HSP110 Regulates the Assembly of the SWI/SNF Complex
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Treatment
2.2. Plasmid and siRNA Transfection
2.3. Total Cell Extraction
2.4. Cytoplasm–Nucleus Fractionation
2.5. Salt Gradient Chromatin Fractionation
2.6. Immunoprecipitation and Pull-Down Assay
2.7. Western Blotting
2.8. Immunofluorescence and Proximity Ligation Assays
2.9. Microscopy and Image Analysis
2.10. Mass Spectrometry Analysis
2.11. Statistics
3. Results
3.1. Proteome Analysis of Nuclear HSP110
3.2. HSP110 Is Recruited into the ATP-Dependent Chromatin-Remodeling SWI–SNF Complex
3.3. HSP110 Can Directly Bind SMARCC2
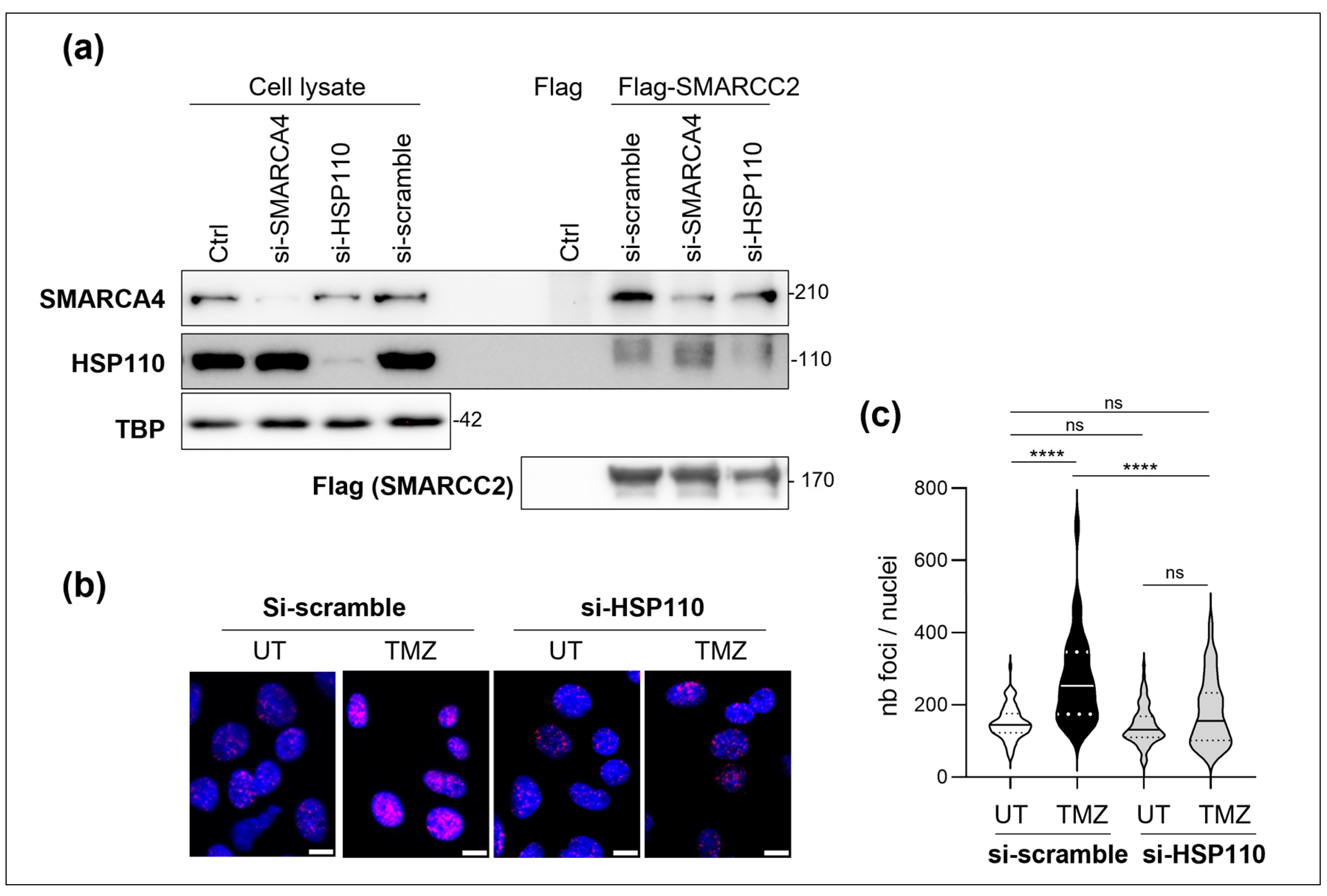
3.4. HSP110 Is Important for the Assembly of the SWI/SNF Complex
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATM | Ataxia-telangiectasia mutated |
| ATR | Ataxia Telangiectasia and Rad3-Related Protein |
| BAF | BRG1 (Brahma homolog 1)/BRM (Brahma)-associated factor |
| BRG1 | Brahma homolog 1 |
| BRM | Brahma |
| cBAF | canonical BAF |
| DNA-PKc | DNA-dependent kinase catalytic subunit |
| DSB | DNA double-strand break |
| HSF | heat shock transcription factor |
| HSP | heat shock proteins non-homologous end joining |
| MSI | microsatellite instability |
| MDPI | Multidisciplinary Digital Publishing Institute |
| ncBAF | non-canonical BAF |
| NHEJ | non-homologous end joining |
| OXA | Oxaliplatin |
| PBAF | polybromo-associated BAF |
| SMARCA4 | SWI/SNF Related BAF Chromatin Remodeling Complex Subunit ATPase 4 |
| SMARCC2 | SWI/SNF Related BAF Chromatin Remodeling Complex Subunit C2 |
| SWI/SNF | Switch/sucrose non-fermentable |
| TMZ | temozolomide |
References
- Mashtalir, N.; Dao, H.T.; Sankar, A.; Liu, H.; Corin, A.J.; Bagert, J.D.; Ge, E.J.; D’Avino, A.R.; Filipovski, M.; Michel, B.C.; et al. Chromatin landscape signals differentially dictate the activities of mSWI/SNF family complexes. Science 2021, 373, 306–315. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, J.; Yu, Z.; Wang, Q.; Li, W.; Ren, Y.; Chen, Z.; He, S.; Xu, Y. Structure of nucleosome-bound human PBAF complex. Nat. Commun. 2022, 13, 7644. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, M.; Takii, R.; Nakai, A. Regulation of HSF1 transcriptional complexes under proteotoxic stress: Mechanisms of heat shock gene transcription involve the stress-induced HSF1 complex formation, changes in chromatin states, and formation of phase-separated condensates. Bioessays 2023, 45, e2300036. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, E.K.; Weirich, C.S.; Guyon, J.R.; Sif, S.; Kingston, R.E. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol. Cell. Biol. 2001, 21, 5826–5837. [Google Scholar] [CrossRef]
- Mikami, H.; Saito, Y.; Okamoto, N.; Kakihana, A.; Kuga, T.; Nakayama, Y. Requirement of Hsp105 in CoCl. Exp. Cell Res. 2017, 352, 225–233. [Google Scholar] [CrossRef]
- Causse, S.Z.; Marcion, G.; Chanteloup, G.; Uyanik, B.; Boudesco, C.; Grigorash, B.B.; Douhard, R.; Dias, A.M.M.; Dumetier, B.; Dondaine, L.; et al. HSP110 translocates to the nucleus upon genotoxic chemotherapy and promotes DNA repair in colorectal cancer cells. Oncogene 2019, 38, 2767–2777. [Google Scholar] [CrossRef]
- Yamane, T.; Saito, Y.; Teshima, H.; Hagino, M.; Kakihana, A.; Sato, S.; Shimada, M.; Kato, Y.; Kuga, T.; Yamagishi, N.; et al. Hsp105α suppresses Adriamycin-induced cell death via nuclear localization signal-dependent nuclear accumulation. J. Cell. Biochem. 2019, 120, 17951–17962. [Google Scholar] [CrossRef]
- Yu, N.; Kakunda, M.; Pham, V.; Lill, J.R.; Du, P.; Wongchenko, M.; Yan, Y.; Firestein, R.; Huang, X. HSP105 recruits protein phosphatase 2A to dephosphorylate β-catenin. Mol. Cell. Biol. 2015, 35, 1390–1400. [Google Scholar] [CrossRef]
- Berthenet, K.; Bokhari, A.; Lagrange, A.; Marcion, G.; Boudesco, C.; Causse, S.; De Thonel, A.; Svrcek, M.; Goloudina, A.R.; Dumont, S.; et al. HSP110 promotes colorectal cancer growth through STAT3 activation. Oncogene 2017, 36, 2328–2336. [Google Scholar] [CrossRef]
- Boudesco, C.; Verhoeyen, E.; Martin, L.; Chassagne-Clement, C.; Salmi, L.; Mhaidly, R.; Pangault, C.; Fest, T.; Ramla, S.; Jardin, F.; et al. HSP110 sustains chronic NF-κB signaling in activated B-cell diffuse large B-cell lymphoma through MyD88 stabilization. Blood 2018, 132, 510–520. [Google Scholar] [CrossRef]
- Gibouin, V.C.; Durand, M.; Boudesco, C.; Hermetet, F.; Nozickova, K.; Chassagne-Clement, C.; Abdelwahed, M.; Klener, P.; Garrido, C.; Jego, G. First-in-class inhibitor of HSP110 blocks BCR activation through SYK phosphorylation in diffuse large B-cell lymphoma. Leukemia 2024, 38, 1742–1750. [Google Scholar] [CrossRef] [PubMed]
- Mashtalir, N.; D’Avino, A.R.; Michel, B.C.; Luo, J.; Pan, J.; Otto, J.E.; Zullow, H.J.; McKenzie, Z.M.; Kubiak, R.L.; St Pierre, R.; et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 2018, 175, 1272–1288. [Google Scholar] [CrossRef]
- Saito, Y.; Yamagishi, N.; Hatayama, T. Different localization of Hsp105 family proteins in mammalian cells. Exp. Cell Res. 2007, 313, 3707–3717. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Ogata, K.; Altan, B.; Yokobori, T.; Ide, M.; Mochiki, E.; Toyomasu, Y.; Kogure, N.; Yanoma, T.; Suzuki, M.; et al. Nuclear heat shock protein 110 expression is associated with poor prognosis and chemotherapy resistance in gastric cancer. Oncotarget 2016, 7, 18415–18423. [Google Scholar] [CrossRef]
- Collura, A.; Lagrange, A.; Svrcek, M.; Marisa, L.; Buhard, O.; Guilloux, A.; Wanherdrick, K.; Dorard, C.; Taieb, A.; Saget, A.; et al. Patients with colorectal tumors with microsatellite instability and large deletions in HSP110 T17 have improved response to 5-fluorouracil–based chemotherapy. Gastroenterology 2014, 146, 401–411. [Google Scholar] [CrossRef]
- Noel, K.; Bokhari, A.; Bertrand, R.; Renaud, F.; Bourgoin, P.; Cohen, R.; Svrcek, M.; Joly, A.C.; Duval, A.; Collura, A. Consequences of the Hsp110DE9 mutation in tumorigenesis and the 5-fluorouracil-based chemotherapy response in Msh2-deficient mice. Cell. Mol. Life Sci. 2022, 79, 332. [Google Scholar] [CrossRef] [PubMed]
- Dorard, C.; de Thonel, A.; Collura, A.; Marisa, L.; Svrcek, M.; Lagrange, A.; Jego, G.; Wanherdrick, K.; Joly, A.L.; Buhard, O.; et al. Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat. Med. 2011, 17, 1283–1289. [Google Scholar] [CrossRef]
- Buhard, O.; Lagrange, A.; Guilloux, A.; Colas, C.; Chouchène, M.; Wanherdrick, K.; Coulet, F.; Guillerm, E.; Dorard, C.; Marisa, L.; et al. HSP110 T17 simplifies and improves the microsatellite instability testing in patients with colorectal cancer. J. Med. Genet. 2016, 53, 377–384. [Google Scholar] [CrossRef]
- Wheat, A.; Yu, C.; Wang, X.; Burke, A.M.; Chemmama, I.E.; Kaake, R.M.; Baker, P.; Rychnovsky, S.D.; Yang, J.; Huang, L. Protein interaction landscapes revealed by advanced in vivo cross-linking-mass spectrometry. Proc. Natl. Acad. Sci. USA 2021, 118, e2023360118. [Google Scholar] [CrossRef]
- Mattoo, R.U.; Sharma, S.K.; Priya, S.; Finka, A.; Goloubinoff, P. Hsp110 is a bona fide chaperone using ATP to unfold stable misfolded polypeptides and reciprocally collaborate with Hsp70 to solubilize protein aggregates. J. Biol. Chem. 2013, 288, 21399–21411. [Google Scholar] [CrossRef]
- Ryu, S.W.; Stewart, R.; Pectol, D.C.; Ender, N.A.; Wimalarathne, O.; Lee, J.H.; Zanini, C.P.; Harvey, A.; Huibregtse, J.M.; Mueller, P.; et al. Proteome-wide identification of HSP70/HSC70 chaperone clients in human cells. PLoS Biol. 2020, 18, e3000606. [Google Scholar] [CrossRef] [PubMed]
- Lynham, J.; Houry, W.A. The Role of Hsp90-R2TP in Macromolecular Complex Assembly and Stabilization. Biomolecules 2022, 12, 1045. [Google Scholar] [CrossRef] [PubMed]
- Dubrez, L.; Causse, S.; Borges Bonan, N.; Dumétier, B.; Garrido, C. Heat-shock proteins: Chaperoning DNA repair. Oncogene 2020, 39, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Harrod, A.; Lane, K.A.; Downs, J.A. The role of the SWI/SNF chromatin remodelling complex in the response to DNA double strand breaks. DNA Repair 2020, 93, 102919. [Google Scholar] [CrossRef]
- Smith-Roe, S.L.; Nakamura, J.; Holley, D.; Chastain, P.D.; Rosson, G.B.; Simpson, D.A.; Ridpath, J.R.; Kaufman, D.G.; Kaufmann, W.K.; Bultman, S.J. SWI/SNF complexes are required for full activation of the DNA-damage response. Oncotarget 2015, 6, 732–745. [Google Scholar] [CrossRef]
- Ribeiro-Silva, C.; Vermeulen, W.; Lans, H. SWI/SNF: Complex complexes in genome stability and cancer. DNA Repair 2019, 77, 87–95. [Google Scholar] [CrossRef]
- Lee, H.S.; Park, J.H.; Kim, S.J.; Kwon, S.J.; Kwon, J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010, 29, 1434–1445. [Google Scholar] [CrossRef]
- Ogiwara, H.; Ui, A.; Otsuka, A.; Satoh, H.; Yokomi, I.; Nakajima, S.; Yasui, A.; Yokota, J.; Kohno, T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 2011, 30, 2135–2146. [Google Scholar] [CrossRef]
- Vélez-Cruz, R.; Manickavinayaham, S.; Biswas, A.K.; Clary, R.W.; Premkumar, T.; Cole, F.; Johnson, D.G. RB localizes to DNA double-strand breaks and promotes DNA end resection and homologous recombination through the recruitment of BRG1. Genes Dev. 2016, 30, 2500–2512. [Google Scholar] [CrossRef]
- Kwon, S.J.; Park, J.H.; Park, E.J.; Lee, S.A.; Lee, H.S.; Kang, S.W.; Kwon, J. ATM-mediated phosphorylation of the chromatin remodeling enzyme BRG1 modulates DNA double-strand break repair. Oncogene 2015, 34, 303–313. [Google Scholar] [CrossRef]
- Peng, G.; Yim, E.K.; Dai, H.; Jackson, A.P.; Burgt, I.; Pan, M.R.; Hu, R.; Li, K.; Lin, S.Y. BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat. Cell Biol. 2009, 11, 865–872. [Google Scholar] [CrossRef] [PubMed]
- Moison, C.; Chagraoui, J.; Caron, M.C.; Gagné, J.P.; Coulombe, Y.; Poirier, G.G.; Masson, J.Y.; Sauvageau, G. Zinc finger protein E4F1 cooperates with PARP-1 and BRG1 to promote DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 2021, 118, e2019408118. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shan, B.; Zou, C.; Wang, H.; Zhang, M.M.; Zhu, H.; Naito, M.G.; Xu, D.; Manuel, V.J.; Mifflin, L.; et al. Nuclear RIPK1 promotes chromatin remodeling to mediate inflammatory response. Cell Res. 2022, 32, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Guo, P.; Hoang, N.; Sanchez, J.; Zhang, E.H.; Rajawasam, K.; Trinidad, K.; Sun, H.; Zhang, H. The assembly of mammalian SWI/SNF chromatin remodeling complexes is regulated by lysine-methylation dependent proteolysis. Nat. Commun. 2022, 13, 6696. [Google Scholar] [CrossRef]
- Shivaswamy, S.; Iyer, V.R. Stress-dependent dynamics of global chromatin remodeling in yeast: Dual role for SWI/SNF in the heat shock stress response. Mol. Cell. Biol. 2008, 28, 2221–2234. [Google Scholar] [CrossRef]
- Erkina, T.Y.; Tschetter, P.A.; Erkine, A.M. Different requirements of the SWI/SNF complex for robust nucleosome displacement at promoters of heat shock factor and Msn2- and Msn4-regulated heat shock genes. Mol. Cell. Biol. 2008, 28, 1207–1217. [Google Scholar] [CrossRef]
- Takii, R.; Fujimoto, M.; Tan, K.; Takaki, E.; Hayashida, N.; Nakato, R.; Shirahige, K.; Nakai, A. ATF1 modulates the heat shock response by regulating the stress-inducible heat shock factor 1 transcription complex. Mol. Cell. Biol. 2015, 35, 11–25. [Google Scholar] [CrossRef]



| Functional Sub-Category | Protein | UniProtKB | Protein Set Score | Function | |
|---|---|---|---|---|---|
| UT | TMZ | ||||
| Chaperones | Nucleophosmin | P06748 | 42.82 | 80.31 | |
| HSP70 | P17066 | - | 105.01 | ||
| CFA/TBCA | O75347 | - | 34.14 | Tubulin-specific chaperone | |
| Chromatin remodeling complexes: | |||||
| SWI/SNC complexes | SMARCA4/BRG1 | P51532 | 35.37 | 108.16 | ATPase |
| SMARCC2/BAF170 | Q8TAQ2 | 39.94 | 211.08 | Core subunit | |
| SMARCB1/SNF5 | Q12824 | - | 34.6 | Core subunit | |
| SMARCE1/BAF57 | Q969G3 | 39.41 | 47.91 | Accessory proteins | |
| PBRM1/BAF180 | Q86U86 | 129.83 | 278.66 | Accessory proteins with | |
| bromo domain | |||||
| ISWI complex | ACF1/BAZ1A | Q9NRL2 | 39.41 | 47.91 | Accessory proteins with bromo domain |
| Histones | H2A | P04908 | - | 43.25 | |
| H4 | P62805 | - | 141.71 | ||
| H3.1 | P68431 | - | 40.9 | ||
| DNA repair | DNA-PKcs/DNPK1 | P78527 | - | 43.34 | NHEJ pathway |
| RNA-binding proteins | FXR1 | 36.17 | - | ||
| ABP1 | P11940 | 34.46 | 42.6 | ||
| Nucleolin | P19338 | 84.39 | 181.27 | ||
| hnRNP A2 | P09651 | 42.8 | 106.34 | ||
| hnRNP H | P31943 | 46.31 | 88.47 | ||
| hnRNP A0 | Q13151 | 45.26 | 38.54 | ||
| hnRNP M | P52272 | - | 84.4 | ||
| hnRNP K | P61978 | - | 35.01 | ||
| hnRNP U | - | 241.27 | |||
| hnRNP D0 | - | 35.39 | |||
| αCP3 | - | 42.52 | |||
| Sam68 | - | 45.49 | |||
| H3.1 | - | 49.31 | |||
| COAA or RNA- | Q96PK6 | - | 184.8 | ||
| BP14 | |||||
| Transcriptional regulators | DDX9 | 49.94 | 146.86 | DNA-dependent RNA | |
| helicase, TF | |||||
| eEF-1α | - | 54.38 | |||
| Prohibitin-2 | - | 36.03 | |||
| GTPases and regulators | Rab-3D, 6A, 15 or 33B | O95716, P20340, P59191, Q9H082, | 33.06 | - | Ras-related protein Rab |
| ASAP2/PAP | O43150 | 38,77 | 40.31 | ||
| GNAT3 or GNA12 | A8MTJ3 or Q03113 | - | 39.75 | ||
| Rac2 | P15153 | - | 51.97 | ||
| Ran | P62826 | - | 56.9G | ||
| Gα12 | - | 39.75 | |||
| KRas | A0A024RAV5 | - | 52.76 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pointeau, O.; Paccagnini, M.; Borges-Bonan, N.; Biziorek, L.; Causse, S.; Garrido, C.; Dubrez, L. HSP110 Regulates the Assembly of the SWI/SNF Complex. Cells 2025, 14, 849. https://doi.org/10.3390/cells14110849
Pointeau O, Paccagnini M, Borges-Bonan N, Biziorek L, Causse S, Garrido C, Dubrez L. HSP110 Regulates the Assembly of the SWI/SNF Complex. Cells. 2025; 14(11):849. https://doi.org/10.3390/cells14110849
Chicago/Turabian StylePointeau, Océane, Manon Paccagnini, Natalia Borges-Bonan, Léo Biziorek, Sébastien Causse, Carmen Garrido, and Laurence Dubrez. 2025. "HSP110 Regulates the Assembly of the SWI/SNF Complex" Cells 14, no. 11: 849. https://doi.org/10.3390/cells14110849
APA StylePointeau, O., Paccagnini, M., Borges-Bonan, N., Biziorek, L., Causse, S., Garrido, C., & Dubrez, L. (2025). HSP110 Regulates the Assembly of the SWI/SNF Complex. Cells, 14(11), 849. https://doi.org/10.3390/cells14110849






