cultures, brain organoids aim to replicate the human brain not just at the cellular level but also regarding overall tissue structure and developmental pathways, creating a unique chance to study human brain development, function, and diseases that are often challenging to investigate through direct experimentation [2]. Throughout the years, brain organoids have been utilized to replicate numerous neurodevelopmental and neurodegenerative disorders, alongside various types of brain tumors, offering promise for the development of new medications. Nevertheless, the application of brain organoids as a model in high-throughput screening (HTS) remains a novel and expanding area of research. Progress in technologies related to hPSC-derived brain organoids for disease modeling will enhance our understanding of these models in HTS, thereby refining the drug discovery and development process that utilizes brain organoids. In this article, we review major stem cells used in the generation of brain organoids and provide an overview of assembloids. We discuss region-specific brain organoids and recent research findings in disease modeling and describe their potential use in drug screening and development.
2. Stem Cells
Stem cells are characterized as cells that can self-renew and differentiate into specialized functional cells [6,7]. Because of their ability to both self-replicate and develop into several types of cells, stem cells are crucial at various stages of development. During the initial stages of embryonic development, pluripotent stem cells serve as progenitors for all tissue types. In contrast, in later developmental stages, tissue-restricted stem cells generate cells with specific functions [8]. The primary features of stem cells include self-renewal (the ability to proliferate extensively), clonality (typically originating from a single cell), and potency (the capacity to develop into various cell types) [9]. Stem cells can be divided into four types based on their origin: embryonic, fetal, adult (which can be resident or tissue-specific), and induced pluripotent stem (iPS) cells. Both embryonic and iPS cells are pluripotent, fetal and perinatal stem cells are generally multipotent, and adult stem cells tend to be either oligopotent or unipotent [9,10]. Pluripotent stem cells can develop into various cell types originating from all three germ layers, the ectoderm, endoderm, and mesoderm, which give rise to all tissues and organs [11]. Multipotent stem cells are present in most tissues and can differentiate into cells from a single germ layer [12], and lastly, oligopotent stem cells can self-renew and create two or more lineages within a specific tissue. In contrast, unipotent stem cells can only transform into one specific cell type and form a single lineage [13,14]. Stem cells have applications in cellular therapies to replace damaged cells or regenerate tissues. Furthermore, research on stem cells has enhanced our comprehension of developmental processes and the origins of diseases [6,8,10]. The most used stem cells for generating brain organoids are embryonic and iPS cells.
2.1. Embryonic Stem Cells
The isolation of human embryonic stem cells (hESCs) in the late 1990s opened new avenues in human developmental biology and regenerative medicine [15]. Embryonic stem cells (ESCs) are obtained from embryos, as indicated by their name. Initially derived from mouse embryos, the 1998 extraction of ESCs from human embryos by James Thomson captured significant attention [16]. ESCs are pluripotent and emerge from the blastocyst’s inner cell mass, a stage of the pre-implantation embryo occurring 5–6 days after fertilization [17]. The blastocyst consists of two layers: the inner cell mass and the outer cell layer, consisting of cells known as trophoblasts. To derive embryonic stem cell (ESC) lines, cells from the inner cell mass are isolated from the trophoblasts and cultured in a controlled environment [18]. The identification of ESCs relies on the presence of specific transcription factors such as Nanog and Oct4 [19,20]. These factors are crucial in keeping the stem cells undifferentiated and enabling self-renewal. ESCs maintained in an undifferentiated condition without genetic abnormalities can be propagated as an ESC line. These cells can be cryopreserved and later thawed for additional cultures and experiments [21]. Proper culture conditions are essential for preserving ESCs in an unperturbed and undifferentiated form [22]. A feeder layer of mouse embryonic fibroblast cells (MEFCs) or a medium containing the anti-differentiation cytokine leukemia inhibitory factor (LIF) can be utilized. The absence of LIF from the medium or the removal of ESCs from the feeder layer can lead to the development of “embryoid bodies”, which contain all three germ layers: the endoderm, mesoderm, and ectoderm [23,24,25,26,27]. Swift advancements in biotechnology have raised several urgent ethical and policy concerns related to the use of embryonic stem cells in stem cell research, primarily due to the use of human embryos. This has resulted in a search for alternatives that can replace the reliance on human embryonic stem cells in stem cell studies [28,29].
2.2. Human Induced Pluripotent Stem Cells
The development of induced pluripotent stem cells (iPSCs) from adult somatic cells was undertaken to address the ethical and immunological concerns related to embryonic stem cells [30]. iPSCs can be characterized as “embryonic stem cell-like” cells produced by reprogramming adult somatic cells by introducing specific pluripotency-related genes. Like embryonic stem cells, iPSCs can extensively proliferate in culture and differentiate into the three primary germ cell layers: endoderm, mesoderm, and ectoderm. This impressive technique of reverting somatic cells to a pluripotent embryonic state while maintaining the same genetic identity is known as somatic cell nuclear transfer (SCNT) [31]. In 2006, Shinya Yamanaka and Kazutoshi Takahashi successfully created mouse iPSCs using a retroviral approach to reprogram mouse fibroblasts by introducing a combination of four transcription factors associated with reprogramming: Oct 3/4 (Octamer-binding transcription factor-3/4), Sox2 (Sex-determining region Y)-box 2, Klf4 (Kruppel Like Factor-4), and c-Myc, collectively referred to as the “OSKM factors” [32]. The following year, in 2007, Yamanaka and his team applied the same reprogramming technique to adult human fibroblasts, resulting in human iPSCs (hiPSCs). Simultaneously, James Thomson’s group also generated hiPSCs using a different delivery method, lentivirus, and an alternative set of four factors: Oct 3/4, Sox2, Nanog, and Lin 28 [33,34]. Significant advancements have been achieved over time to enhance reprogramming efficiency and mitigate the risks associated with this method, such as low reprogramming efficiency, chromosomal instability, and insertional mutagenesis. Innovative strategies employed to enhance reprogramming include the inhibition of obstacles to reprogramming, the use of non-integrative delivery techniques, the overexpression of beneficial genes, and the application of specific molecules that promote reprogramming [35,36,37,38,39,40]. The potential of iPSC technology is substantial for personalized cell-based therapies, human disease modeling, and drug development and testing.
2.3. Neural Progenitor or Stem Cells
Neural progenitor cells (NPCs) are multipotent neural stem cells (NSCs) capable of self-renewal and differentiation into neurons and glial cells [41]. However, NPCs do not give rise to non-neuronal cells present in the central nervous system (CNS), such as immune cells. NPCs are found throughout the CNS at various stages of development, including during embryogenesis, in neonatal brains, and in mature adults, indicating that NPCs are not solely embryonic stem cells [42]. NPCs are identified by their specific brain localization, morphological characteristics, gene expression patterns, developmental timing, and functional roles [43]. Additionally, NPCs can be generated in vitro through the differentiation of ESCs or iPSCs using specific neural induction growth factors [44].
3. Brain Organoids
Brain organoids derived from hESCs or hiPSCs can be categorized as cerebral, also known as whole brain organoids or, by consensus, unguided neural organoids [45]. These are usually generated through unguided or minimally guided techniques that depend on self-organizing development and the inherent differentiation abilities of hPSC aggregates. Alternatively, there are region-specific organoids, or regionalized neural organoids, such as those from the forebrain, midbrain, and hindbrain, which are formed using guided methods that involve adding external patterning factors to encourage hPSCs to differentiate into specific lineages [45,46,47]. This section briefly discusses these several types of brain organoids (Figure 1).
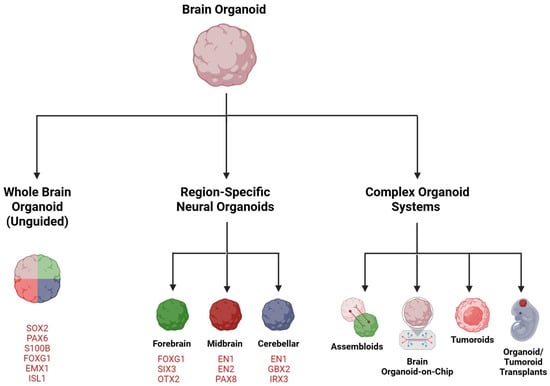
Figure 1.
Brain organoids—an overview. Embryonic or induced human pluripotent stem cells (hPSCs) can be differentiated into self-organizing, unguided neural organoids (whole brain organoids) or, by using established differentiation protocols, into neural organoids resembling specific brain regions (guided). Commonly used organoid-specific markers are indicated. Organoids can be combined with other organoids, additional cell types, or hPSCs of various origin into assembloids to construct complex, functional brain structures. Organoids-on-a-chip use bioengineering and microfluidic technologies to enhance cytoarchitecture and long-term organoid culture. Tumoroids are specialized brain organoids mimicking brain tumors with applications in drug discovery, to understand tumorigenesis, or as assembloids, in cancer neuroscience. Organoids and tumoroids are suitable as transplants in mouse brains. Created in https://BioRender.com.
3.1. Whole Brain or Cerebral Organoid
The groundbreaking achievement of creating cerebral organoids began with discoveries made in 2005 by a research team headed by Yoshiki Sasai [48]. This team was the first to successfully generate 3D neural tissues in vitro that resemble the mouse cerebrum using a cell aggregation suspension culture with mouse embryonic stem cells (mESCs). By 2008, Sasai’s group had refined a differentiation method for cerebral nerve tissues, enabling the 3D differentiation of brain tissues using human ESCs [49]. In the years following the initial reports on neural organoids, the field has witnessed a remarkable increase in their application and ongoing enhancement. Protocols for whole brain organoids depend on the cells’ intrinsic signaling capabilities within the culture, forming various brain regions and CNS structures [4,46,50,51,52]. Although whole brain organoids do not encompass all areas of the brain, they can evolve into multiple neural structures. Since the precise sequence and timing of the necessary signals for specific developmental processes remain unknown, this method allows for observing the patterning of neighboring neural structures, interactions between cells across different brain regions, and characterizing cell morphologies and synapse development. Whole brain organoids, which frequently differ in size and include various cell types with distinct regional identities in a mixed arrangement, have generated interest in utilizing guided differentiation protocols to shape neural structures [53].
3.2. Forebrain Organoid
The forebrain represents the largest section of the brain and includes the cerebral cortex, white matter, and corpus callosum. Cortical organoids that replicate the structure of the cerebral cortex have been more thoroughly studied and are used more frequently than other types of brain organoids. These cortical organoids have garnered significant attention due to the unique nature and evolutionary expansion of the cerebral cortex in humans compared to other species, as it is also commonly affected by numerous neurological disorders [54,55,56]. Over the years, various protocols have been developed to generate cortical organoids from stem cells [3,57,58,59,60,61]. Forebrain organoids have been utilized in modeling diseases, such as neurodevelopmental and neurodegenerative disorders and infections like the Zika virus. This article will cover disease modeling using organoids in a later section.
3.3. Midbrain Organoid
The midbrain is the uppermost section of the brainstem and includes many significant nuclei and white matter pathways, primarily associated with motor regulation and auditory and visual pathways [62]. Midbrain organoids feature spatially organized clusters of dopaminergic neurons (mDA) that exhibit active synapses, with astrocytes and oligodendrocytes. Several midbrain organoid generation systems have been developed and improved over the past decade [63,64,65,66,67,68,69,70,71]. Organoids that model the human midbrain are particularly noteworthy because this area is explicitly affected by neurodegenerative processes in Parkinson’s disease [72], which will be discussed in detail in the later disease-modeling section.
3.4. Cerebellar Organoid
The cerebellum is the largest part of the hindbrain and is home to the highest number of neurons in the human brain. It plays a crucial role in coordinating voluntary movements, motor skills, balance, posture, and speech, as well as contributing to more complex neurological functions like cognition and emotional regulation [73]. This region of the brain may be involved in various human developmental disorders, including autism spectrum disorder, epilepsy, ADHD, and cerebral palsy, and it is recognized as the origin of medulloblastoma, the most prevalent brain tumor in children [74,75,76,77,78,79,80,81,82,83]. Research utilizing brain organoids to investigate cerebellar development has not been extensively pursued. The first comprehensive protocol for creating cerebellar organoids was introduced in 2015 [84], and subsequent studies based on this methodology have been conducted [85,86,87,88,89]. The most recent studies [54,90] succeeded in producing cerebellar organoids that include functional Purkinje cells and enable the prolonged cultivation of all primary cell types found in the cerebellum.
4. Complex Organoid Systems
4.1. Assembloids
Organoids have enhanced our understanding of organ assembly and how disease can disrupt normal physiology. Nevertheless, they frequently fail to represent the interactions between different tissue types and/or lineages that lead to the emergence of tissue properties during development. Assembloids are fusions of organoids from specific brain regions that aim to replicate the interactions between different regions and cells and the development of neural circuitry, by merging various brain areas and/or cell types. Consequently, assembloids can serve as a tool for modeling nuanced functional disruptions that represent intricate neurodevelopmental, neuropsychiatric, and neurodegenerative disorders [45,91,92,93,94]. Multiple organoids can be combined to create “multi-region assembloids”. These organoids may originate from healthy sources, patient-derived cells, genetically modified hiPSCs, hESCs, or primary tissue sources such as isolated adult stem cells, excised tumors that retain growth potential, or fetal tissue. These assembloids can incorporate two, three, or even more components [93]. Assembloids can be used to study rare diseases, and this will be discussed further in a later chapter.
4.2. Brain Organoid-on-a-Chip
Despite the considerable progress made with brain organoids, several limitations remain that impede their acceptance and use by the pharmaceutical industry for modeling neurological disorders and conducting drug tests. One of the key challenges with organoids is the significant variability that can occur, even among those derived from the same stem cells and cultured under identical conditions. Additionally, brain organoids often develop a necrotic core as they mature. This phenomenon arises from an insufficient vascular supply, which hinders the delivery of essential oxygen and nutrients for proper growth [2,95]. In light of these constraints, microfluidic devices are alternative culture systems that have the potential to enhance culture conditions, thereby promoting more consistent organoid development [96,97]. As such, microfluidic cell cultures leverage engineering and technological methods to address biological questions.
Brain organoid-on-chip systems have emerged as a new study area in recent years. These systems combine brain organoid cultivation with microfluidic technologies to enhance culture conditions, increase physiological relevance, ensure reproducibility, and support the industrial applicability of brain organoids. Brain organoids-on-chips are viewed as beneficial for improving culture conditions by minimizing the shear stress that cells endure, enhancing oxygen supply and distribution, facilitating nutrient and waste exchange, and promoting the establishment of chemical gradients [98]. The chips are typically designed in microcolumn, single-channel, or multichannel configurations to provide the physiological, biochemical, and physical signals required to develop brain organoids, thereby promoting their long-term survival and intricate architecture [99].
5. Brain Tumoroids
Advancing personalized medicine and addressing treatment resistance in brain tumors have become crucial for effectively translating laboratory research findings into clinical practice [100]. iPSC- or ESC-derived brain organoids themselves may be valuable for studying significant stages of tumor development; however, they do not accurately reflect the unique genetic makeup and cellular and molecular diversity associated with specific tumors and individual patients. Progress in stem cell culture has led to the development of innovative organoid technology, which produces 3D tissues from patient samples in vitro that closely resemble the structures, unique functions, molecular traits, genomic changes, expression patterns, and tumor microenvironments of primary tumors [101]. Tumor organoids, also known as tumoroids, are 3D structures composed of diverse cell types derived from patient samples, replicating the original tumor’s essential histopathological, genetic, and phenotypic features [100]. Brain tumoroids replicate the brain’s structure and function while preserving their histopathological characteristics, genetic makeup, mutation patterns, and responses to treatment [102,103]. Glioblastoma tumoroids are the most used brain tumoroids. Several studies have demonstrated that these models provide several advantages, including the ability to conduct molecular investigations. Additionally, they serve as promising tools for cost-effective studies on new anticancer drugs, which can be applied in precision medicine [104].
6. Functional Assays and Omics Used in Brain Organoid Characterization
Defining the 3D structural arrangement of cells, including the shape, size, and distribution of neurons within organoids, is crucial for identifying similarities with the human brain [105]. The initial characterizations of organoids utilized standard immunohistological markers primarily for identifying cell types, alongside qPCR to evaluate gene expression and determine the composition of cell types; however, comprehensive characterization remained limited [4,51] (Figure 2).
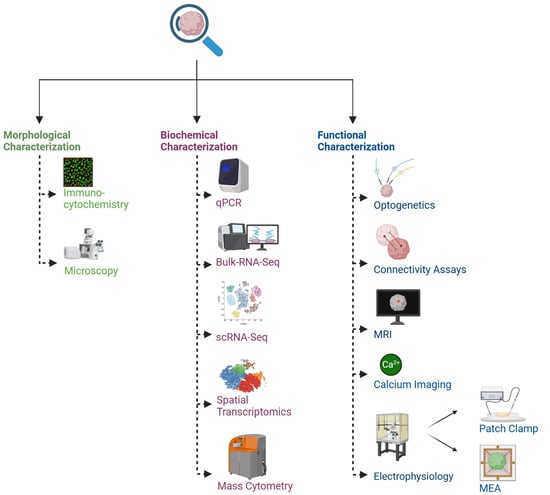
Figure 2.
Common methodologies for studying brain organoids and assembloids. Reproducibility, reliability, and translational applicability of neural organoids and assembloids require thorough quality control, standardization, and molecular and functional validation. Created in https://BioRender.com.
Single-cell RNA sequencing (scRNA-seq) has recently uncovered a notable diversity of cell types present in brain organoids [52]. Understanding this diversity is crucial for enhancing the development and characterization of organoids, particularly in the context of disease modeling [94,106]. Initial organoid characterization lacked depth. Without a clear understanding of neuronal maturity and subtypes, cellular interactions, and functional development, assessing the effectiveness of brain organoids as accurate physiological disease models remained challenging [46,107].
Electrophysiology is the primary method for analyzing the functionality of neural cells and tissues, including brain organoids. The capability to monitor neuronal activity is vital for numerous applications of brain organoids, particularly in disease modeling and drug development [108], and can be achieved using various electrophysiology techniques. The patch-clamping technique enables recording individual neurons’ activity within a brain organoid with high temporal precision, facilitating an in-depth examination of specific neurons [3,108,109]. This high temporal precision is particularly beneficial for assessing how specific disturbances, such as drug treatment or optogenetic stimulation, affect neuronal responses. However, since only single neurons are evaluated, there is a lack of information regarding network connectivity or dynamics that are critical to the overall function of the organoid [106]. On the other hand, calcium imaging addresses the shortcomings of patch clamping by enabling the real-time imaging of neural activity in small neuron clusters. This method is advantageous for investigating particular areas of brain organoids and for analyzing synaptic activity and neural circuits [110]. Research employing confocal or multi-photon microscopy has demonstrated changes in fluorescence resulting from the interaction of Ca2+ ions with genetically encoded calcium indicators derived from the aminopolycarboxylic acid BAPTA (1,2-bis(o-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid) [111]. While calcium imaging enables the functional characterization of individual neurons and small groups, it falls short of comprehensively analyzing dynamic processes occurring at the network level [105].
Microelectrode arrays (MEAs) are becoming more popular for screening applications and various studies because they combine the precise timing of patch clamping with the broader network perspective offered by calcium imaging [112,113,114]. MEAs can simultaneously track responses to stimulation protocols, utilizing a range of electrodes from dozens to thousands [115,116]. Investigating large areas of brain organoids has uncovered significant connectivity between different regions within the organoids, indicating the presence of long-range neural circuits and connectivity between regions. [117]. As brain organoids increase in complexity and are employed to model intricate developmental processes and diseases, the capacity to identify and analyze connectivity and neural circuits between different regions over substantial distances becomes essential, making MEAs valuable tools for understanding these circuits. Recently developed 3D MEAs [118,119] that integrate electrodes into flexible, hinged probes allow for extracellular recordings from 3D neural networks, including those found in organoids. Notably, these devices work with various existing recording systems promoting quick adoption among researchers studying brain organoids.
Optogenetics and the previously mentioned techniques have enabled precise stimulation and mechanistic investigations [120]. Recently, all-optical electrophysiology has become a technique for manipulating and recording neural activity with high spatiotemporal resolution [121,122,123]. The capability to stimulate and record using this approach facilitates network-level recordings of neural circuits. Although it may be slightly less accurate than patch clamping, optical electrophysiology retains much of the resolution offered by patch clamping while enhancing throughput, which is crucial for the extensive analysis required for brain organoids [106]. Recently, a technique utilizing MRI for the deep tissue imaging of whole brain organoids has been developed. This high-resolution imaging has been applied to visualize human brain organoids as small as 2 mm in diameter. MRI and diffusion tractography present a powerful method for imaging organoids. This has the potential to significantly contribute to the investigation of microstructural alterations in brain organoids that model various conditions affecting the human brain and to be used to evaluate neurotoxicity in drug screening [124].
A recent review by pioneers in the field highlighted methods for characterizing brain organoids [94]. This consensus article provides a detailed framework to enhance the reproducibility, reliability, and translational applicability of neural organoid and assembloid models. It specifies recommended practices for generating and characterizing neural cultures derived from hPSCs, such as the thorough quality control of induced iPSC lines, the standardization of differentiation protocols, and suggestions for functional and molecular validation. The authors address the difficulties associated with 3D phenotyping, transplantation into animal models, and ethical considerations, offering essential experimental guidelines for accurately modeling human brain development, disease, and evolution. This work is a key reference for researchers utilizing stem cell-based models in neuroscience, highlighting the importance of transparency, data sharing, and rigorous statistical methods.
7. Brain Organoids as Disease Models—Cancer
The emergence of brain organoids derived from human stem cells provides a flexible platform for understanding the molecular and cellular processes involved in brain development, neurodevelopmental disorders, and cancer, thereby connecting fundamental research to therapeutic advancements or personalized medicine [46,125,126,127].
7.1. Gliomas
Gliomas are the most prevalent primary brain tumors in adults. Histologically they are similar to normal glial cells and these tumors are commonly named based on these similarities [128,129]. Glioblastoma (GB) is the most common and aggressive type of primary brain tumor in adults. It exhibits distinct histopathological features, including necrosis and endothelial proliferation which contribute to its classification as grade IV according to the World Health Organization (WHO) brain tumor classification [130]. In children, most gliomas are low-grade tumors, typically associated with favorable survival rates [131]. However, approximately 30% of these tumors fall into the high-grade category, which is associated with poorer outcomes and a limited number of long-term survivors [132]. Pediatric high-grade glioma (pHGG) develops in the cerebral hemispheres, known as glioblastoma multiforme (GBM), or in midline structures like the thalamus or brainstem and named diffuse intrinsic pontine glioma (DIPG). Recent molecular studies of pHGG have revealed genetic and epigenetic characteristics that differ from those found in adult gliomas [133]. Existing GBM model systems are inadequate in capturing the complex characteristics of GBM and have not led to sufficiently effective therapies. In the search for new preclinical models for GBM, a variety of 3D organoid-based systems using human cells have been created [134] (Table 1).

Table 1.
Brain organoid disease models.
Two primary types of brain organoid-based GBM models are derived from clinically relevant stem cells. One of these models enables the examination of GBM initiation and responses to treatments without explicitly accounting for the impact of surrounding tissues or other cell types. This model typically involves the genetic modification of stem cells to incorporate driver genetic changes associated with GBM at a particular point during the differentiation of the cultured organoids. The second model type is often more appropriate for investigating advanced GBM, as it considers its invasion, proliferation, and interactions with other cell populations in the brain. These models can be created from a blend of glioblastoma stem-like cells and organoids derived from PSCs. Alternatively, they may consist of co-cultures of GBM cells, which can be either patient-derived primary cells or established GBM cell lines, prepared in single-cell suspensions or as spheroids [167,168]. A recent study produced engineered GBM brain organoids using genetically altered stem cells with mutations from both the proneural and mesenchymal subtypes of GBM. The research revealed that mutations linked to GBM disrupt neural development within brain organoids. Furthermore, when cells from engineered GBM brain organoids were transplanted into mice, they formed tumors that mimicked the specific characteristics of human GBM tumors. By utilizing integrated single-cell transcriptomic data from engineered GBM brain organoids and patient-derived GBM tumors, this research team uncovered shifting gene expression patterns in developmental cell states contributing to tumor progression [135]. A different study effectively generated patient-derived GBM organoids that maintained the pathological characteristics of the parental GBM. The patient-derived GBM organoid–neuron co-culture system was also explored, and it revealed interactions between neurons labeled with GFP and patient-derived GBM organoids labeled with mCherry. The in situ stereotaxic implantation of patient-derived GBM organoids into the brains of nude mice established intracranial orthotopic GBM models [136].
Deligne and colleagues developed an integrated method to produce, expand, and thoroughly analyze 3D pediatric high-grade glioma-derived organoids from fresh tissues for precision and personalized medicine assays. They generated 3D patient-derived tumor organoids from diverse primary and relapsed histological subtypes and tumor grades of pHGG. The generated 3D models were biobanked and revived for additional molecular, cellular, and preclinical studies, achieving a success rate of 91% [169]. A different study revealed the successful creation of a neuroimmune-competent brain organoid model composed entirely of human cells that effectively represents multiple critical diffuse midline glioma (DMG) features. They generated human microglia-containing brain organoids derived from genetically modified hPSCs. By fusing these microglia-containing organoids with DMG spheroids, they established a manageable and modular platform for investigating interactions between human tumors and the brain within a 3D microenvironment. The microglia brain organoids contained microglia exhibiting a mature morphology and motility, alongside various neuron subtypes, simulating the intricate microenvironment in brain tumors. Subsequently, these organoids were combined with tumor spheroids harboring H3K27M mutations and TP53P27R/K132R alterations to form a microglia-containing brain organoid-tumor fusion model. The results of this study suggested that DMG cells interacting with another DMG spheroid show restricted infiltrative activity, preserving a distinct cellular boundary over a prolonged period [137]. A recent review extensively discussed organoid models of glioblastoma [167].
7.2. Medulloblastoma
Medulloblastoma is a highly aggressive brain tumor that originates from the cerebellum, primarily affects children, and is responsible for a significant proportion of morbidity and mortality among cancer patients [170,171]. Medulloblastoma exhibits biological and clinical diversity, comprising several subgroups [171,172]. Over the years, continuous efforts have been made to develop models of this disease that accurately replicate in vivo tumors. This has led to the growing use of cerebellar organoids for modeling medulloblastoma. A recent investigation utilizing the CRISPR/Cas9 gene editing of Patched-1 (PTCH1) in hiPSCs led to the generation of a medulloblastoma-cerebellar organoid [138]. PTCH1 serves as the primary receptor for the Sonic Hedgehog (SHH) ligand and plays a crucial role in inhibiting SHH signaling, which is vital for human embryonic development. Mutations that result in its loss of function have been linked to disrupted neuronal growth and the aggressive brain tumor, medulloblastoma. The resulting organoids were employed to explore the initial molecular and cellular effects of PTCH1 mutations on the development of the human cerebellum. The results from these studies indicated that the developmental processes occurring in cerebellar organoids mirror the in vivo mechanisms of regionalization and SHH signaling, providing new perspectives on the early pathophysiological stages involved in the tumorigenesis of medulloblastoma without relying on animal models [138,139].
One study aimed at understanding the phenotypic traits of SHH medulloblastoma in a nonmalignant tumor microenvironment using SHH medulloblastoma cells co-cultured with cerebellar organoids to promote a malignant phenotype. Tumor spheroids from two different SHH medulloblastoma cell lines were grown alongside control organoids. The investigators analyzed the transcriptional profiles of the co-cultured tumor cells and compared them to those from mono-cultures, patient tumors, and patient-derived orthotopic xenograft (PDX) mouse models. The co-cultured SHH medulloblastoma cells displayed phenotypes akin to actual patient tumors and showed greater heterogeneity compared to mono-cultures. Some cell subpopulations activated NEUROD1, a marker for differentiating granule cells, which was not present in mono-cultures. Other subpopulations demonstrated traits of a cancer stem-like state like those found in vivo. This study introduced a novel in vitro model of SHH medulloblastoma with promising translational potential [139].
Another study used cerebellar organoids to model the Group 3 medulloblastoma subtype, which among medulloblastoma has the poorest prognosis. The investigators overexpressed c-MYC and Otx2, which are recognized as significant promoters of this subtype. Initially, they pre-differentiated the cerebellar organoids until day 35, ensuring all progenitors were present, and subsequently transfected the organoids with the oncogenes. This research employed a transposon system to enable the ongoing overexpression of the targeted genes over time, replicating the oncogene amplification and overexpression seen in patients. The resulting medulloblastoma-like organoids exhibited a DNA methylation profile that aligns with human Group 3 tumors. Furthermore, this study demonstrated that SMARCA4 can mitigate the tumorigenic effects of Otx2 and c-MYC in vivo and that treatment with Tazemetostat, an EZH2-specific inhibitor, diminishes Otx2/c-MYC tumorigenesis both in ex vivo cultures and human cerebellar organoids [140].
8. Brain Organoids as Disease Models—Neurodevelopmental Disorders
8.1. Autism Spectrum Disorder
Autism spectrum disorder, known as ASD, is a complex neurodevelopmental condition that arises in developing fetuses and young children during their early growth stages. It is a multifactorial condition that in part is attributed to changes in the expression of specific genes [173,174]. ASD affects more than 2% of the preschool population and encompasses a significant range of conditions that contribute to the heterogeneity of autism [175]. The absence of a suitable human disease model hampers research efforts to effectively replicate human pathological features and reflect the genetic diversity associated with ASD [147,176,177,178,179]. Extensive studies have been conducted using brain organoids to model and investigate the molecular basis of autism (Table 1). One of the most recent studies utilized iPSCs and cerebral organoids derived from three individuals with idiopathic non-regressive ASD who presented with normocephaly [141]. This study demonstrated that neural stem cells in the ASD organoids exhibited premature differentiation and impaired expression of fatty acid binding protein 7 (FABP7). Furthermore, it was determined that FABP7 operated through the Mitogen-Activated Protein Kinase Kinase1/2 (MEK1/2) pathway to facilitate early neurogenesis. This discovery offers new insights into the molecular mechanisms involved in the pathology of idiopathic normocephalic ASD with non-regression and encourages the development of therapeutic drugs aimed at ASD treatment by targeting the FABP7/MEK axis [141].
A different study established a human cerebral organoid model using induced pluripotent stem cells, applying targeted genome editing to eliminate the protein expression of the CNTNAP2 gene. Homozygous mutations leading to the loss of function in CNTNAP2 are linked to cortical dysplasia focal epilepsy syndrome, which includes features of autism spectrum disorder (ASD) and intellectual disability [180]. In CNTNAP2−/− cerebral organoids, an accelerated cell cycle, disorganized ventricular zones, and increased cortical folding were shown. Proteomic studies revealed disruptions in glutamatergic and GABAergic synaptic pathways, while transcriptomic analysis showed altered gene expression related to inhibitory neuron networks. Additionally, there was heightened AKT/mTOR signaling observed in these organoids. A spatial transcriptomic analysis of the CNTNAP2−/− ventricular-like zones revealed gene expression alterations, indicating the upregulation of pathways related to cell cycle regulation, synapses, and glutamatergic/GABAergic activity. A significant overlap was discovered among the differentially expressed genes from day-30 organoid omics datasets and those from idiopathic ASD (macrocephaly) iPSC-derived telencephalic organoids [142].
Tanabe’s group utilized patients-derived organoids to investigate whether the onset of ASD is linked to an underdeveloped choroid plexus [143]. The choroid plexus consists of multiciliated epithelial cells that differentiate from ependymal cells lining the ventricular surfaces [181]. It generates protein-rich cerebrospinal fluid and completes its maturation before brain development. It is believed that dysfunction in the choroid plexus significantly impacts ASD. Over time, an increase in the expression of transthyretin (TTR), a marker gene for the choroid plexus, was detected in choroid plexus organoids sourced from healthy individuals. Additionally, epithelial cells that expressed TTR exhibited an aggregated signal at the apical surface, resembling the intrinsic multicilia found in choroid plexus epithelial cells. Conversely, choroid plexus organoids derived from ASD patients displayed a decrease in TTR expression for one patient. The expression levels of the neural progenitor cell marker Pax6 and the neuronal marker β3-tubulin showed no significant difference, suggesting that the developmental abnormalities in the choroid plexus emerged later in the differentiation process. When evaluating organoid size at the choroid plexus differentiation stage (day 50), the surface area of organoids from ASD patients was significantly smaller, half the size. The assessment of ciliogenesis and protein expression (TTR, Otx2, AQP1, and ZO-1) in organoids from ASD patients showed a marked reduction in these genes. These findings suggested that a choroid plexus exhibiting impaired multiciliogenesis and a reduced expression of key differentiation genes affects blood cerebrospinal fluid barrier integrity and contributes to dysfunction in organoids from individuals with ASD. Overall, this study highlighted that an underdeveloped choroid plexus is a prevalent characteristic in ASD pathology [143].
SHANK (SH3 and multiple ankyrin repeat domains) proteins act as scaffolds at synapses and are primarily found in the post-synaptic density of excitatory synapses. Changes in SHANK3 have been closely linked to non-syndromic ASD. Research using cerebral organoids to observe the developmental changes in SHANK3 expression identified the binding site of the upstream Early Growth Response 1 (EGR1) in the promoter region of the SHANK3 gene. The validation of this interaction through chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assays (EMSAs) confirmed that EGR1 interacts with the promoter region of SHANK3, and alterations in EGR1 levels affect SHANK3 transcription. This highlights the important link between EGR1 and SHANK3 in the context of ASD [144].
SCN2A encodes the Nav1.2 voltage-gated sodium channel and is significantly reduced in individuals with autism [182]. Wu et al. [145] explored the role of microglial phagocytosis in synapses and found that microglia are integral to the pathogenesis of autism in SCN2A-deficient mouse and human cerebral organoids models involving excessive phagocytic activity. The capacity of microglia to eliminate surplus synapses from affected neurons is evolutionarily preserved and has been seen in rodent and human cellular models with SCN2A deficiency [145].
A separate investigation utilized forebrain organoids to delve into germline research. Mutations in the PTEN gene account for 0.2–1% of all cases of ASD and around 17% of ASD patients with microcephaly. This study used forebrain organoids from gene-edited isogenic human iPSCs containing PTEN mutant alleles linked to ASD or cancer to investigate how these mutations interfere with neurodevelopmental processes. The results indicated that the ASD allele hindered early neuroectoderm formation, leading to impaired electrophysiological function. Organoids derived from cancer-related iPSCs exhibited disturbances in neuronal differentiation, radial glia positioning, and cortical layering after 72 days of development. Perifosine, an AKT inhibitor, diminished the overly activated AKT and partially rectified the cellular organization abnormalities observed in the ASD organoids. ScRNA-seq analyses of early-stage organoids indicated a gene reduction associated with neural cell fate in the ASD mutant organoids [146].
Genes associated with chromatin remodeling frequently show mutations in individuals with ASD. One of the most affected and penetrant genes is chromodomain helicase DNA-binding protein 8 (CHD8). It is characterized by a SFN2-like ATPase and contains two chromo-domains. Most ASD-associated de novo mutations in CHD8 result in the loss of function, leading to gene haploinsufficiency. CHD8 haploinsufficiency is associated with the abnormal development of the cerebral cortex. Using cerebral organoids, it was found that CHD8 haploinsufficiency disrupted neurodevelopmental pathways, causing the accelerated generation of inhibitory neurons and the delayed generation of excitatory neurons, resulting in opposing proportional expansions at days 60 and 120 [147]. This imbalance correlated with an increase in the size of the cerebral organoids, serving as an in vitro representation of macrocephaly observed in patients [183]. Another group investigated changes in forebrain development by utilizing cortical organoids from boys with idiopathic ASD and their unaffected fathers across a study involving 13 families. Thus, the development of ASD in both macrocephalic and normocephalic individuals involves the imbalance of excitatory neurons in the dorsal cortical plate and other cell types, stemming from divergent transcription factor expression during early cortical development. Although no genetic variants were found in the probands linked to the gene expression changes, a significant overlap with known ASD risk genes suggested a genetic convergence between rare ASD types and idiopathic cases [148].
A study by Birtele et al. [149] utilized cortical organoids to investigate the non-synaptic roles of the ASD-related gene SYNGAP1 (synaptic Ras GTPase-activating protein 1). Evidence suggests that SYNGAP1 may have a significant function during the early phases of cortical neurogenesis. The study revealed abnormal cytoskeletal dynamics that disrupt the scaffolding and division orientation of human radial glial cells, leading to impaired lamination and the hastened maturation of cortical projection neurons in cortical organoids with SYNGAP1 haploinsufficiency [149].
ASD is complex and multifaceted. Currently, over 800 genes and dozens of genetic syndromes have been associated with ASD, with environmental and epigenetic factors further adding to this complexity [184]. The work outlined above and additional studies [179,185,186,187,188,189,190,191,192,193,194,195,196] extensively summarized in [183] demonstrate the immense value brain organoids bring to modeling autism, thereby greatly enhancing our understanding of the molecular underpinnings of this neurodevelopmental condition.
8.2. Epilepsy
Epilepsy is a condition that includes various underlying disorders [197]. The defining characteristic of epilepsy is seizures [198]; it manifests in different types. In focal epilepsy, seizures originate from a specific region of the brain, while in generalized epilepsy, seizures arise simultaneously on both sides of the brain. Epilepsies are classified according to their causes, which can be either acquired or genetic [199]. Roughly 30–40% of individuals with epilepsy do not achieve the total control of their symptoms through antiepileptic medication [200]. The introduction of next-generation sequencing has led to the discovery of over 140 genetic loci linked to various forms of epilepsy. Genes linked to epilepsy encode various proteins, ranging from ion channels to transcription factors. While the specific pathways through which mutations lead to seizures are not fully understood, they ultimately enhance brain excitability [201]. Conventional methods for modeling genetic forms of epilepsy have relied on rodent models created through genetic alteration or by selecting spontaneous mutations in rodent populations. These models often fail to represent the human condition and its underlying mechanisms [202,203]. The use of brain organoids for studying epilepsy and similar disorders is becoming more prevalent, underscoring their increasing relevance in epilepsy research.
Focal cortical dysplasia (FCD) stems from abnormal neural progenitor cell proliferation and tissue organization during brain development, and is linked with developmental delays, reduced cognitive function, and persistent epilepsy [204,205]. The primary pathogenic factor for FCD II is thought to be somatic mosaicism affecting genes involved in the mTOR pathway, occurring in cells such as cortical neurons in the brain, which leads to localized abnormal cortex growth [206]. Loss of function in specific genes, which includes GATOR1 complex subunits, including DEPDC5, NPRL2, and NPRL3, leads to the activation of the mTOR pathway [207,208]. Cortical organoids from FCD II patients were larger than those from the control. This finding aligns with the hyperactivation of the mTOR signaling pathway and abnormal growth observed in patients with FCD [150].
Another recent study focused on PCDH19-clustering epilepsy (PCE). This arises from pathogenic variants in the Protocadherin-19 (PCDH19) gene, located on the X chromosome [209]. This condition primarily affects females and mosaic males. The mosaic expression of the cell adhesion molecule PCDH19 is believed to disrupt interactions between cells expressing mutant and wild-type PCDH19, leading to the development of the disorder. In this study, the investigators used genome editing to create isogenic female human ESCs featuring HA-FLAG-tagged PCDH19 (wild-type) or homozygous PCDH19 knockout. They generated human cortical organoids from mixed GFP-labeled wild-type and RFP-labeled knockout cells. The study found that PCDH19 was present in early wild-type neural rosettes (days 20–35) and co-localized with N-Cadherin in areas like the ventricular zone. In mosaic PCE, abnormal cell sorting in the ventricular zone was noted, which continued even with different ratios of wild-type to knockout cells. PCE cortical organoids exhibited variations in PCDH19 and N-Cadherin expression and irregular deep-layer neurogenesis. These issues were absent in organoids made only from wild-type or knockout cells, simulating male carrier scenarios. The outcomes from these mosaic PCE human cortical organoid models suggest that PCDH19 is crucial for the organization of radial glial cells in the human ventricular zone and for early cortical development. Thus, this model may serve as an essential platform for investigating the mechanisms behind PCE-related cortical hyperexcitability and evaluating potential precision therapies [151].
Glucose Transporter 1-Deficiency Syndrome (GLUT1-DS) is a rare genetic condition caused by mutations in the GLUT1 gene leading to reduced brain metabolism and symptoms including epilepsy, motor impairments, and cognitive challenges [210,211]. Investigators generated brain organoids from iPSCs derived from a GLUT1-DS patient and healthy control. The functional assay results indicated a comparable distribution of neurons and astrocytes in both organoids. However, the GLUT1-DS organoids displayed lower cellular density and smaller sizes. Both organoids also showed characteristic functional spikes, but the GLUT1-DS organoids exhibited unique epileptiform activity and increased sensitivity to glucose deprivation. Overall, the results from this study supported the application of brain organoids as a model for investigating GLUT1-DS and emphasized their potential for evaluating new therapeutic approaches to enhance glucose metabolism and address epilepsy in patients [152].
Tuberous sclerosis (TSC) is categorized among cortical development malformations arising from mutations in either TSC1 (hamartin) or TSC2 (tuberin) that are components of the mTOR pathway [212]. Patients experience debilitating neuropsychiatric symptoms that are frequently resistant to treatment, including uncontrollable epileptic seizures and intellectual disability. The key features of TSC include cortical tubers—abnormal neurons and glial cells in the cortex associated with seizures. More than three out of four individuals with TSC develop subependymal nodules (SEN). Research indicates that cortical tubers and SEN/SEGAs (subependymal giant cell astrocytoma) typically arise from similar genetic alterations, often characterized by the loss of the second allele of either TSC1 or TSC2 somatic mutations leading to TSC onset that have been observed in both mouse and human models, highlighting the critical role of mutations in NPCs. This randomness may explain the variability in tuber number and size in TSC patients. A higher tuber load correlates with more severe epilepsy and intellectual disability, indicating that somatic mosaicism might significantly impact the differences in neurological symptoms among TSC individuals [213]. Genetic analyses reveal that SEN/SEGAs typically lose the second allele, whereas this is less common in cortical tubers, which raises questions about the two-hit hypothesis. These differences could stem from specific human brain cell types and processes crucial for disease development. In one study, Eichmuller et al. [153] created human cerebral organoids from patients’ iPSCs and compared these to primary human tissues. The study identified a type of neural stem cells called CLIP cells, which proliferate excessively in TSC cases, resulting in excessive interneurons, brain tumors, and cortical malformations. The investigators further explored various therapeutic strategies for TSC and related disorders, including blocking the EGF receptor, which has shown promise in reducing tumor burden. This work also showed that examining cortical malformation can yield valuable insights into unique aspects of human brain development [153]. Other studies [214,215,216] that utilized brain organoids to study disorders characterized by epilepsy explore the fundamental mechanisms and possible treatment strategies related to these conditions.
8.3. Cerebral Palsy
Cerebral palsy (CP) affects movement and posture, reducing activity and functionality due to non-progressive damage to the developing brain [217]. It is the most prevalent physical disability affecting children in the United States [218,219]. Over the years efforts have been made to develop more realistic models for studying CP, including brain organoids. However, using brain organoids in CP research presents significant challenges, particularly because the specific cause of CP remains unknown in about 80% of patients.
TYW1, an enzyme responsible for tRNAphe modifications, regulates the proliferation and migration of neurons during early neural development [220]. A deficiency in TYW1 could disrupt the accurate pairing of tRNAphe with its respective codons, resulting in reduced levels of proteins rich in phenylalanine. The accurate translation of proteins is essential for neural development [221]. The dysfunction of TYW1 has been associated with CP associated with significant intellectual disabilities. Using cerebral organoids derived from TYW1−/− hESCs, studies showed that the depletion of TYW1 led to the impaired differentiation of neurons. The loss of TYWI led to the increased expression of human-specific endogenous retrovirus-K (HERVK) in this model due to the decreased levels of SMARCAD1, thereby hindering neuron differentiation [154].
Another study focused on neonatal hypoxic injury (NHI), a severe condition impacting over 60% of infants born with a very low birth weight. The condition leads to long-term neurological issues such as seizures, cerebral palsy, and intellectual disabilities [222,223]. The investigators demonstrated the effectiveness of minocycline for treating human NHI using a cerebral organoid model of hypoxia where they examined the development of an in vitro cerebral organoid model under varying oxygen levels. The findings indicated that hypoxia reduced the expression of critical gene markers associated with the development of the forebrain, oligodendrocytes, glial cells, and cortical layers, as well as genes essential for migrating cortical neurons. Conversely, ventral markers showed no change and even increased under hypoxic conditions. The detrimental impact of hypoxia on dorsal brain genes, and on oligodendrocytes and neuronal progenitors, could be alleviated by administering minocycline, a small molecule approved by the FDA [155].
One study developed a GPAM knockout line; GPAM (glycerol-3-phosphate acyltransferase 1, mitochondrial) is an enzyme in the mitochondria that synthesizes glycerolphospholipids and triacylglycerols. Mutations that result in the loss of GPAM function have been linked to hypomyelination in the corticospinal tract of patients with cerebral palsy [220]. The investigators created a GPAM knockout human ESC line, SYSUe-008-A, using CRISPR/Cas9 technology to investigate this rare disorder in human brain organoids. The GPAM knockout cell line retained a normal karyotype and exhibited pluripotent stem cell markers and differentiation capabilities similar to wild-type hESCs, making it a valuable resource for modeling the disease in brain organoids and conducting drug screening for cerebral palsy [224].
8.4. Attention Deficit Hyperactivity Disorders
Attention-deficit/hyperactivity disorder (ADHD) is a neurodevelopmental condition marked by inattention, impulsivity, and hyperactivity. It is often identified in early childhood and persists into adulthood [225,226]. ADHD ranks among the most genetically inheritable neuropsychiatric conditions [227,228]. One study utilized brain organoids to investigate the origins of ADHD within the early cerebral cortex, particularly the telencephalon. The investigators created an ADHD organoid using patient-derived iPSCs from an 18-year-old male with ADHD according to DSM-IV-TR criteria. It was found that telencephalon organoids exhibited less growth in their layer structures than control organoids with a higher neuron count but reduced cell proliferation between days 35 and 56, resulting in a notable difference. These alterations in neural stem cell characteristics and cortical structure development may be significant for understanding ADHD’s pathogenesis and align with findings from neuroimaging studies [156].
9. Assembloids and Rare Diseases of the Nervous System
Dysfunctions in the neural circuits of the cortico-striatal pathway are implicated in neurodevelopmental disorders such as ASD, schizophrenia, and obsessive–compulsive disorder [229,230,231,232]. To test this, investigators created brain organoids that mimic the developing human striatum and combined them with cerebral cortical organoids in a 3D culture to form cortico-striatal assembloids. The results showed cortical neurons extending axonal projections into striatal organoids and establishing synaptic connections. Following the assembly, medium spiny neurons exhibited maturation in their electrophysiological properties and showed calcium activity when cortical neurons were stimulated optogenetically. The investigators also generated cortico-striatal assembloids from individuals with a neurodevelopmental disorder linked to a deletion on chromosome 22q13.3. This work highlighted disease-associated abnormalities in calcium activity, suggesting that this approach facilitates the exploration of cortico-striatal connectivity development using patient-derived assembloids [157].
Similarly, a recent study generated a striato-nigral (Str-SN) circuit, consisting of medium spiny neuronal projections from the striatum to the midbrain’s substantia nigra (SN). Impairments in the Str-SN circuitry can lead to various motor disabilities linked to neurodegenerative conditions, such as Huntington’s disease. The investigators created 3D Str-SN assembloids by combining striatum-like organoids with midbrain SN-like organoids. Significant medium spiny neuron (MSN) projections from the striatum formed synaptic connections with GABAergic neurons in SN organoids and demonstrated optically evoked inhibitory post-synaptic currents. This study’s results indicated that treatment with brain-derived neurotrophic factors could improve the defects in reciprocal projections within HD-iPSC-derived assembloids. Thus, Str-SN assembloids may be utilized to identify defects in MSN projections and serve as potential platforms for drug testing in Huntington’s disease [158].
Neuronal connections facilitate muscle activation and movement generation. Andersen et al. [159] created 3D cortico-motor assembloids by combining organoids that replicate the cerebral cortex or hindbrain/spinal cord with skeletal muscle spheroids. Techniques such as rabies tracing, calcium imaging, and patch-clamp recordings revealed that corticofugal neurons established links to spinal spheroids, while spinal motor neurons connected with muscle tissue. The uncaging of glutamate or optogenetic activation of cortical spheroids elicited significant contractions in 3D muscle, and these assembloids remained morphologically and functionally intact for as long as 10 weeks post-fusion. This approach highlights the remarkable self-assembly of 3D cultures to form functional circuits for studying development and diseases [159].
Assembloids have been combined with CRISPR screening to explore the roles of the significant number of genes related to neurodevelopmental disorders in the development of human interneurons. The first screen aimed at interneuron generation revealed thirteen candidate genes. A follow-up screen for interneuron migration in over 1000 forebrain assembloids pinpointed 33 potential candidates comprising cytoskeleton-associated genes and the endoplasmic reticulum gene LNPK. The results demonstrated that LNPK deletion led to the irregular migration of interneurons. These findings further emphasized the effectiveness of the CRISPR-assembloid platform in systematically linking neurodevelopmental disorder genes to human development and uncovering the mechanisms behind diseases [160].
Two other studies highlighting the importance of interneurons were conducted by Birey at al. [233,234] in a model of Timothy syndrome, a neurodevelopmental condition associated with mutations in the CaV1.2 calcium channel. Nervous system development involves a coordinated series of events, particularly the movement of GABAergic neurons from the ventral to the dorsal forebrain and their integration into cortical circuits. The investigators created forebrain organoids that included cortical glutamatergic and GABAergic neurons to replicate the migration patterns of interneurons seen during fetal development. They found that interneurons displayed unusual migratory behavior in Timothy syndrome and demonstrated that, post-migration, these interneurons successfully integrated with glutamatergic neurons, forming a functional microphysiological system [233]. Notably, the abnormality in saltation length was associated with irregular actomyosin and myosin light chain (MLC) phosphorylation, while the alteration in saltation frequency was influenced by heightened sensitivity to GABA, which could be corrected by GABA-A receptor antagonists. The study further demonstrated an increase in hypersynchronous hCS network activity in Timothy syndrome that was intensified by interneuron migration. Collectively, these studies highlighted the intricate role of L-type calcium channel Cav1.2 function in the migration of human cortical interneurons and identified new approaches to remediating deficiencies in the context of Timothy syndrome [234].
Many organoid protocols still overlook microglia, which are essential for brain development and have been connected to various neuroinflammatory conditions. One study introduced an innovative method to create neuroimmune assembloids using iPSC-derived cortical organoids and microglia. The authors proposed two approaches: one that directly incorporated microglia progenitors into the organoids and another that combined microglia with neuronal progenitors in a specific ratio [161]. Another study highlighting the need to incorporate immune cells into assembloids was conducted by Goldberg et al. [235]. Mutations in the Three Prime Repair Exonuclease 1 (TREX1) gene have been linked to Aicardi–Goutières Syndrome (AGS). In this study, the investigators employed three-month-old assembloids that included microglia and oligodendrocyte-containing organoids to examine the roles of TREX1 in human microglia. Functionally, TREX1 was essential in microglia during the shift from gliogenic intermediate progenitors referred to as pre-oligodendrocyte precursor cells to precursors in the oligodendrocyte lineage, which hampers oligodendrogenesis while favoring astrogliogenesis in human assembloids. The findings further revealed that TREX1 affects cholesterol metabolism, resulting in a more active microglial morphology and heightened phagocytosis when TREX1 is absent. Regulating the cholesterol metabolism using atorvastatin, an FDA-approved HMG-CoA reductase inhibitor, restored microglial characteristics, suggesting potential therapeutic strategies for diseases like AGS [235].
Studies on Parkinson’s disease (PD) highlight some of the challenges associated with using organoids to study neurodegenerative diseases. Traditional brain organoids from iPSCs require lengthy maturation periods and may not accurately represent the older brain’s epigenetics, limiting their use in modeling late-onset diseases like PD. Kim et al. [236] presented a method for creating 3D-induced dopaminergic (iDA) neuron organoids directly from human fibroblasts, which better mimic the aging brain. The 3D iDA neuron–astrocyte assembloids demonstrated the importance of glial cells in neurodegenerative processes. In PD assembloids, control astrocytes alongside A53T mutant iDAs protected neurons, while A53T mutant astrocytes led to neuronal degeneration and synucleinopathy. The findings suggested that 3D-iDA organoids and assembloids are valuable models for studying PD’s pathogenesis and potential therapies [236]. Another study that generated an assembloid model of PD presented a model of the nigro-striatal pathway using midbrain–striatum assembloids with inducible aging. The findings demonstrated that these assembloids can establish nigro-striatal connectivity, characterized by catecholamine release from the midbrain to the striatum and the formation of synapses between the midbrain and striatal neurons. In addition, assembloids that overexpress progerin exhibited aging characteristics that lead to early signs of neurodegeneration. Thus this model may aid in elucidating the roles of aging and nigro-striatal connectivity in the onset and progression of PD [162].
Synaptic plasticity mechanisms, such as long-term potentiation (LTP) and long-term depression (LTD), are essential for learning and memory. A study by Patton et al. [163] created thalamocortical assembloids by merging hiPSC-derived organoids from the thalamus and cortex. Single-nucleus RNA sequencing revealed that over 80% of thalamic organoid cells were glutamatergic neurons. These assembloids formed reciprocal axonal projections and synapses, displaying short-term plasticity like animal models. While both long-term potentiation (LTP) and long-term depression (LTD) were induced, the underlying mechanisms varied from those observed in rodents. Therefore, thalamocortical assembloids serve as a useful model for investigating synaptic plasticity within human neural circuits [163]. Dysfunctions in thalamocortical communication may play a role in neuropsychiatric conditions. Variants in CACNA1G, which encodes a T-type calcium channel in the thalamus, are associated with absence seizures, intellectual disabilities, and schizophrenia. Research revealed that the M1531V CACNA1G variant linked to seizures caused changes in T-type currents in thalamic neurons and increased activity in both thalamic and cortical neurons. Conversely, the loss of CACNA1G, associated with a higher risk of schizophrenia, resulted in abnormal connectivity in the thalamocortical pathway and heightened spontaneous activity in the thalamus. Overall, these studies highlighted the potential of multicellular systems to study genetic risk variants for diseases at the cellular and circuit levels [46,164].
Assembloids representing different organ systems underscore the future potential of complex assembloid systems. For example, cortical–blood vessel assembloids created by merging cortical organoids with blood vessel organoids enable vascular support for brain organoids. This led to a notable increase in the expression of microglia and astrocytes within the brain organoids in a study aimed at finding the correlation between COVID-19 and Alzheimer’s disease (AD) [165]. With a focus on the neurotrophic effects of SARS-CoV-2, the research identified AD pathologies, such as β-amyloid plaques, which were influenced by the inflammatory responses triggered by SARS-CoV-2 infection. Overall, the assembloid system offered a sophisticated platform for examining human neurotrophic diseases, such as COVID-19, and implied that the neuroinflammation resulting from viral infections plays a role in the advancement of AD [165].
New research into the capacity of the Zika virus (ZIKV) to infect and eliminate CNS tumors such as GBM, medulloblastoma, and Atypical Teratoid Rhabdoid Tumors (ATRTs) highlights the virus’ potential for oncolytic therapy. There is limited evidence demonstrating the safety of ZIKV for treating malignant CNS tumors, which is necessary for advancing this strategy to clinical trials especially in children. Ferreira et al. [237] created a co-culture model using mature human cerebral organoids combined with tumor cells from GBM, medulloblastoma, or ATRT. These models provided the means to monitor the growth and invasion of tumor cells within cerebral organoids and utilized the assembloids to assess the oncolytic effects of ZIKV, its replication abilities, and its preferential targeting of cancer cells over normal cells. The results suggested that ZIKV replicates more rapidly in aggressive CNS tumor cells than in normal cells found in cerebral organoids. However, the elevated replication of ZIKV in tumor cells did not directly correlate with oncolytic effects, indicating the involvement of internal and external cellular factors in tumor cell death by ZIKV [237]. Overall, these studies not only highlight how assembloids aid in investigating rare nervous system diseases but also emphasize the immense potential of merging brain organoid methodology with brain cancer research in the newly emerging field of cancer neuroscience.
10. Brain Organoids in Drug Discovery
The application of brain organoids has advanced basic research, especially in modeling diseases and identifying targets related to neurodevelopmental, neuropsychiatric, and neurological conditions [238]. Despite the promise of organoid technology in drug screening, several challenges have emerged concerning its reproducibility, scalability, and relevance to human diseases. To improve the utility of organoids in drug discovery, several technical strategies can be considered, such as using CRISPR to create isogenic models, applying single-cell RNA sequencing for detailed cellular profiling, and leveraging machine learning to process intricate datasets. Additionally, tools like high-content imaging, automated liquid handling, and standardized assays are crucial in facilitating this goal [239]. Robots and software designed to identify organoids in microwells and conduct quantitative imaging analysis facilitated the phenotyping of organoids. This lays a promising foundation for high-quality HTS and potential use as a tool for therapeutic screening [238]. For example, Renner et al. implemented a fully automated HTS workflow for the creation and assessment of midbrain organoids. The researchers utilized a liquid-handling robot capable of executing processes such as seeding, maintenance, fixation, whole-mount staining, and clearing in a fully scalable automated manner within standard 96-well plates. The midbrain organoids generated showed minimal differences in size distribution, morphology, and cellular composition within and between batches, displaying comparable morphological characteristics to those reported in other studies. Further analysis through RNA sequencing and quantitative whole-mount high-content imaging (HCI) validated the reproducibility of these organoids [69]. Another investigation presented a compelling, network-based platform for drug screening, developed by merging mathematical modeling and the pathological characteristics of Alzheimer’s disease with human iPSC-derived cerebral organoids, including isogenic lines modified using CRISPR-Cas9. They utilized 1300 organoids sourced from 11 individuals to establish a high-content screening (HCS) system and evaluate FDA-approved drugs that can penetrate the blood–brain barrier. This research offers a pathway for precision medicine by integrating mathematical modeling with a small-scale pathological brain model using cortical organoids [240]. However, while advances in integrating organoids into drug discovery setups have been made (Table 2), significant challenges in standardization, scalability, and cost remain even beyond those of standard 3D cell culture [241,242].

Table 2.
Brain organoids in drug discovery.
11. Discussion
Brain organoids have become valuable tools for modeling neurodevelopmental disorders, brain tumors, and the development of new therapeutics. However, several issues must be addressed to unlock their full potential for clinical applications. One notable challenge with current organoid models is that they often lack genetic diversity [248]. Many researchers rely on iPSCs from a limited pool of donors, which overlooks inter-individual variability in disease manifestation and drug response. By generating organoids from a wider variety of iPSC sources, including those tailored to specific patients and reflecting diverse populations, one can enhance the accuracy of preclinical results. This approach would also pave the way for more personalized treatment options, ultimately leading to better patient outcomes. Brain organoids exhibit significant potential for modeling rare neurological and neurodevelopmental disorders yet are underutilized. While animal models often struggle to replicate the subtle characteristics of these conditions, organoids generated from patient iPSCs offer a distinct opportunity to investigate disease mechanisms and evaluate potential drug candidates in a relevant cellular environment. Establishing brain organoid biobanks, comprising organized collections of patient-derived and genetically modified organoids, will be essential for facilitating large-scale, disease-oriented research initiatives.
Despite their potential, brain organoids face substantial challenges regarding standardization [94]. Variability in differentiation protocols, culture conditions, and batch-to-batch reproducibility impedes their widespread adoption. Therefore, establishing standardized protocols and robust quality control measures is imperative to ensure consistent organoid generation, maturation, and phenotypic stability across research laboratories. Another essential consideration is integrating the microenvironment with the ECM. Current organoid models lack a comprehensive array of brain-resident and supportive cell types, including vascular, immune, and glial cells. Incorporating these components through co-culture systems or bioengineering techniques can enhance physiological relevance, particularly in modeling diseases characterized by complex cell–cell interactions.
The use of brain organoids in studying immunoneurological disorders is an exciting application [249]. Conditions like multiple sclerosis and autoimmune encephalitis involve complex interactions between the immune system and the nervous system, which are difficult to replicate with conventional models. Progress in incorporating microglia, peripheral immune cells, and cytokine environments into organoids and assembloids could lead to more accurate representations of neuroinflammation and neuroimmune signaling [250]. Additionally, organoid models are starting to explore neuropsychiatric disorders like addiction [251], schizophrenia [252], and bipolar disorder [253], which have traditionally been difficult to model using conventional methods. While the complex nature and circuit-level dysfunction of these conditions pose challenges for modeling, organoids could aid in revealing intrinsic cellular factors and act as platforms for testing new psychoactive or neuroprotective substances [253]. Lastly, the ability to utilize brain organoids to model lysosomal storage diseases and other genetic metabolic disorders [254] paves the way for understanding mechanisms and testing therapies. These conditions typically encompass intricate pathways of multicellular degeneration, which align well with the multicellular structure of organoid systems.
12. Conclusions
Advances in stem cell biology, CRISPR technology, and the development of complex brain organoids and assembloids, combined with advanced analysis technologies such as omics approaches and electrophysiological microelectrode arrays, are expanding our understanding of the disease mechanisms underlying a vast array of neurological and neuropsychiatric disorders. It can be expected that over the next few years, the integration of technological advances in the generation of more homogenous brain organoids with screening robotics and the expanded development of accurate disease models for a wide array of brain disorders will advance the development of drugs for neurological disorders, neuropsychiatric diseases, substance abuse disorders, rare brain diseases, and other brain disorders previously thought impossible to treat.
Author Contributions
Conceptualization, A.O.A. and S.A.L.; Writing—original draft, A.O.A. and S.A.L.; Writing—review and editing, A.O.A. and S.A.L.; Visualization, A.O.A. and S.A.L.; Funding acquisition, S.A.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Institutes of Health grant R01 CA263216 and the Nemours Foundation. This study was further supported by a supplement to a Center of Biomedical Research Excellence (COBRE) award from the National Institute of General Medical Science of the National Institutes of Health under grant number P30GM145765.
Data Availability Statement
No new data were created or analyzed in this study.
Acknowledgments
The authors thank Karen Sperle for her thoughtful suggestions and reading of the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Corro, C.; Novellasdemunt, L.; Li, V.S.W. A brief history of organoids. Am. J. Physiol. Cell Physiol. 2020, 319, C151–C165. [Google Scholar] [CrossRef]
- Qian, X.; Song, H.; Ming, G.L. Brain organoids: Advances, applications and challenges. Development 2019, 146, dev166074. [Google Scholar] [CrossRef] [PubMed]
- Pasca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Bagley, J.A.; Reumann, D.; Bian, S.; Levi-Strauss, J.; Knoblich, J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 2017, 14, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.; Lebkowski, J.; McNiece, I. Stem Cells. Biol. Blood Marrow Transplant. 2010, 16, S115–S118. [Google Scholar] [CrossRef] [PubMed]
- Poliwoda, S.; Noor, N.; Downs, E.; Schaaf, A.; Cantwell, A.; Ganti, L.; Kaye, A.D.; Mosel, L.I.; Carroll, C.B.; Viswanath, O.; et al. Stem cells: A comprehensive review of origins and emerging clinical roles in medical practice. Orthop. Rev. 2022, 14, 37498. [Google Scholar] [CrossRef]
- Daley, G.Q. Stem cells and the evolving notion of cellular identity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140376. [Google Scholar] [CrossRef]
- Kolios, G.; Moodley, Y. Introduction to stem cells and regenerative medicine. Respiration 2013, 85, 3–10. [Google Scholar] [CrossRef]
- Ilic, D.; Polak, J.M. Stem cells in regenerative medicine: Introduction. Br. Med. Bull. 2011, 98, 117–126. [Google Scholar] [CrossRef]
- De Miguel, M.P.; Fuentes-Julian, S.; Alcaina, Y. Pluripotent stem cells: Origin, maintenance and induction. Stem Cell Rev. Rep. 2010, 6, 633–649. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, M.Z.; Zuba-Surma, E.; Kucia, M.; Poniewierska, A.; Suszynska, M.; Ratajczak, J. Pluripotent and multipotent stem cells in adult tissues. Adv. Med. Sci. 2012, 57, 1–17. [Google Scholar] [CrossRef]
- Majo, F.; Rochat, A.; Nicolas, M.; Jaoude, G.A.; Barrandon, Y. Oligopotent stem cells are distributed throughout the mammalian ocular surface. Nature 2008, 456, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Bentzinger, C.F.; Wang, Y.X.; von Maltzahn, J.; Rudnicki, M.A. The emerging biology of muscle stem cells: Implications for cell-based therapies. Bioessays 2013, 35, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Golchin, A.; Chatziparasidou, A.; Ranjbarvan, P.; Niknam, Z.; Ardeshirylajimi, A. Embryonic Stem Cells in Clinical Trials: Current Overview of Developments and Challenges. Adv. Exp. Med. Biol. 2021, 1312, 19–37. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef]
- Bongso, A. Blastocyst culture for deriving human embryonic stem cells. Methods Mol. Biol. 2006, 331, 13–22. [Google Scholar] [CrossRef]
- Hambiliki, F.; Strom, S.; Zhang, P.; Stavreus-Evers, A. Co-localization of NANOG and OCT4 in human pre-implantation embryos and in human embryonic stem cells. J. Assist. Reprod. Genet. 2012, 29, 1021–1028. [Google Scholar] [CrossRef]
- Wang, Z.; Oron, E.; Nelson, B.; Razis, S.; Ivanova, N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 2012, 10, 440–454. [Google Scholar] [CrossRef]
- Baharvand, H.; Ashtiani, S.K.; Valojerdi, M.R.; Shahverdi, A.; Taee, A.; Sabour, D. Establishment and in vitro differentiation of a new embryonic stem cell line from human blastocyst. Differentiation 2004, 72, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Lezmi, E.; Jung, J.; Benvenisty, N. High prevalence of acquired cancer-related mutations in 146 human pluripotent stem cell lines and their differentiated derivatives. Nat. Biotechnol. 2024, 42, 1667–1671. [Google Scholar] [CrossRef]
- Doetschman, T.C.; Eistetter, H.; Katz, M.; Schmidt, W.; Kemler, R. The in vitro development of blastocyst-derived embryonic stem cell lines: Formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 1985, 87, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Hamazaki, T.; Iiboshi, Y.; Oka, M.; Papst, P.J.; Meacham, A.M.; Zon, L.I.; Terada, N. Hepatic maturation in differentiating embryonic stem cells in vitro. FEBS Lett. 2001, 497, 15–19. [Google Scholar] [CrossRef]
- Thoma, E.C.; Maurus, K.; Wagner, T.U.; Schartl, M. Parallel differentiation of embryonic stem cells into different cell types by a single gene-based differentiation system. Cell Reprogram. 2012, 14, 106–111. [Google Scholar] [CrossRef]
- Shiroi, A.; Ueda, S.; Ouji, Y.; Saito, K.; Moriya, K.; Sugie, Y.; Fukui, H.; Ishizaka, S.; Yoshikawa, M. Differentiation of embryonic stem cells into insulin-producing cells promoted by Nkx2.2 gene transfer. World J. Gastroenterol. 2005, 11, 4161–4166. [Google Scholar] [CrossRef]
- Heydarkhan-Hagvall, S.; Gluck, J.M.; Delman, C.; Jung, M.; Ehsani, N.; Full, S.; Shemin, R.J. The effect of vitronectin on the differentiation of embryonic stem cells in a 3D culture system. Biomaterials 2012, 33, 2032–2040. [Google Scholar] [CrossRef]
- Ludwig, T.E.; Andrews, P.W.; Barbaric, I.; Benvenisty, N.; Bhattacharyya, A.; Crook, J.M.; Daheron, L.M.; Draper, J.S.; Healy, L.E.; Huch, M.; et al. ISSCR standards for the use of human stem cells in basic research. Stem Cell Rep. 2023, 18, 1744–1752. [Google Scholar] [CrossRef] [PubMed]
- Medicine National Academies of Sciences, Engineering; Policy and Global Affairs; Law Committee on Science, Technology; Regulatory Issues Associated with Neural Chimeras and Organoids Committee on Ethical. The National Academies Collection: Reports funded by National Institutes of Health. In The Emerging Field of Human Neural Organoids, Transplants, and Chimeras: Science, Ethics, and Governance; National Academies Press (US): Washington, DC, USA, 2021. [Google Scholar]
- Sohn, Y.D.; Han, J.W.; Yoon, Y.S. Generation of induced pluripotent stem cells from somatic cells. Prog. Mol. Biol. Transl. Sci. 2012, 111, 1–26. [Google Scholar] [CrossRef]
- Omole, A.E.; Fakoya, A.O.J. Ten years of progress and promise of induced pluripotent stem cells: Historical origins, characteristics, mechanisms, limitations, and potential applications. PeerJ 2018, 6, e4370. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Judson, R.L.; Babiarz, J.E.; Venere, M.; Blelloch, R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat. Biotechnol. 2009, 27, 459–461. [Google Scholar] [CrossRef]
- Subramanyam, D.; Lamouille, S.; Judson, R.L.; Liu, J.Y.; Bucay, N.; Derynck, R.; Blelloch, R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Rais, Y.; Zviran, A.; Geula, S.; Gafni, O.; Chomsky, E.; Viukov, S.; Mansour, A.A.; Caspi, I.; Krupalnik, V.; Zerbib, M.; et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 2013, 502, 65–70. [Google Scholar] [CrossRef]
- Onder, T.T.; Kara, N.; Cherry, A.; Sinha, A.U.; Zhu, N.; Bernt, K.M.; Cahan, P.; Mancarci, B.O.; Unternaehrer, J.; Gupta, P.B.; et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature 2012, 483, 598–602. [Google Scholar] [CrossRef]
- Edel, M.J.; Menchon, C.; Menendez, S.; Consiglio, A.; Raya, A.; Izpisua Belmonte, J.C. Rem2 GTPase maintains survival of human embryonic stem cells as well as enhancing reprogramming by regulating p53 and cyclin D1. Genes. Dev. 2010, 24, 561–573. [Google Scholar] [CrossRef]
- Li, H.; Collado, M.; Villasante, A.; Strati, K.; Ortega, S.; Cañamero, M.; Blasco, M.A.; Serrano, M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 2009, 460, 1136–1139. [Google Scholar] [CrossRef]
- Temple, S. The development of neural stem cells. Nature 2001, 414, 112–117. [Google Scholar] [CrossRef]
- Merkle, F.T.; Tramontin, A.D.; Garcia-Verdugo, J.M.; Alvarez-Buylla, A. Radial glia give rise to adult neural stem cells in the subventricular zone. Proc. Natl. Acad. Sci. USA 2004, 101, 17528–17532. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Cerdeno, V.; Noctor, S.C. Neural Progenitor Cell Terminology. Front. Neuroanat. 2018, 12, 104. [Google Scholar] [CrossRef]
- Galiakberova, A.A.; Dashinimaev, E.B. Neural Stem Cells and Methods for Their Generation From Induced Pluripotent Stem Cells in vitro. Front. Cell Dev. Biol. 2020, 8, 815. [Google Scholar] [CrossRef]
- Pașca, S.P.; Arlotta, P.; Bateup, H.S.; Camp, J.G.; Cappello, S.; Gage, F.H.; Knoblich, J.A.; Kriegstein, A.R.; Lancaster, M.A.; Ming, G.L.; et al. A nomenclature consensus for nervous system organoids and assembloids. Nature 2022, 609, 907–910. [Google Scholar] [CrossRef] [PubMed]
- Birtele, M.; Lancaster, M.; Quadrato, G. Modelling human brain development and disease with organoids. Nat. Rev. Mol. Cell Biol. 2025, 26, 389–412. [Google Scholar] [CrossRef] [PubMed]
- Pașca, S.P. The rise of three-dimensional human brain cultures. Nature 2018, 553, 437–445. [Google Scholar] [CrossRef]
- Watanabe, K.; Kamiya, D.; Nishiyama, A.; Katayama, T.; Nozaki, S.; Kawasaki, H.; Watanabe, Y.; Mizuseki, K.; Sasai, Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 2005, 8, 288–296. [Google Scholar] [CrossRef]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef]
- Camp, J.G.; Badsha, F.; Florio, M.; Kanton, S.; Gerber, T.; Wilsch-Brauninger, M.; Lewitus, E.; Sykes, A.; Hevers, W.; Lancaster, M.; et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 2015, 112, 15672–15677. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Knoblich, J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014, 9, 2329–2340. [Google Scholar] [CrossRef]
- Quadrato, G.; Nguyen, T.; Macosko, E.Z.; Sherwood, J.L.; Min Yang, S.; Berger, D.R.; Maria, N.; Scholvin, J.; Goldman, M.; Kinney, J.P.; et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 2017, 545, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Andrews, M.G.; Nowakowski, T.J. Human brain development through the lens of cerebral organoid models. Brain Res. 2019, 1725, 146470. [Google Scholar] [CrossRef] [PubMed]
- Atamian, A.; Birtele, M.; Hosseini, N.; Nguyen, T.; Seth, A.; Del Dosso, A.; Paul, S.; Tedeschi, N.; Taylor, R.; Coba, M.P.; et al. Human cerebellar organoids with functional Purkinje cells. Cell Stem Cell 2024, 31, 39–51.e36. [Google Scholar] [CrossRef] [PubMed]
- Bystron, I.; Blakemore, C.; Rakic, P. Development of the human cerebral cortex: Boulder Committee revisited. Nat. Rev. Neurosci. 2008, 9, 110–122. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.V.; Lui, J.H.; Parker, P.R.; Kriegstein, A.R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 2010, 464, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Velasco, S.; Kedaigle, A.J.; Simmons, S.K.; Nash, A.; Rocha, M.; Quadrato, G.; Paulsen, B.; Nguyen, L.; Adiconis, X.; Regev, A.; et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 2019, 570, 523–527. [Google Scholar] [CrossRef]
- Sloan, S.A.; Andersen, J.; Pasca, A.M.; Birey, F.; Pasca, S.P. Generation and assembly of human brain region-specific three-dimensional cultures. Nat. Protoc. 2018, 13, 2062–2085. [Google Scholar] [CrossRef]
- Renner, M.; Lancaster, M.A.; Bian, S.; Choi, H.; Ku, T.; Peer, A.; Chung, K.; Knoblich, J.A. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017, 36, 1316–1329. [Google Scholar] [CrossRef]
- Kadoshima, T.; Sakaguchi, H.; Nakano, T.; Soen, M.; Ando, S.; Eiraku, M.; Sasai, Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. USA 2013, 110, 20284–20289. [Google Scholar] [CrossRef]
- Revah, O.; Gore, F.; Kelley, K.W.; Andersen, J.; Sakai, N.; Chen, X.; Li, M.Y.; Birey, F.; Yang, X.; Saw, N.L.; et al. Maturation and circuit integration of transplanted human cortical organoids. Nature 2022, 610, 319–326. [Google Scholar] [CrossRef]
- Ruchalski, K.; Hathout, G.M. A medley of midbrain maladies: A brief review of midbrain anatomy and syndromology for radiologists. Radiol. Res. Pract. 2012, 2012, 258524. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Jacob, F.; Song, M.M.; Nguyen, H.N.; Song, H.; Ming, G.L. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 2018, 13, 565–580. [Google Scholar] [CrossRef]
- Jo, J.; Xiao, Y.; Sun, A.X.; Cukuroglu, E.; Tran, H.D.; Goke, J.; Tan, Z.Y.; Saw, T.Y.; Tan, C.P.; Lokman, H.; et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell 2016, 19, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Monzel, A.S.; Smits, L.M.; Hemmer, K.; Hachi, S.; Moreno, E.L.; van Wuellen, T.; Jarazo, J.; Walter, J.; Bruggemann, I.; Boussaad, I.; et al. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Rep. 2017, 8, 1144–1154. [Google Scholar] [CrossRef] [PubMed]
- Smits, L.M.; Reinhardt, L.; Reinhardt, P.; Glatza, M.; Monzel, A.S.; Stanslowsky, N.; Rosato-Siri, M.D.; Zanon, A.; Antony, P.M.; Bellmann, J.; et al. Modeling Parkinson’s disease in midbrain-like organoids. npj Park. Disease 2019, 5, 5. [Google Scholar] [CrossRef]
- Sabate-Soler, S.; Nickels, S.L.; Saraiva, C.; Berger, E.; Dubonyte, U.; Barmpa, K.; Lan, Y.J.; Kouno, T.; Jarazo, J.; Robertson, G.; et al. Microglia integration into human midbrain organoids leads to increased neuronal maturation and functionality. Glia 2022, 70, 1267–1288. [Google Scholar] [CrossRef]
- Yao, X.; Kang, J.H.; Kim, K.P.; Shin, H.; Jin, Z.L.; Guo, H.; Xu, Y.N.; Li, Y.H.; Hali, S.; Kwon, J.; et al. Production of Highly Uniform Midbrain Organoids from Human Pluripotent Stem Cells. Stem Cells Int. 2023, 2023, 3320211. [Google Scholar] [CrossRef]
- Renner, H.; Grabos, M.; Becker, K.J.; Kagermeier, T.E.; Wu, J.; Otto, M.; Peischard, S.; Zeuschner, D.; TsyTsyura, Y.; Disse, P.; et al. A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. Elife 2020, 9, e52904. [Google Scholar] [CrossRef]
- Chen, M.; Niclis, J.C.; Denham, M. Protocol for generating reproducible miniaturized controlled midbrain organoids. STAR Protoc. 2023, 4, 102451. [Google Scholar] [CrossRef]
- Pavlinov, I.; Tambe, M.; Abbott, J.; Nguyen, H.N.; Xu, M.; Pradhan, M.; Farkhondeh, A.; Zheng, W. In depth characterization of midbrain organoids derived from wild type iPSC lines. PLoS ONE 2023, 18, e0292926. [Google Scholar] [CrossRef]
- Berger, E.; Magliaro, C.; Paczia, N.; Monzel, A.S.; Antony, P.; Linster, C.L.; Bolognin, S.; Ahluwalia, A.; Schwamborn, J.C. Millifluidic culture improves human midbrain organoid vitality and differentiation. Lab Chip 2018, 18, 3172–3183. [Google Scholar] [CrossRef] [PubMed]
- Melka, N.; Pszczolinska, A.; Klejbor, I.; Morys, J. The cerebellum: The ‘little’ brain and its big role. Folia Morphol. 2024, 83, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Kloth, A.D.; Badura, A. The cerebellum, sensitive periods, and autism. Neuron 2014, 83, 518–532. [Google Scholar] [CrossRef] [PubMed]
- D’Mello, A.M.; Crocetti, D.; Mostofsky, S.H.; Stoodley, C.J. Cerebellar gray matter and lobular volumes correlate with core autism symptoms. Neuroimage Clin. 2015, 7, 631–639. [Google Scholar] [CrossRef]
- Weinschutz Mendes, H.; Neelakantan, U.; Liu, Y.; Fitzpatrick, S.E.; Chen, T.; Wu, W.; Pruitt, A.; Jin, D.S.; Jamadagni, P.; Carlson, M.; et al. High-throughput functional analysis of autism genes in zebrafish identifies convergence in dopaminergic and neuroimmune pathways. Cell Rep. 2023, 42, 112243. [Google Scholar] [CrossRef]
- Ahmadi, A.; Saadatmand, M.; Wallois, F. Evaluation of potential alterations related to ADHD in the effective connectivity between the default mode network and cerebellum, hippocampus, thalamus, and primary visual cortex. Cereb. Cortex 2024, 34, bhae335. [Google Scholar] [CrossRef]
- Parkkinen, S.; Radua, J.; Andrews, D.S.; Murphy, D.; Dell’Acqua, F.; Parlatini, V. Cerebellar network alterations in adult attention-deficit/hyperactivity disorder. J. Psychiatry Neurosci. 2024, 49, E233–E241. [Google Scholar] [CrossRef]
- Tse, N.Y.; Chen, Y.; Irish, M.; Cordato, N.J.; Landin-Romero, R.; Hodges, J.R.; Piguet, O.; Ahmed, R.M. Cerebellar contributions to cognition in corticobasal syndrome and progressive supranuclear palsy. Brain Commun. 2020, 2, fcaa194. [Google Scholar] [CrossRef]
- Millard, N.E.; De Braganca, K.C. Medulloblastoma. J. Child. Neurol. 2016, 31, 1341–1353. [Google Scholar] [CrossRef]
- Haldipur, P.; Millen, K.J.; Aldinger, K.A. Human Cerebellar Development and Transcriptomics: Implications for Neurodevelopmental Disorders. Annu. Rev. Neurosci. 2022, 45, 515–531. [Google Scholar] [CrossRef]
- Stoodley, C.J. The Cerebellum and Neurodevelopmental Disorders. Cerebellum 2016, 15, 34–37. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, M.E.; Gill, J.S.; Sillitoe, R.V. Abnormal Cerebellar Development in Autism Spectrum Disorders. Dev. Neurosci. 2021, 43, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Muguruma, K.; Nishiyama, A.; Kawakami, H.; Hashimoto, K.; Sasai, Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015, 10, 537–550. [Google Scholar] [CrossRef]
- Watson, L.M.; Wong, M.M.K.; Vowles, J.; Cowley, S.A.; Becker, E.B.E. A Simplified Method for Generating Purkinje Cells from Human-Induced Pluripotent Stem Cells. Cerebellum 2018, 17, 419–427. [Google Scholar] [CrossRef]
- Silva, T.P.; Sousa-Luis, R.; Fernandes, T.G.; Bekman, E.P.; Rodrigues, C.A.V.; Vaz, S.H.; Moreira, L.M.; Hashimura, Y.; Jung, S.; Lee, B.; et al. Transcriptome profiling of human pluripotent stem cell-derived cerebellar organoids reveals faster commitment under dynamic conditions. Biotechnol. Bioeng. 2021, 118, 2781–2803. [Google Scholar] [CrossRef]
- Nayler, S.; Agarwal, D.; Curion, F.; Bowden, R.; Becker, E.B.E. High-resolution transcriptional landscape of xeno-free human induced pluripotent stem cell-derived cerebellar organoids. Sci. Rep. 2021, 11, 12959. [Google Scholar] [CrossRef]
- Hua, T.T.; Bejoy, J.; Song, L.; Wang, Z.; Zeng, Z.; Zhou, Y.; Li, Y.; Sang, Q.A. Cerebellar Differentiation from Human Stem Cells Through Retinoid, Wnt, and Sonic Hedgehog Pathways. Tissue Eng. Part A 2021, 27, 881–893. [Google Scholar] [CrossRef]
- Chen, Y.; Bury, L.A.; Chen, F.; Aldinger, K.A.; Miranda, H.C.; Wynshaw-Boris, A. Generation of advanced cerebellar organoids for neurogenesis and neuronal network development. Hum. Mol. Genet. 2023, 32, 2832–2841. [Google Scholar] [CrossRef] [PubMed]
- Atamian, A.; Birtele, M.; Hosseini, N.; Quadrato, G. Generation and long-term culture of human cerebellar organoids from pluripotent stem cells. Nat. Protoc. 2024. [Google Scholar] [CrossRef]
- Makrygianni, E.A.; Chrousos, G.P. From Brain Organoids to Networking Assembloids: Implications for Neuroendocrinology and Stress Medicine. Front. Physiol. 2021, 12, 621970. [Google Scholar] [CrossRef]
- Marton, R.M.; Pasca, S.P. Organoid and Assembloid Technologies for Investigating Cellular Crosstalk in Human Brain Development and Disease. Trends Cell Biol. 2020, 30, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Kanton, S.; Pasca, S.P. Human assembloids. Development 2022, 149, dev201120. [Google Scholar] [CrossRef] [PubMed]
- Pașca, S.P.; Arlotta, P.; Bateup, H.S.; Camp, J.G.; Cappello, S.; Gage, F.H.; Knoblich, J.A.; Kriegstein, A.R.; Lancaster, M.A.; Ming, G.L.; et al. A framework for neural organoids, assembloids and transplantation studies. Nature 2025, 639, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Cho, H. Human brain organoids in Alzheimer’s disease. Organoid 2021, 1, e5. [Google Scholar] [CrossRef]
- Holloway, P.M.; Willaime-Morawek, S.; Siow, R.; Barber, M.; Owens, R.M.; Sharma, A.D.; Rowan, W.; Hill, E.; Zagnoni, M. Advances in microfluidic in vitro systems for neurological disease modeling. J. Neurosci. Res. 2021, 99, 1276–1307. [Google Scholar] [CrossRef]
- Park, S.E.; Georgescu, A.; Huh, D. Organoids-on-a-chip. Science 2019, 364, 960–965. [Google Scholar] [CrossRef]
- Castiglione, H.; Vigneron, P.A.; Baquerre, C.; Yates, F.; Rontard, J.; Honegger, T. Human Brain Organoids-on-Chip: Advances, Challenges, and Perspectives for Preclinical Applications. Pharmaceutics 2022, 14, 2301. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, Y.; Li, Z.; Wang, H.; Li, N.; Deng, Y. Microfluidic Brain-on-a-Chip: From Key Technology to System Integration and Application. Small 2023, 19, e2304427. [Google Scholar] [CrossRef]
- Soubéran, A.; Jiguet-Jiglaire, C.; Toutain, S.; Morando, P.; Baeza-Kallee, N.; Appay, R.; Boucard, C.; Graillon, T.; Meyer, M.; Farah, K.; et al. Brain tumoroids: Treatment prediction and drug development for brain tumors with fast, reproducible, and easy-to-use personalized models. Neuro Oncol. 2025, 27, 415–429. [Google Scholar] [CrossRef]
- Xu, H.; Jiao, D.; Liu, A.; Wu, K. Tumor organoids: Applications in cancer modeling and potentials in precision medicine. J. Hematol. Oncol. 2022, 15, 58. [Google Scholar] [CrossRef]
- Xu, H.; Lyu, X.; Yi, M.; Zhao, W.; Song, Y.; Wu, K. Organoid technology and applications in cancer research. J. Hematol. Oncol. 2018, 11, 116. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Manfrin, A.; Lutolf, M.P. Progress and potential in organoid research. Nat. Rev. Genet. 2018, 19, 671–687. [Google Scholar] [CrossRef] [PubMed]
- Soubéran, A.; Tchoghandjian, A. Practical Review on Preclinical Human 3D Glioblastoma Models: Advances and Challenges for Clinical Translation. Cancers 2020, 12, 2347. [Google Scholar] [CrossRef]
- Poli, D.; Magliaro, C.; Ahluwalia, A. Experimental and Computational Methods for the Study of Cerebral Organoids: A Review. Front. Neurosci. 2019, 13, 162. [Google Scholar] [CrossRef]
- Passaro, A.P.; Stice, S.L. Electrophysiological Analysis of Brain Organoids: Current Approaches and Advancements. Front. Neurosci. 2020, 14, 622137. [Google Scholar] [CrossRef]
- Quadrato, G.; Brown, J.; Arlotta, P. The promises and challenges of human brain organoids as models of neuropsychiatric disease. Nat. Med. 2016, 22, 1220–1228+1078–8956. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Sun, L.; Fang, A.; Li, P.; Wu, Q.; Wang, X. Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein Cell 2017, 8, 823–833. [Google Scholar] [CrossRef]
- Di Lullo, E.; Kriegstein, A.R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 2017, 18, 573–584. [Google Scholar] [CrossRef]
- Sakaguchi, H.; Ozaki, Y.; Ashida, T.; Matsubara, T.; Oishi, N.; Kihara, S.; Takahashi, J. Self-Organized Synchronous Calcium Transients in a Cultured Human Neural Network Derived from Cerebral Organoids. Stem Cell Rep. 2019, 13, 458–473. [Google Scholar] [CrossRef]
- Romoser, V.A.; Hinkle, P.M.; Persechini, A. Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence. A new class of fluorescent indicators. J. Biol. Chem. 1997, 272, 13270–13274. [Google Scholar] [CrossRef]
- Shafer, T.J.; Brown, J.P.; Lynch, B.; Davila-Montero, S.; Wallace, K.; Friedman, K.P. Evaluation of Chemical Effects on Network Formation in Cortical Neurons Grown on Microelectrode Arrays. Toxicol. Sci. 2019, 169, 436–455. [Google Scholar] [CrossRef] [PubMed]
- Frank, C.L.; Brown, J.P.; Wallace, K.; Mundy, W.R.; Shafer, T.J. From the Cover: Developmental Neurotoxicants Disrupt Activity in Cortical Networks on Microelectrode Arrays: Results of Screening 86 Compounds During Neural Network Formation. Toxicol. Sci. 2017, 160, 121–135. [Google Scholar] [CrossRef] [PubMed]
- Cotterill, E.; Hall, D.; Wallace, K.; Mundy, W.R.; Eglen, S.J.; Shafer, T.J. Characterization of Early Cortical Neural Network Development in Multiwell Microelectrode Array Plates. J. Biomol. Screen. 2016, 21, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Poli, D.; Wheeler, B.C.; Demarse, T.B.; Brewer, G.J. Pattern separation and completion of distinct axonal inputs transmitted via micro-tunnels between co-cultured hippocampal dentate, CA3, CA1 and entorhinal cortex networks. J. Neural Eng. 2018, 15, 046009. [Google Scholar] [CrossRef]
- Berdondini, L.; Imfeld, K.; Maccione, A.; Tedesco, M.; Neukom, S.; Koudelka-Hep, M.; Martinoia, S. Active pixel sensor array for high spatio-temporal resolution electrophysiological recordings from single cell to large scale neuronal networks. Lab Chip 2009, 9, 2644–2651. [Google Scholar] [CrossRef]
- Giandomenico, S.L.; Mierau, S.B.; Gibbons, G.M.; Wenger, L.M.D.; Masullo, L.; Sit, T.; Sutcliffe, M.; Boulanger, J.; Tripodi, M.; Derivery, E.; et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 2019, 22, 669–679. [Google Scholar] [CrossRef]
- Soscia, D.A.; Lam, D.; Tooker, A.C.; Enright, H.A.; Triplett, M.; Karande, P.; Peters, S.K.G.; Sales, A.P.; Wheeler, E.K.; Fischer, N.O. A flexible 3-dimensional microelectrode array for in vitro brain models. Lab Chip 2020, 20, 901–911. [Google Scholar] [CrossRef]
- Shin, H.; Jeong, S.; Lee, J.H.; Sun, W.; Choi, N.; Cho, I.J. 3D high-density microelectrode array with optical stimulation and drug delivery for investigating neural circuit dynamics. Nat. Commun. 2021, 12, 492. [Google Scholar] [CrossRef]
- Shiri, Z.; Simorgh, S.; Naderi, S.; Baharvand, H. Optogenetics in the Era of Cerebral Organoids. Trends Biotechnol. 2019, 37, 1282–1294. [Google Scholar] [CrossRef]
- Hochbaum, D.R.; Zhao, Y.; Farhi, S.L.; Klapoetke, N.; Werley, C.A.; Kapoor, V.; Zou, P.; Kralj, J.M.; Maclaurin, D.; Smedemark-Margulies, N.; et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat. Methods 2014, 11, 825–833. [Google Scholar] [CrossRef]
- Werley, C.A.; Brookings, T.; Upadhyay, H.; Williams, L.A.; McManus, O.B.; Dempsey, G.T. All-Optical Electrophysiology for Disease Modeling and Pharmacological Characterization of Neurons. Curr. Protoc. Pharmacol. 2017, 11, 825–833. [Google Scholar] [CrossRef]
- Kiskinis, E.; Kralj, J.M.; Zou, P.; Weinstein, E.N.; Zhang, H.; Tsioras, K.; Wiskow, O.; Ortega, J.A.; Eggan, K.; Cohen, A.E. All-Optical Electrophysiology for High-Throughput Functional Characterization of a Human iPSC-Derived Motor Neuron Model of ALS. Stem Cell Rep. 2018, 10, 1991–2004. [Google Scholar] [CrossRef] [PubMed]
- Versace, A.; Hitchens, T.K.; Wallace, C.T.; Watkins, S.C.; D’Aiuto, L. 11.7T Diffusion Magnetic Resonance Imaging and Tractography to Probe Human Brain Organoid Microstructure. Biol. Psychiatry Glob. Open Sci. 2024, 4, 100344. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, C.A.; Muotri, A.R. Brain Organoids and the Study of Neurodevelopment. Trends Mol. Med. 2018, 24, 982–990. [Google Scholar] [CrossRef]
- Gordon, A.; Geschwind, D.H. Human in vitro models for understanding mechanisms of autism spectrum disorder. Mol. Autism 2020, 11, 26. [Google Scholar] [CrossRef]
- Ardhanareeswaran, K.; Mariani, J.; Coppola, G.; Abyzov, A.; Vaccarino, F.M. Human induced pluripotent stem cells for modelling neurodevelopmental disorders. Nat. Rev. Neurol. 2017, 13, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Weller, M.; Wen, P.Y.; Chang, S.M.; Dirven, L.; Lim, M.; Monje, M.; Reifenberger, G. Glioma. Nat. Rev. Dis. Primers 2024, 10, 33. [Google Scholar] [CrossRef]
- Gusyatiner, O.; Hegi, M.E. Glioma epigenetics: From subclassification to novel treatment options. Semin. Cancer Biol. 2018, 51, 50–58. [Google Scholar] [CrossRef]
- Wirsching, H.G.; Galanis, E.; Weller, M. Glioblastoma. Handb. Clin. Neurol. 2016, 134, 381–397. [Google Scholar] [CrossRef]
- Bandopadhayay, P.; Bergthold, G.; London, W.B.; Goumnerova, L.C.; Morales La Madrid, A.; Marcus, K.J.; Guo, D.; Ullrich, N.J.; Robison, N.J.; Chi, S.N.; et al. Long-term outcome of 4,040 children diagnosed with pediatric low-grade gliomas: An analysis of the Surveillance Epidemiology and End Results (SEER) database. Pediatr. Blood Cancer 2014, 61, 1173–1179. [Google Scholar] [CrossRef]
- Qaddoumi, I.; Sultan, I.; Gajjar, A. Outcome and prognostic features in pediatric gliomas: A review of 6212 cases from the Surveillance, Epidemiology, and End Results database. Cancer 2009, 115, 5761–5770. [Google Scholar] [CrossRef] [PubMed]
- Mackay, A.; Burford, A.; Carvalho, D.; Izquierdo, E.; Fazal-Salom, J.; Taylor, K.R.; Bjerke, L.; Clarke, M.; Vinci, M.; Nandhabalan, M.; et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017, 32, 520–537.e5. [Google Scholar] [CrossRef] [PubMed]
- Rybin, M.J.; Ivan, M.E.; Ayad, N.G.; Zeier, Z. Organoid Models of Glioblastoma and Their Role in Drug Discovery. Front. Cell Neurosci. 2021, 15, 605255. [Google Scholar] [CrossRef]
- Ishahak, M.; Han, R.H.; Annamalai, D.; Woodiwiss, T.; McCornack, C.; Cleary, R.T.; DeSouza, P.A.; Qu, X.; Dahiya, S.; Kim, A.H.; et al. Genetically Engineered Brain Organoids Recapitulate Spatial and Developmental States of Glioblastoma Progression. Adv. Sci. 2025, 12, e2410110. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, J.; Wang, S.; Guo, P.; Liao, K.; Shi, Z.; Zhao, J.; Lin, S.; Yang, M.; Cai, G.; et al. Generation and banking of patient-derived glioblastoma organoid and its application in cancer neuroscience. Am. J. Cancer Res. 2024, 14, 5000–5010. [Google Scholar] [CrossRef]
- Sarnow, K.; Majercak, E.; Qurbonov, Q.; Cruzeiro, G.A.V.; Jeong, D.; Haque, I.A.; Khalil, A.; Baird, L.C.; Filbin, M.G.; Tang, X. Neuroimmune-competent human brain organoid model of diffuse midline glioma. Neuro Oncol. 2025, 27, 369–382. [Google Scholar] [CrossRef] [PubMed]
- van Essen, M.J.; Apsley, E.J.; Riepsaame, J.; Xu, R.; Northcott, P.A.; Cowley, S.A.; Jacob, J.; Becker, E.B.E. PTCH1-mutant human cerebellar organoids exhibit altered neural development and recapitulate early medulloblastoma tumorigenesis. Dis. Model. Mech. 2024, 17, dmm050323. [Google Scholar] [CrossRef]
- van Essen, M.J.; Nicheperovich, A.; Schuster-Bockler, B.; Becker, E.B.E.; Jacob, J. Sonic hedgehog medulloblastoma cells in co-culture with cerebellar organoids converge towards in vivo malignant cell states. Neurooncol. Adv. 2025, 7, vdae218. [Google Scholar] [CrossRef]
- Ballabio, C.; Anderle, M.; Gianesello, M.; Lago, C.; Miele, E.; Cardano, M.; Aiello, G.; Piazza, S.; Caron, D.; Gianno, F.; et al. Modeling medulloblastoma in vivo and with human cerebellar organoids. Nat. Commun. 2020, 11, 583. [Google Scholar] [CrossRef]
- Han, X.; He, Y.; Wang, Y.; Hu, W.; Chu, C.; Huang, L.; Hong, Y.; Han, L.; Zhang, X.; Gao, Y.; et al. Deficiency of FABP7 Triggers Premature Neural Differentiation in Idiopathic Normocephalic Autism Organoids. Adv. Sci. 2025, 12, e2406849. [Google Scholar] [CrossRef]
- Chalkiadaki, K.; Statoulla, E.; Zafeiri, M.; Voudouri, G.; Amvrosiadis, T.; Typou, A.; Theodoridou, N.; Moschovas, D.; Avgeropoulos, A.; Samiotaki, M.; et al. GABA/Glutamate Neuron Differentiation Imbalance and Increased AKT/mTOR Signaling in CNTNAP2(-/-) Cerebral Organoids. Biol. Psychiatry Glob. Open Sci. 2025, 5, 100413. [Google Scholar] [CrossRef] [PubMed]
- Tanabe, M.; Saito, Y.; Takasaki, A.; Nakano, K.; Yamamoto, S.; Suzuki, C.; Kawamura, N.; Hattori, A.; Oikawa, M.; Nagashima, S.; et al. Role of immature choroid plexus in the pathology of model mice and human iPSC-derived organoids with autism spectrum disorder. Cell Rep. 2025, 44, 115133. [Google Scholar] [CrossRef] [PubMed]
- Juan, C.X.; Mao, Y.; Han, X.; Qian, H.Y.; Chu, K.K. EGR1 Regulates SHANK3 Transcription at Different Stages of Brain Development. Neuroscience 2024, 540, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Zhang, J.; Chen, X.; Wettschurack, K.; Que, Z.; Deming, B.A.; Olivero-Acosta, M.I.; Cui, N.; Eaton, M.; Zhao, Y.; et al. Microglial over-pruning of synapses during development in autism-associated SCN2A-deficient mice and human cerebral organoids. Mol. Psychiatry 2024, 29, 2424–2437. [Google Scholar] [CrossRef]
- Kang, S.C.; Sarn, N.B.; Venegas, J.; Tan, Z.; Hitomi, M.; Eng, C. Germline PTEN genotype-dependent phenotypic divergence during the early neural developmental process of forebrain organoids. Mol. Psychiatry 2024, 29, 1767–1781. [Google Scholar] [CrossRef]
- Villa, C.E.; Cheroni, C.; Dotter, C.P.; Lopez-Tobon, A.; Oliveira, B.; Sacco, R.; Yahya, A.C.; Morandell, J.; Gabriele, M.; Tavakoli, M.R.; et al. CHD8 haploinsufficiency links autism to transient alterations in excitatory and inhibitory trajectories. Cell Rep. 2022, 39, 110615. [Google Scholar] [CrossRef]
- Jourdon, A.; Wu, F.; Mariani, J.; Capauto, D.; Norton, S.; Tomasini, L.; Amiri, A.; Suvakov, M.; Schreiner, J.D.; Jang, Y.; et al. Modeling idiopathic autism in forebrain organoids reveals an imbalance of excitatory cortical neuron subtypes during early neurogenesis. Nat. Neurosci. 2023, 26, 1505–1515. [Google Scholar] [CrossRef]
- Birtele, M.; Del Dosso, A.; Xu, T.; Nguyen, T.; Wilkinson, B.; Hosseini, N.; Nguyen, S.; Urenda, J.P.; Knight, G.; Rojas, C.; et al. Non-synaptic function of the autism spectrum disorder-associated gene SYNGAP1 in cortical neurogenesis. Nat. Neurosci. 2023, 26, 2090–2103. [Google Scholar] [CrossRef]
- Lu, R.; Xu, Y.; Li, H.; Xiong, M.; Zhou, W.; Feng, W.; Zhao, R. Identifying the Pathogenicity of a Novel NPRL3 Missense Mutation Using Personalized Cortical Organoid Model of Focal Cortical Dysplasia. J. Mol. Neurosci. 2024, 75, 3. [Google Scholar] [CrossRef]
- Niu, W.; Deng, L.; Mojica-Perez, S.P.; Tidball, A.M.; Sudyk, R.; Stokes, K.; Parent, J.M. Abnormal cell sorting and altered early neurogenesis in a human cortical organoid model of Protocadherin-19 clustering epilepsy. Front. Cell Neurosci. 2024, 18, 1339345. [Google Scholar] [CrossRef]
- Muller, Y.; Lengacher, L.; Friscourt, F.; Quairiaux, C.; Stoppini, L.; Magistretti, P.J.; Lengacher, S.; Finsterwald, C. Epileptiform activity in brain organoids derived from patient with Glucose Transporter 1 Deficiency Syndrome. Front. Neurosci. 2024, 18, 1498801. [Google Scholar] [CrossRef] [PubMed]
- Eichmuller, O.L.; Corsini, N.S.; Vertesy, A.; Morassut, I.; Scholl, T.; Gruber, V.E.; Peer, A.M.; Chu, J.; Novatchkova, M.; Hainfellner, J.A.; et al. Amplification of human interneuron progenitors promotes brain tumors and neurological defects. Science 2022, 375, eabf5546. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Guo, R.; Ye, X.; Tang, S.; Chen, M.; Zhou, P.; Yang, M.; Liao, C.; Li, H.; Lin, B.; et al. Wybutosine hypomodification of tRNAphe activates HERVK and impairs neuronal differentiation. iScience 2024, 27, 109748. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, E.M.; Means, R.E.; Michaud, M.; Madri, J.A.; Katz, S.G. Minocycline mitigates the effect of neonatal hypoxic insult on human brain organoids. Cell Death Dis. 2019, 10, 325. [Google Scholar] [CrossRef]
- Zhang, D.; Eguchi, N.; Okazaki, S.; Sora, I.; Hishimoto, A. Telencephalon Organoids Derived from an Individual with ADHD Show Altered Neurodevelopment of Early Cortical Layer Structure. Stem Cell Rev. Rep. 2023, 19, 1482–1491. [Google Scholar] [CrossRef]
- Miura, Y.; Li, M.Y.; Birey, F.; Ikeda, K.; Revah, O.; Thete, M.V.; Park, J.Y.; Puno, A.; Lee, S.H.; Porteus, M.H.; et al. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol. 2020, 38, 1421–1430. [Google Scholar] [CrossRef]
- Wu, S.; Hong, Y.; Chu, C.; Gan, Y.; Li, X.; Tao, M.; Wang, D.; Hu, H.; Zheng, Z.; Zhu, Q.; et al. Construction of human 3D striato-nigral assembloids to recapitulate medium spiny neuronal projection defects in Huntington’s disease. Proc. Natl. Acad. Sci. USA 2024, 121, e2316176121. [Google Scholar] [CrossRef]
- Andersen, J.; Revah, O.; Miura, Y.; Thom, N.; Amin, N.D.; Kelley, K.W.; Singh, M.; Chen, X.; Thete, M.V.; Walczak, E.M.; et al. Generation of Functional Human 3D Cortico-Motor Assembloids. Cell 2020, 183, 1913–1929.e26. [Google Scholar] [CrossRef]
- Meng, X.; Yao, D.; Imaizumi, K.; Chen, X.; Kelley, K.W.; Reis, N.; Thete, M.V.; Arjun McKinney, A.; Kulkarni, S.; Panagiotakos, G.; et al. Assembloid CRISPR screens reveal impact of disease genes in human neurodevelopment. Nature 2023, 622, 359–366. [Google Scholar] [CrossRef]
- Kalpana, K.; Rao, C.; Semrau, S.; Zhang, B.; Noggle, S.; Fossati, V. Generating Neuroimmune Assembloids Using Human Induced Pluripotent Stem Cell (iPSC)-Derived Cortical Organoids and Microglia. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2024. [Google Scholar] [CrossRef]
- Barmpa, K.; Saraiva, C.; Lopez-Pigozzi, D.; Gomez-Giro, G.; Gabassi, E.; Spitz, S.; Brandauer, K.; Rodriguez Gatica, J.E.; Antony, P.; Robertson, G.; et al. Modeling early phenotypes of Parkinson’s disease by age-induced midbrain-striatum assembloids. Commun. Biol. 2024, 7, 1561. [Google Scholar] [CrossRef]
- Patton, M.H.; Thomas, K.T.; Bayazitov, I.T.; Newman, K.D.; Kurtz, N.B.; Robinson, C.G.; Ramirez, C.A.; Trevisan, A.J.; Bikoff, J.B.; Peters, S.T.; et al. Synaptic plasticity in human thalamocortical assembloids. Cell Rep. 2024, 43, 114503. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Miura, Y.; Li, M.Y.; Revah, O.; Selvaraj, S.; Birey, F.; Meng, X.; Thete, M.V.; Pavlov, S.D.; Andersen, J.; et al. Human assembloids reveal the consequences of CACNA1G gene variants in the thalamocortical pathway. Neuron 2024, 112, 4048–4059.e4047. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Park, K.H.; Kim, D.H.; Kim, N.G.; Lee, S.E.; Shin, N.; Kook, M.G.; Kim, Y.B.; Kang, K.S. Cortical-blood vessel assembloids exhibit Alzheimer’s disease phenotypes by activating glia after SARS-CoV-2 infection. Cell Death Discov. 2023, 9, 32. [Google Scholar] [CrossRef]
- Kim, J.I.; Imaizumi, K.; Jurjut, O.; Kelley, K.W.; Wang, D.; Thete, M.V.; Hudacova, Z.; Amin, N.D.; Levy, R.J.; Scherrer, G.; et al. Human assembloid model of the ascending neural sensory pathway. Nature 2025. [Google Scholar] [CrossRef]
- Correia, C.D.; Calado, S.M.; Matos, A.; Esteves, F.; De Sousa-Coelho, A.L.; Campinho, M.A.; Fernandes, M.T. Advancing Glioblastoma Research with Innovative Brain Organoid-Based Models. Cells 2025, 14, 292. [Google Scholar] [CrossRef] [PubMed]
- Khamis, Z.I.; Sarker, D.B.; Xue, Y.; Al-Akkary, N.; James, V.D.; Zeng, C.; Li, Y.; Sang, Q.A. Modeling Human Brain Tumors and the Microenvironment Using Induced Pluripotent Stem Cells. Cancers 2023, 15, 1253. [Google Scholar] [CrossRef]
- Deligne, C.; Tourbez, A.; Bénard, F.; Meyer, S.; Curt, A.; Gianesello, M.; Hamadou, M.; Clavier, L.; Coquet, C.; Bocquet, C.; et al. Establishing a living biobank of pediatric high-grade glioma and ependymoma suitable for cancer pharmacology. Neuro Oncol. 2025. [Google Scholar] [CrossRef]
- Rutkowski, S.; von Hoff, K.; Emser, A.; Zwiener, I.; Pietsch, T.; Figarella-Branger, D.; Giangaspero, F.; Ellison, D.W.; Garre, M.L.; Biassoni, V.; et al. Survival and prognostic factors of early childhood medulloblastoma: An international meta-analysis. J. Clin. Oncol. 2010, 28, 4961–4968. [Google Scholar] [CrossRef]
- Northcott, P.A.; Robinson, G.W.; Kratz, C.P.; Mabbott, D.J.; Pomeroy, S.L.; Clifford, S.C.; Rutkowski, S.; Ellison, D.W.; Malkin, D.; Taylor, M.D.; et al. Medulloblastoma. Nat. Rev. Dis. Primers 2019, 5, 11. [Google Scholar] [CrossRef]
- Northcott, P.A.; Jones, D.T.; Kool, M.; Robinson, G.W.; Gilbertson, R.J.; Cho, Y.J.; Pomeroy, S.L.; Korshunov, A.; Lichter, P.; Taylor, M.D.; et al. Medulloblastomics: The end of the beginning. Nat. Rev. Cancer 2012, 12, 818–834. [Google Scholar] [CrossRef]
- Eyring, K.W.; Geschwind, D.H. Three decades of ASD genetics: Building a foundation for neurobiological understanding and treatment. Hum. Mol. Genet. 2021, 30, R236–R244. [Google Scholar] [CrossRef]
- Parikshak, N.N.; Swarup, V.; Belgard, T.G.; Irimia, M.; Ramaswami, G.; Gandal, M.J.; Hartl, C.; Leppa, V.; Ubieta, L.T.; Huang, J.; et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 2016, 540, 423–427. [Google Scholar] [CrossRef]
- Lamanna, J.; Meldolesi, J. Autism Spectrum Disorder: Brain Areas Involved, Neurobiological Mechanisms, Diagnoses and Therapies. Int. J. Mol. Sci. 2024, 25, 2423. [Google Scholar] [CrossRef] [PubMed]
- Abraham, J.R.; Szoko, N.; Barnard, J.; Rubin, R.A.; Schlatzer, D.; Lundberg, K.; Li, X.; Natowicz, M.R. Proteomic Investigations of Autism Brain Identify Known and Novel Pathogenetic Processes. Sci. Rep. 2019, 9, 13118. [Google Scholar] [CrossRef] [PubMed]
- Parenti, I.; Rabaneda, L.G.; Schoen, H.; Novarino, G. Neurodevelopmental Disorders: From Genetics to Functional Pathways. Trends Neurosci. 2020, 43, 608–621. [Google Scholar] [CrossRef] [PubMed]
- Leblond, C.S.; Le, T.L.; Malesys, S.; Cliquet, F.; Tabet, A.C.; Delorme, R.; Rolland, T.; Bourgeron, T. Operative list of genes associated with autism and neurodevelopmental disorders based on database review. Mol. Cell Neurosci. 2021, 113, 103623. [Google Scholar] [CrossRef]
- Paulsen, B.; Velasco, S.; Kedaigle, A.J.; Pigoni, M.; Quadrato, G.; Deo, A.J.; Adiconis, X.; Uzquiano, A.; Sartore, R.; Yang, S.M.; et al. Autism genes converge on asynchronous development of shared neuron classes. Nature 2022, 602, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Strauss, K.A.; Puffenberger, E.G.; Huentelman, M.J.; Gottlieb, S.; Dobrin, S.E.; Parod, J.M.; Stephan, D.A.; Morton, D.H. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N. Engl. J. Med. 2006, 354, 1370–1377. [Google Scholar] [CrossRef] [PubMed]
- Imayoshi, I.; Shimogori, T.; Ohtsuka, T.; Kageyama, R. Hes genes and neurogenin regulate non-neural versus neural fate specification in the dorsal telencephalic midline. Development 2008, 135, 2531–2541. [Google Scholar] [CrossRef]
- Sanders, S.J.; Campbell, A.J.; Cottrell, J.R.; Moller, R.S.; Wagner, F.F.; Auldridge, A.L.; Bernier, R.A.; Catterall, W.A.; Chung, W.K.; Empfield, J.R.; et al. Progress in Understanding and Treating SCN2A-Mediated Disorders. Trends Neurosci. 2018, 41, 442–456. [Google Scholar] [CrossRef]
- Santos, J.L.S.; Araújo, C.A.; Rocha, C.A.G.; Costa-Ferro, Z.S.M.; Souza, B.S.F. Modeling Autism Spectrum Disorders with Induced Pluripotent Stem Cell-Derived Brain Organoids. Biomolecules 2023, 13, 260. [Google Scholar] [CrossRef] [PubMed]
- Genovese, A.; Butler, M.G. The Autism Spectrum: Behavioral, Psychiatric and Genetic Associations. Genes 2023, 14, 677. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.T.; Chan, Y.; Dawes, P.; Guo, X.; Erdin, S.; Tai, D.J.C.; Liu, S.; Reichert, J.M.; Burns, M.J.; Chan, Y.K.; et al. Orgo-Seq integrates single-cell and bulk transcriptomic data to identify cell type specific-driver genes associated with autism spectrum disorder. Nat. Commun. 2022, 13, 3243. [Google Scholar] [CrossRef]
- Mariani, J.; Coppola, G.; Zhang, P.; Abyzov, A.; Provini, L.; Tomasini, L.; Amenduni, M.; Szekely, A.; Palejev, D.; Wilson, M.; et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 2015, 162, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Mellios, N.; Feldman, D.A.; Sheridan, S.D.; Ip, J.P.K.; Kwok, S.; Amoah, S.K.; Rosen, B.; Rodriguez, B.A.; Crawford, B.; Swaminathan, R.; et al. MeCP2-regulated miRNAs control early human neurogenesis through differential effects on ERK and AKT signaling. Mol. Psychiatry 2018, 23, 1051–1065. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Zhang, W.; Wang, X.; Jiao, C.; Xu, S.; Liu, C.; Tang, B.; Chen, C. Human forebrain organoids reveal connections between valproic acid exposure and autism risk. Transl. Psychiatry 2022, 12, 130. [Google Scholar] [CrossRef]
- Modafferi, S.; Zhong, X.; Kleensang, A.; Murata, Y.; Fagiani, F.; Pamies, D.; Hogberg, H.T.; Calabrese, V.; Lachman, H.; Hartung, T.; et al. Gene-Environment Interactions in Developmental Neurotoxicity: A Case Study of Synergy between Chlorpyrifos and CHD8 Knockout in Human BrainSpheres. Environ. Health Perspect. 2021, 129, 77001. [Google Scholar] [CrossRef]
- Pearson, G.; Song, C.; Hohmann, S.; Prokhorova, T.; Sheldrick-Michel, T.M.; Knöpfel, T. DNA Methylation Profiles of GAD1 in Human Cerebral Organoids of Autism Indicate Disrupted Epigenetic Regulation during Early Development. Int. J. Mol. Sci. 2022, 23, 9188. [Google Scholar] [CrossRef]
- Trujillo, C.A.; Adams, J.W.; Negraes, P.D.; Carromeu, C.; Tejwani, L.; Acab, A.; Tsuda, B.; Thomas, C.A.; Sodhi, N.; Fichter, K.M.; et al. Pharmacological reversal of synaptic and network pathology in human MECP2-KO neurons and cortical organoids. EMBO Mol. Med. 2021, 13, e12523. [Google Scholar] [CrossRef]
- Urresti, J.; Zhang, P.; Moran-Losada, P.; Yu, N.K.; Negraes, P.D.; Trujillo, C.A.; Antaki, D.; Amar, M.; Chau, K.; Pramod, A.B.; et al. Cortical organoids model early brain development disrupted by 16p11.2 copy number variants in autism. Mol. Psychiatry 2021, 26, 7560–7580. [Google Scholar] [CrossRef]
- Wang, P.; Mokhtari, R.; Pedrosa, E.; Kirschenbaum, M.; Bayrak, C.; Zheng, D.; Lachman, H.M. CRISPR/Cas9-mediated heterozygous knockout of the autism gene CHD8 and characterization of its transcriptional networks in cerebral organoids derived from iPS cells. Mol. Autism 2017, 8, 11. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chiola, S.; Yang, G.; Russell, C.; Armstrong, C.J.; Wu, Y.; Spampanato, J.; Tarboton, P.; Ullah, H.M.A.; Edgar, N.U.; et al. Modeling human telencephalic development and autism-associated SHANK3 deficiency using organoids generated from single neural rosettes. Nat. Commun. 2022, 13, 5688. [Google Scholar] [CrossRef] [PubMed]
- Wegscheid, M.L.; Anastasaki, C.; Hartigan, K.A.; Cobb, O.M.; Papke, J.B.; Traber, J.N.; Morris, S.M.; Gutmann, D.H. Patient-derived iPSC-cerebral organoid modeling of the 17q11.2 microdeletion syndrome establishes CRLF3 as a critical regulator of neurogenesis. Cell Rep. 2021, 36, 109315. [Google Scholar] [CrossRef]
- Zhang, W.; Ma, L.; Yang, M.; Shao, Q.; Xu, J.; Lu, Z.; Zhao, Z.; Chen, R.; Chai, Y.; Chen, J.F. Cerebral organoid and mouse models reveal a RAB39b-PI3K-mTOR pathway-dependent dysregulation of cortical development leading to macrocephaly/autism phenotypes. Genes Dev. 2020, 34, 580–597. [Google Scholar] [CrossRef]
- Murray, C.J.; Vos, T.; Lozano, R.; Naghavi, M.; Flaxman, A.D.; Michaud, C.; Ezzati, M.; Shibuya, K.; Salomon, J.A.; Abdalla, S.; et al. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: A systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012, 380, 2197–2223. [Google Scholar] [CrossRef]
- Grone, B.P.; Baraban, S.C. Animal models in epilepsy research: Legacies and new directions. Nat. Neurosci. 2015, 18, 339–343. [Google Scholar] [CrossRef]
- Nolan, D.; Fink, J. Genetics of epilepsy. Handb. Clin. Neurol. 2018, 148, 467–491. [Google Scholar] [CrossRef]
- Brodie, M.J.; Barry, S.J.; Bamagous, G.A.; Norrie, J.D.; Kwan, P. Patterns of treatment response in newly diagnosed epilepsy. Neurology 2012, 78, 1548–1554. [Google Scholar] [CrossRef] [PubMed]
- Holmes, G.L. Effect of Seizures on the Developing Brain and Cognition. Semin. Pediatr. Neurol. 2016, 23, 120–126. [Google Scholar] [CrossRef]
- Miller, D.J.; Bhaduri, A.; Sestan, N.; Kriegstein, A. Shared and derived features of cellular diversity in the human cerebral cortex. Curr. Opin. Neurobiol. 2019, 56, 117–124. [Google Scholar] [CrossRef]
- Zeng, H.; Shen, E.H.; Hohmann, J.G.; Oh, S.W.; Bernard, A.; Royall, J.J.; Glattfelder, K.J.; Sunkin, S.M.; Morris, J.A.; Guillozet-Bongaarts, A.L.; et al. Large-scale cellular-resolution gene profiling in human neocortex reveals species-specific molecular signatures. Cell 2012, 149, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Barkovich, A.J.; Guerrini, R.; Kuzniecky, R.I.; Jackson, G.D.; Dobyns, W.B. A developmental and genetic classification for malformations of cortical development: Update 2012. Brain 2012, 135, 1348–1369. [Google Scholar] [CrossRef]
- Semah, F.; Picot, M.C.; Adam, C.; Broglin, D.; Arzimanoglou, A.; Bazin, B.; Cavalcanti, D.; Baulac, M. Is the underlying cause of epilepsy a major prognostic factor for recurrence? Neurology 1998, 51, 1256–1262. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.S.; Gopalappa, R.; Kim, S.H.; Ramakrishna, S.; Lee, M.; Kim, W.I.; Kim, J.; Park, S.M.; Lee, J.; Oh, J.H.; et al. Somatic Mutations in TSC1 and TSC2 Cause Focal Cortical Dysplasia. Am. J. Hum. Genet. 2017, 100, 454–472. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, M.; Saitsu, H.; Takei, N.; Tohyama, J.; Kato, M.; Kitaura, H.; Shiina, M.; Shirozu, H.; Masuda, H.; Watanabe, K.; et al. Somatic Mutations in the MTOR gene cause focal cortical dysplasia type IIb. Ann. Neurol. 2015, 78, 375–386. [Google Scholar] [CrossRef]
- Ribierre, T.; Deleuze, C.; Bacq, A.; Baldassari, S.; Marsan, E.; Chipaux, M.; Muraca, G.; Roussel, D.; Navarro, V.; Leguern, E.; et al. Second-hit mosaic mutation in mTORC1 repressor DEPDC5 causes focal cortical dysplasia-associated epilepsy. J. Clin. Investig. 2018, 128, 2452–2458. [Google Scholar] [CrossRef]
- Dibbens, L.M.; Tarpey, P.S.; Hynes, K.; Bayly, M.A.; Scheffer, I.E.; Smith, R.; Bomar, J.; Sutton, E.; Vandeleur, L.; Shoubridge, C.; et al. X-linked protocadherin 19 mutations cause female-limited epilepsy and cognitive impairment. Nat. Genet. 2008, 40, 776–781. [Google Scholar] [CrossRef]
- Leen, W.G.; Klepper, J.; Verbeek, M.M.; Leferink, M.; Hofste, T.; van Engelen, B.G.; Wevers, R.A.; Arthur, T.; Bahi-Buisson, N.; Ballhausen, D.; et al. Glucose transporter-1 deficiency syndrome: The expanding clinical and genetic spectrum of a treatable disorder. Brain 2010, 133, 655–670. [Google Scholar] [CrossRef]
- Pearson, T.S.; Akman, C.; Hinton, V.J.; Engelstad, K.; De Vivo, D.C. Phenotypic spectrum of glucose transporter type 1 deficiency syndrome (Glut1 DS). Curr. Neurol. Neurosci. Rep. 2013, 13, 342. [Google Scholar] [CrossRef]
- Thiele, E.A. Managing and understanding epilepsy in tuberous sclerosis complex. Epilepsia 2010, 51 (Suppl. S1), 90–91. [Google Scholar] [CrossRef]
- Blair, J.D.; Hockemeyer, D.; Bateup, H.S. Genetically engineered human cortical spheroid models of tuberous sclerosis. Nat. Med. 2018, 24, 1568–1578. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Yao, H.; Negraes, P.D.; Wang, J.; Trujillo, C.A.; de Souza, J.S.; Muotri, A.R.; Haddad, G.G. Neuronal hyperexcitability and ion channel dysfunction in CDKL5-deficiency patient iPSC-derived cortical organoids. Neurobiol. Dis. 2022, 174, 105882. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.X.; Yuan, Q.; Fukuda, M.; Yu, W.; Yan, H.; Lim, G.G.Y.; Nai, M.H.; D’Agostino, G.A.; Tran, H.D.; Itahana, Y.; et al. Potassium channel dysfunction in human neuronal models of Angelman syndrome. Science 2019, 366, 1486–1492. [Google Scholar] [CrossRef]
- Repudi, S.; Steinberg, D.J.; Elazar, N.; Breton, V.L.; Aquilino, M.S.; Saleem, A.; Abu-Swai, S.; Vainshtein, A.; Eshed-Eisenbach, Y.; Vijayaragavan, B.; et al. Neuronal deletion of Wwox, associated with WOREE syndrome, causes epilepsy and myelin defects. Brain 2021, 144, 3061–3077. [Google Scholar] [CrossRef] [PubMed]
- Schiariti, V.; Shierk, A.; Stashinko, E.E.; Sukal-Moulton, T.; Feldman, R.S.; Aman, C.; Mendoza-Puccini, M.C.; Brandenburg, J.E.; National Institute of Neurological Disorders; Stroke Cerebral Palsy Common Data Elements Oversight Committee. Cerebral palsy pain instruments: Recommended tools for clinical research studies by the National Institute of Neurological Disorders and Stroke Cerebral Palsy Common Data Elements project. Dev. Med. Child. Neurol. 2024, 66, 610–622. [Google Scholar] [CrossRef]
- Vitrikas, K.; Dalton, H.; Breish, D. Cerebral Palsy: An Overview. Am. Fam. Physician 2020, 101, 213–220. [Google Scholar]
- Sadowska, M.; Sarecka-Hujar, B.; Kopyta, I. Cerebral Palsy: Current Opinions on Definition, Epidemiology, Risk Factors, Classification and Treatment Options. Neuropsychiatr. Dis. Treat. 2020, 16, 1505–1518. [Google Scholar] [CrossRef]
- Li, N.; Zhou, P.; Tang, H.; He, L.; Fang, X.; Zhao, J.; Wang, X.; Qi, Y.; Sun, C.; Lin, Y.; et al. In-depth analysis reveals complex molecular aetiology in a cohort of idiopathic cerebral palsy. Brain 2022, 145, 119–141. [Google Scholar] [CrossRef]
- Harnett, D.; Ambrozkiewicz, M.C.; Zinnall, U.; Rusanova, A.; Borisova, E.; Drescher, A.N.; Couce-Iglesias, M.; Villamil, G.; Dannenberg, R.; Imami, K.; et al. A critical period of translational control during brain development at codon resolution. Nat. Struct. Mol. Biol. 2022, 29, 1277–1290. [Google Scholar] [CrossRef]
- Curristin, S.M.; Cao, A.; Stewart, W.B.; Zhang, H.; Madri, J.A.; Morrow, J.S.; Ment, L.R. Disrupted synaptic development in the hypoxic newborn brain. Proc. Natl. Acad. Sci. USA 2002, 99, 15729–15734. [Google Scholar] [CrossRef]
- Ment, L.R.; Vohr, B.; Allan, W.; Katz, K.H.; Schneider, K.C.; Westerveld, M.; Duncan, C.C.; Makuch, R.W. Change in cognitive function over time in very low-birth-weight infants. JAMA 2003, 289, 705–711. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Li, B.; Yang, M.; Guo, R.; Yuan, S.; Wang, J.; Hu, H. Generation of GPAM knockout human embryonic stem cell line SYSUe-008-A using CRISPR/Cas9. Stem Cell Res. 2021, 53, 102303. [Google Scholar] [CrossRef] [PubMed]
- Agnew-Blais, J.C.; Polanczyk, G.V.; Danese, A.; Wertz, J.; Moffitt, T.E.; Arseneault, L. Evaluation of the Persistence, Remission, and Emergence of Attention-Deficit/Hyperactivity Disorder in Young Adulthood. JAMA Psychiatry 2016, 73, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, S.P. Attention Deficit Hyperactivity Disorder (ADHD): Controversy, Developmental Mechanisms, and Multiple Levels of Analysis. Annu. Rev. Clin. Psychol. 2018, 14, 291–316. [Google Scholar] [CrossRef]
- Thapar, A. Discoveries on the Genetics of ADHD in the 21st Century: New Findings and Their Implications. Am. J. Psychiatry 2018, 175, 943–950. [Google Scholar] [CrossRef]
- Lee, P.H.; Anttila, V.; Won, H.; Feng, Y.-C.A.; Rosenthal, J.; Zhu, Z.; Tucker-Drob, E.M.; Nivard, M.G.; Grotzinger, A.D.; Posthuma, D.; et al. Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell 2019, 179, 1469–1482.e11. [Google Scholar] [CrossRef]
- Shepherd, G.M. Corticostriatal connectivity and its role in disease. Nat. Rev. Neurosci. 2013, 14, 278–291. [Google Scholar] [CrossRef]
- Milad, M.R.; Rauch, S.L. Obsessive-compulsive disorder: Beyond segregated cortico-striatal pathways. Trends Cogn. Sci. 2012, 16, 43–51. [Google Scholar] [CrossRef]
- Peca, J.; Feliciano, C.; Ting, J.T.; Wang, W.; Wells, M.F.; Venkatraman, T.N.; Lascola, C.D.; Fu, Z.; Feng, G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 2011, 472, 437–442. [Google Scholar] [CrossRef]
- Welch, J.M.; Lu, J.; Rodriguiz, R.M.; Trotta, N.C.; Peca, J.; Ding, J.D.; Feliciano, C.; Chen, M.; Adams, J.P.; Luo, J.; et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature 2007, 448, 894–900. [Google Scholar] [CrossRef]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N.; et al. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Birey, F.; Li, M.Y.; Gordon, A.; Thete, M.V.; Valencia, A.M.; Revah, O.; Pasca, A.M.; Geschwind, D.H.; Pasca, S.P. Dissecting the molecular basis of human interneuron migration in forebrain assembloids from Timothy syndrome. Cell Stem Cell 2022, 29, 248–264.e247. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, G.; Coelho, L.; Mo, G.; Adang, L.A.; Patne, M.; Chen, Z.; Garcia-Bassets, I.; Mesci, P.; Muotri, A.R. TREX1 is required for microglial cholesterol homeostasis and oligodendrocyte terminal differentiation in human neural assembloids. Mol. Psychiatry 2024, 29, 566–579. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kang, S.; Cho, B.; An, S.; Kim, Y.; Kim, J. Parkinson’s Disease Modeling Using Directly Converted 3D Induced Dopaminergic Neuron Organoids and Assembloids. Adv. Sci. 2025, 12, e2412548. [Google Scholar] [CrossRef]
- Ferreira, R.S.; Jandrey, E.H.F.; Granha, I.; Endo, A.K.; Ferreira, R.O.; Araujo, B.H.S.; Zatz, M.; Okamoto, O.K. Differential Replication and Oncolytic Effects of Zika Virus in Aggressive CNS Tumor Cells: Insights from Organoid and Tumoroid Models. Viruses 2024, 16, 1764. [Google Scholar] [CrossRef]
- Costamagna, G.; Comi, G.P.; Corti, S. Advancing Drug Discovery for Neurological Disorders Using iPSC-Derived Neural Organoids. Int. J. Mol. Sci. 2021, 22, 2659. [Google Scholar] [CrossRef]
- Li, C.; Fleck, J.S.; Martins-Costa, C.; Burkard, T.R.; Themann, J.; Stuempflen, M.; Peer, A.M.; Vertesy, A.; Littleboy, J.B.; Esk, C.; et al. Single-cell brain organoid screening identifies developmental defects in autism. Nature 2023, 621, 373–380. [Google Scholar] [CrossRef]
- Park, J.C.; Jang, S.Y.; Lee, D.; Lee, J.; Kang, U.; Chang, H.; Kim, H.J.; Han, S.H.; Seo, J.; Choi, M.; et al. A logical network-based drug-screening platform for Alzheimer’s disease representing pathological features of human brain organoids. Nat. Commun. 2021, 12, 280. [Google Scholar] [CrossRef]
- Langhans, S.A. Three-Dimensional in Vitro Cell Culture Models in Drug Discovery and Drug Repositioning. Front. Pharmacol. 2018, 9, 6. [Google Scholar] [CrossRef]
- Langhans, S.A. Using 3D in vitro cell culture models in anti-cancer drug discovery. Expert. Opin. Drug Discov. 2021, 16, 841–850. [Google Scholar] [CrossRef]
- Durens, M.; Nestor, J.; Williams, M.; Herold, K.; Niescier, R.F.; Lunden, J.W.; Phillips, A.W.; Lin, Y.C.; Dykxhoorn, D.M.; Nestor, M.W. High-throughput screening of human induced pluripotent stem cell-derived brain organoids. J. Neurosci. Methods 2020, 335, 108627. [Google Scholar] [CrossRef] [PubMed]
- Acharya, P.; Shrestha, S.; Joshi, P.; Choi, N.Y.; Lekkala, V.K.R.; Kang, S.Y.; Ni, G.; Lee, M.Y. Dynamic culture of cerebral organoids using a pillar/perfusion plate for the assessment of developmental neurotoxicity. Biofabrication 2024, 17, 015001. [Google Scholar] [CrossRef]
- Acharya, P.; Joshi, P.; Shrestha, S.; Choi, N.Y.; Jeong, S.; Lee, M.Y. Uniform cerebral organoid culture on a pillar plate by simple and reproducible spheroid transfer from an ultralow attachment well plate. Biofabrication 2024, 16, 025005. [Google Scholar] [CrossRef]
- Ramani, A.; Pasquini, G.; Gerkau, N.J.; Jadhav, V.; Vinchure, O.S.; Altinisik, N.; Windoffer, H.; Muller, S.; Rothenaigner, I.; Lin, S.; et al. Reliability of high-quantity human brain organoids for modeling microcephaly, glioma invasion and drug screening. Nat. Commun. 2024, 15, 10703. [Google Scholar] [CrossRef] [PubMed]
- Abedellatif, S.E.; Hosni, R.; Waha, A.; Gielen, G.H.; Banat, M.; Hamed, M.; Guresir, E.; Frohlich, A.; Sirokay, J.; Wulf, A.L.; et al. Melanoma Brain Metastases Patient-Derived Organoids: An In Vitro Platform for Drug Screening. Pharmaceutics 2024, 16, 1042. [Google Scholar] [CrossRef] [PubMed]
- Bhaduri, A. Chimeric brain organoids capture human genetic diversity. Nature 2024, 631, 32–33. [Google Scholar] [CrossRef]
- Shalita, R.; Amit, I. The industrial genomic revolution: A new era in neuroimmunology. Neuron 2022, 110, 3429–3443. [Google Scholar] [CrossRef]
- Simões-Abade, M.B.C.; Patterer, M.; Nicaise, A.M.; Pluchino, S. Brain organoid methodologies to explore mechanisms of disease in progressive multiple sclerosis. Front. Cell Neurosci. 2024, 18, 1488691. [Google Scholar] [CrossRef]
- Li, K.; Gu, L.; Cai, H.; Lu, H.C.; Mackie, K.; Guo, F. Human brain organoids for understanding substance use disorders. Drug Metab. Pharmacokinet. 2025, 60, 101036. [Google Scholar] [CrossRef]
- Villanueva, R. Advances in the knowledge and therapeutics of schizophrenia, major depression disorder, and bipolar disorder from human brain organoid research. Front. Psychiatry 2023, 14, 1178494. [Google Scholar] [CrossRef]
- Cheng, K.; Kshirsagar, A.; Nixon, J.; Lau, J.; Yang, K.; Sawa, A.; Kathuria, A. Model systems for emulating human tissue and physiology in psychiatric research. Front. Neurosci. 2025, 19, 1527826. [Google Scholar] [CrossRef] [PubMed]
- Gaudioso, Á.; Silva, T.P.; Ledesma, M.D. Models to study basic and applied aspects of lysosomal storage disorders. Adv. Drug Deliv. Rev. 2022, 190, 114532. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).