Efficient Generation of Induced Pluripotent Stem Cell-Derived Definitive Endoderm Cells with Growth Factors and Small Molecules
Abstract
1. Introduction
2. Materials and Methods
2.1. Small Molecules, Chemicals, Growth Factors, and Antibodies
2.2. Human Pluripotent Stem Cells
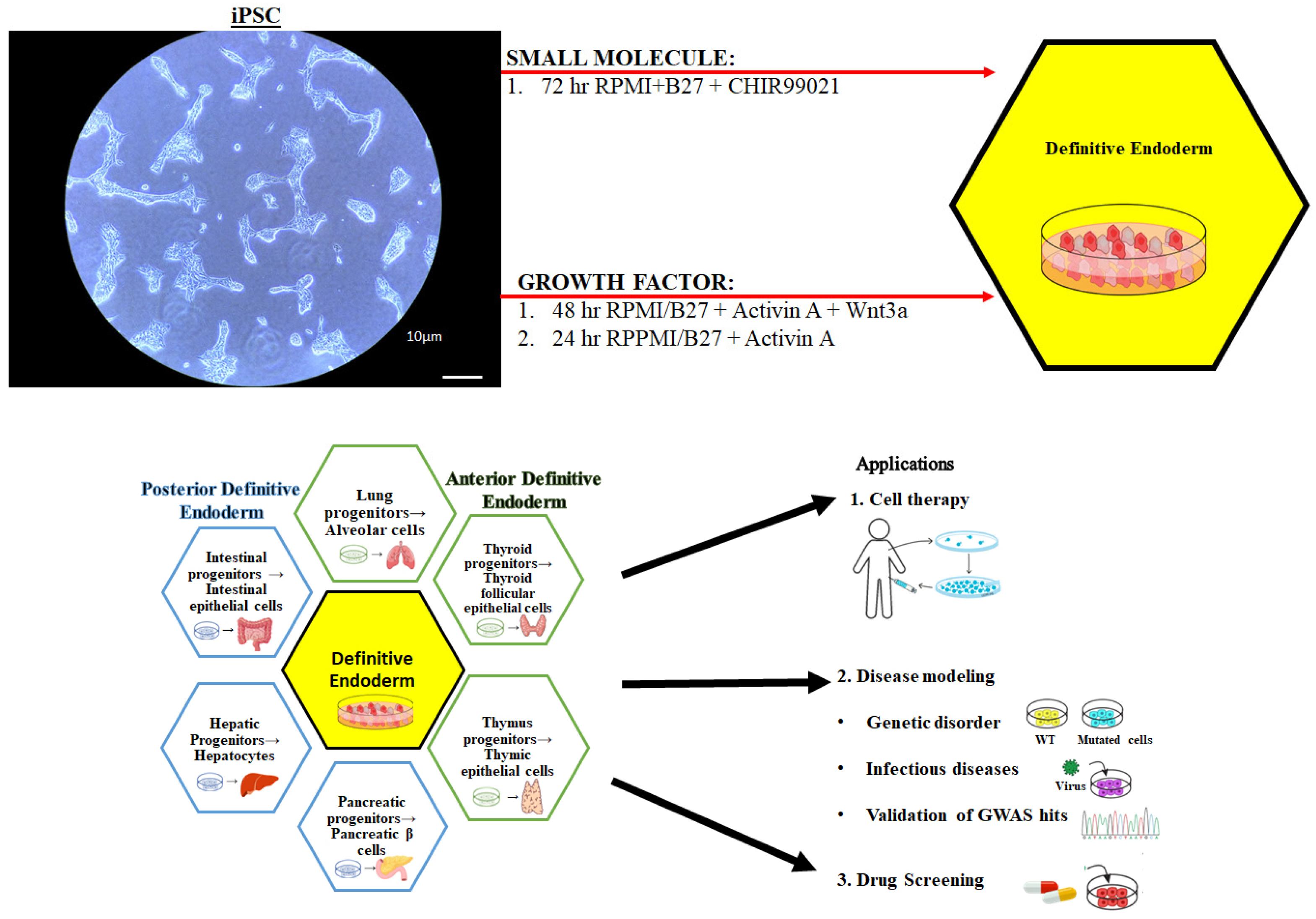
2.3. Small Molecule Definitive Endoderm Protocol
2.4. Growth Factor Definitive Endoderm Protocol
2.5. Small Molecule Hepatoblast Specification Protocol
2.6. Growth Factor Hepatoblast Specification Protocol
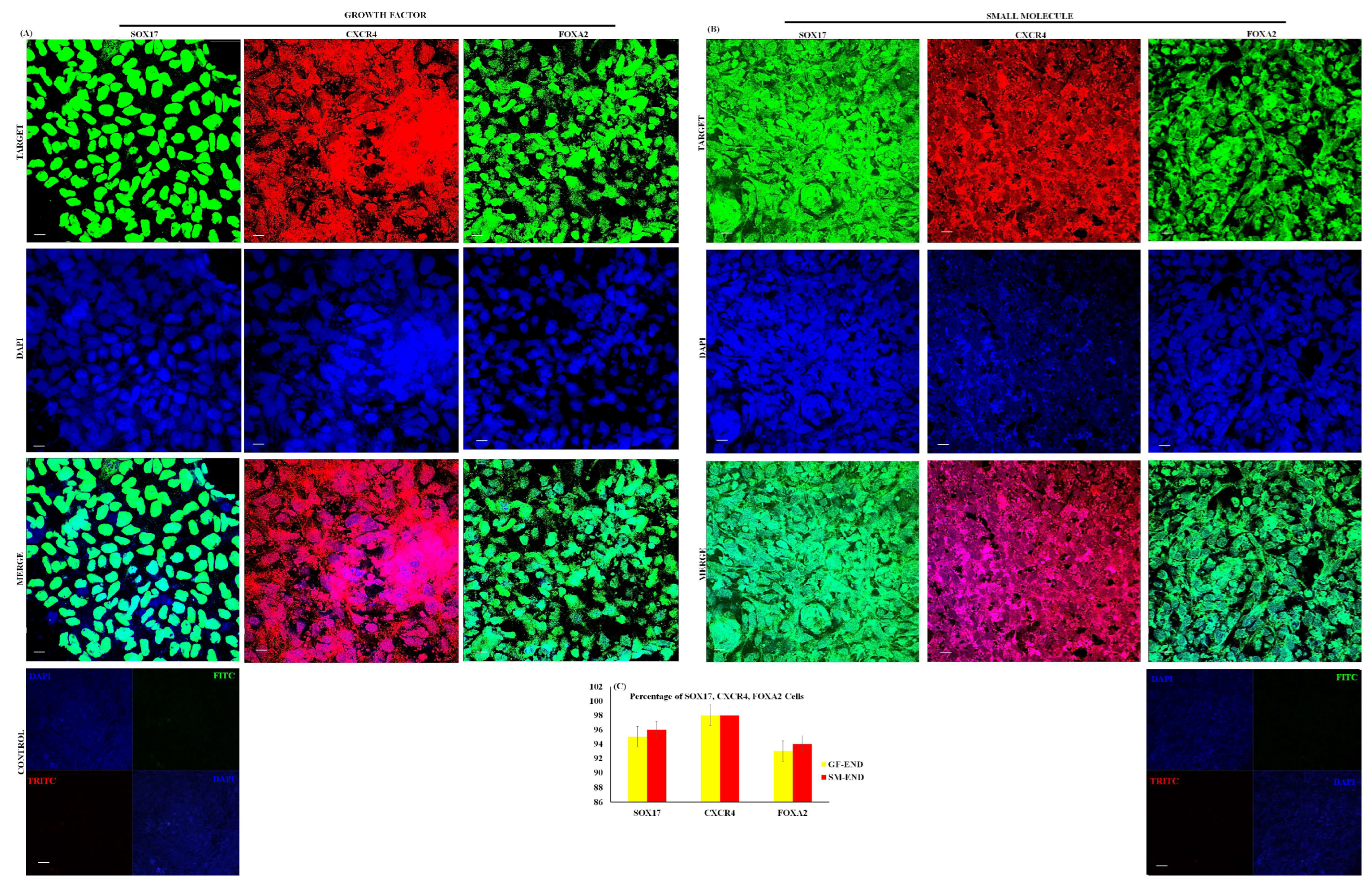
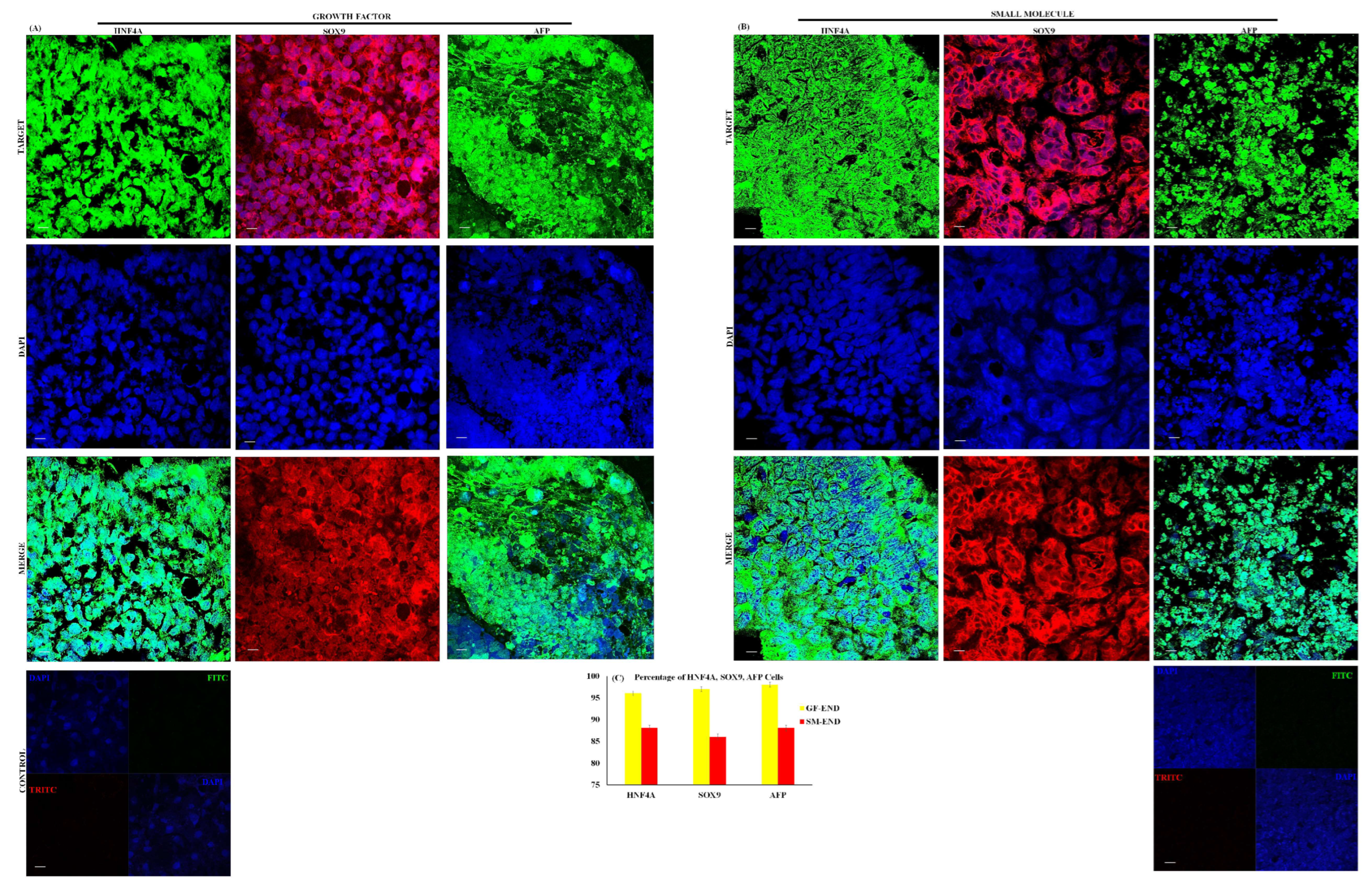
2.7. Immunocytochemistry
2.8. RNA Isolation, cDNA Synthesis, and RT-PCR Analysis
2.9. Proteomics, Metabolomics, and LC-MS/MS Analysis
2.9.1. Bioinformatics Analysis
2.9.2. Pathway Analysis
2.9.3. STRING Analysis
2.9.4. Imaging
2.9.5. Statistical Analysis
3. Results
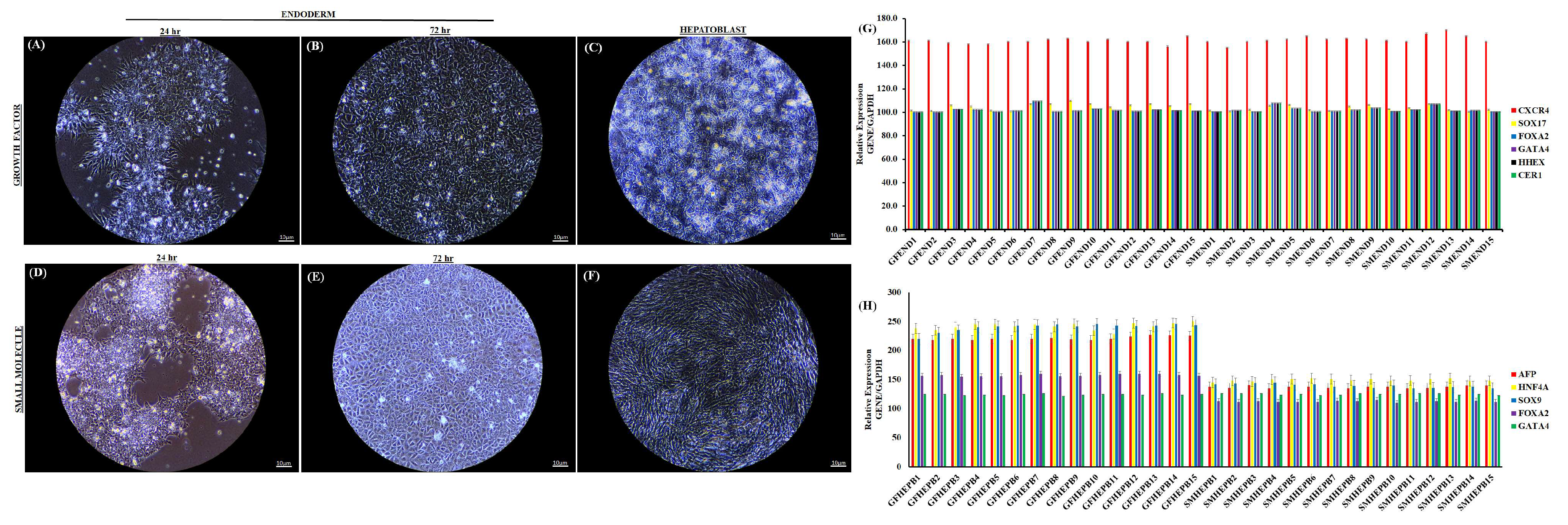
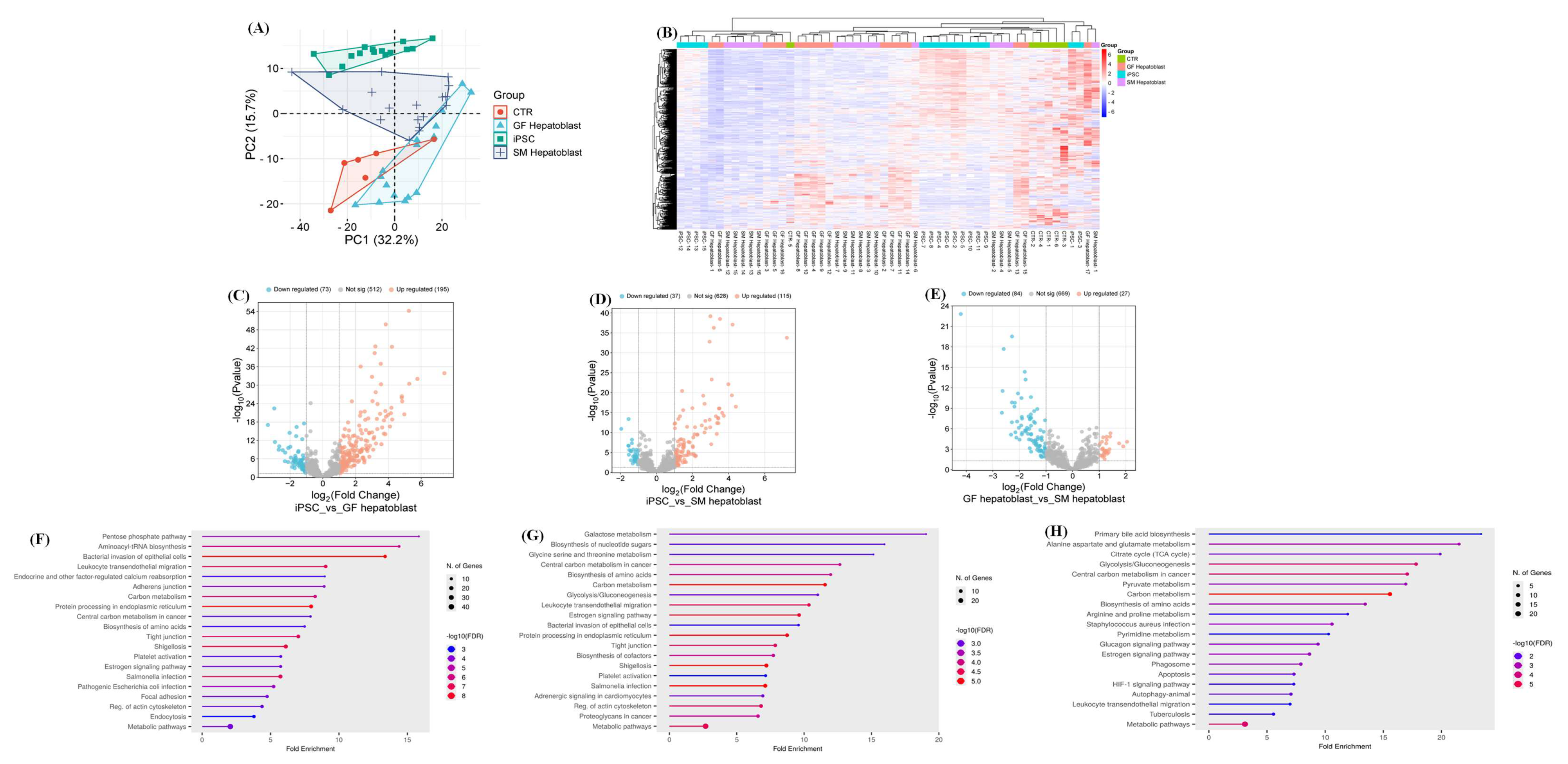
3.1. Characterization of Definitive Endoderm Cells
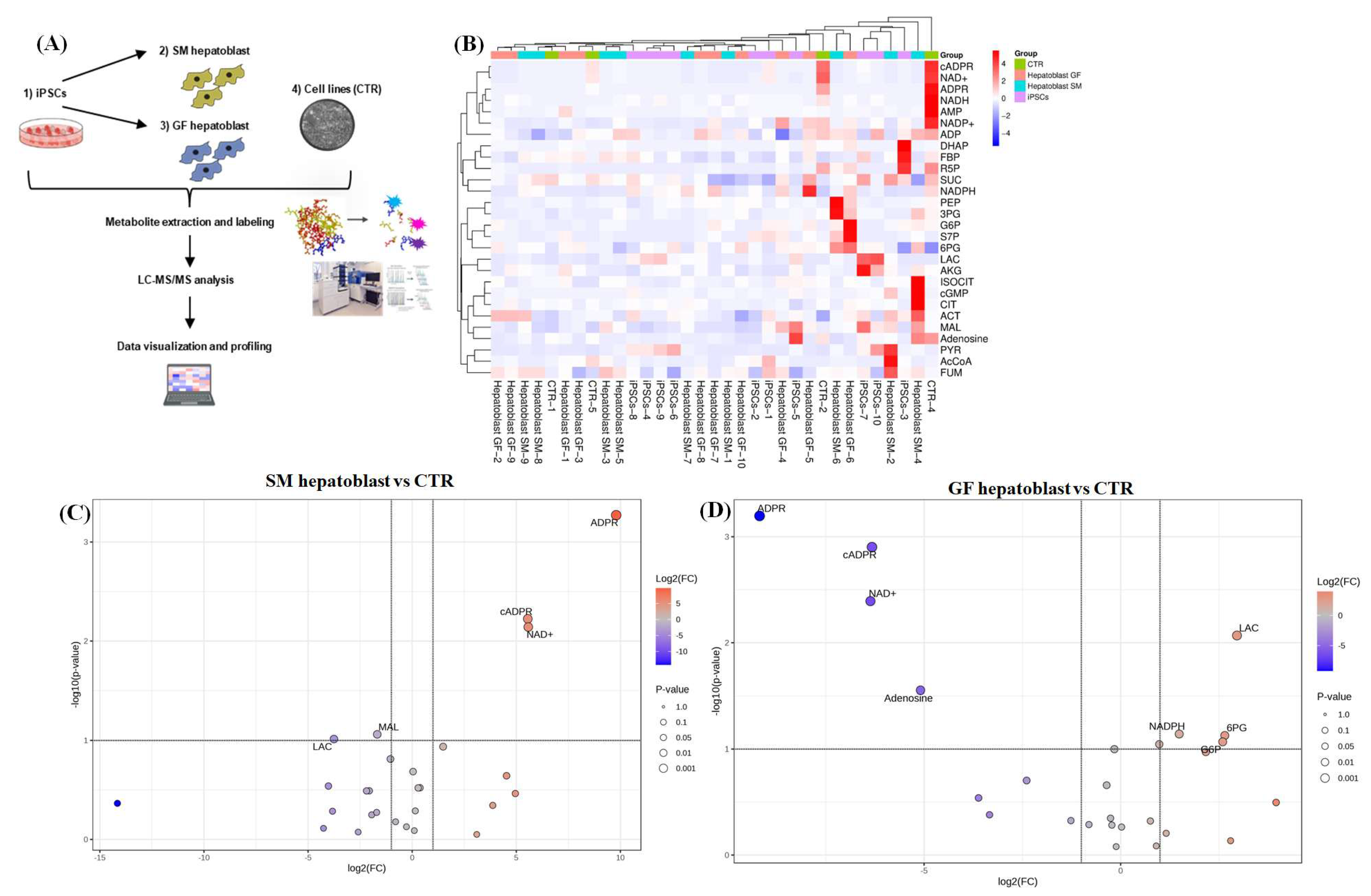
3.2. Proteomics of Endoderm and Hepatoblast Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Declarations
Abbreviations
References
- Nowotschin, S.; Hadjantonakis, A.K.; Campbell, K. The endoderm: A divergent cell lineage with many commonalities. Development 2019, 146, dev150920. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.M.; Melton, D.A. Vertebrate endoderm development. Annu. Rev. Cell Dev. Biol. 1999, 15, 393–410. [Google Scholar] [CrossRef] [PubMed]
- Nowotschin, S.; Setty, M.; Kuo, Y.Y.; Liu, V.; Garg, V.; Sharma, R.; Simon, C.S.; Saiz, N.; Gardner, R.; Boutet, S.C.; et al. The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 2019, 569, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xu, C.; Ma, T. In vitro organogenesis from pluripotent stem cells. Organogenesis 2014, 10, 159–163. [Google Scholar] [CrossRef]
- Molè, M.A.; Coorens, T.H.H.; Shahbazi, M.N.; Weberling, A.; Weatherbee, B.A.T.; Gantner, C.W.; Sancho-Serra, C.; Richardson, L.; Drinkwater, A.; Syed, N.; et al. A single cell characterization of human embryogenesis identifies pluripotency transitions and putative anterior hypoblast center. Nat. Commun. 2021, 12, 3679. [Google Scholar] [CrossRef]
- Loh, K.M.; Ang, L.T.; Zhang, J.; Kumar, V.; Ang, J.; Auyeong, J.Q.; Lee, K.L.; Choo, S.H.; Lim, C.Y.; Nichane, M.; et al. Efficient endoderm induction from human pluripotent stem cells by logically directing signals controlling lineage bifurcations. Cell Stem Cell 2014, 14, 237–252. [Google Scholar] [CrossRef]
- Sahabian, A.; Dahlmann, J.; Martin, U.; Olmer, R. Production and cryopreservation of definitive endoderm from human pluripotent stem cells under defined and scalable culture conditions. Nat. Protoc. 2021, 16, 1581–1599. [Google Scholar] [CrossRef]
- D’Amour, K.; Agulnick, A.; Eliazer, S.; Kelly, G.O.; Kroon, E.; Baetge, E.E. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat. Biotechnol. 2005, 23, 1534–1541. [Google Scholar] [CrossRef]
- Kopper, O.; Benvenisty, N. Stepwise differentiation of human embryonic stem cells into early endoderm derivatives and their molecular characterization. Stem Cell Res. 2012, 8, 335–345. [Google Scholar] [CrossRef]
- Pauklin, S.; Vallier, L. Activin/Nodal signaling in stem cells. Development 2015, 142, 607–619. [Google Scholar] [CrossRef]
- Sullivan, G.J.; Hay, D.C.; Park, I.H.; Fletcher, J.; Hannoun, Z.; Payne, C.M.; Dalgetty, D.; Black, J.R.; Ross, J.A.; Samuel, K.; et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology 2010, 51, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Hay, D.C.; Fletcher, J.; Payne, C.; Terrace, J.D.; Gallagher, R.C.; Snoeys, J.; Black, J.R.; Wojtacha, D.; Samuel, K.; Hannoun, Z.; et al. Highly efficient differentiation of hESCs to functional hepatic endoderm requires Activin A and Wnt3a signaling. Proc. Natl. Acad. Sci. USA 2008, 105, 12301–12306. [Google Scholar] [CrossRef] [PubMed]
- Zaret, K.S. Using small molecules to great effect in stem cell differentiation. Cell Stem Cell 2009, 4, 373–374. [Google Scholar] [CrossRef] [PubMed]
- Bogacheva, M.S.; Khan, S.; Kanninen, L.K.; Yliperttula, M.; Leung, A.W.; Lou, Y.R. Differences in definitive endoderm induction approaches using growth factors and small molecules. J. Cell Physiol. 2018, 233, 3578–3589. [Google Scholar] [CrossRef]
- Borowiak, M.; Maehr, R.; Chen, S.; Chen, A.E.; Tang, W.; Fox, J.L.; Schreiber, S.L.; Melton, D.A. Small molecules efficiently direct endodermal differentiation of mouse and human embryonic stem cells. Cell Stem Cell 2009, 3, 348–358. [Google Scholar] [CrossRef]
- Bone, H.K.; Nelson, A.S.; Goldring, C.E.; Tosh, D.; Welham, M.J. A novel chemically directed route for the generation of definitive endoderm from human embryonic stem cells based on inhibition of GSK-3. J. Cell Sci. 2011, 15, 1992–2000. [Google Scholar] [CrossRef]
- Du, C.; Feng, Y.; Qiu, D.; Xu, Y.; Pang, M.; Cai, N.; Xiang, A.P.; Zhang, Q. Highly efficient and expedited hepatic differentiation from human pluripotent stem cells by pure small-molecule cocktails. Stem Cell Res. Ther. 2018, 9, 58. [Google Scholar] [CrossRef]
- Li, Q.; Li, J.; Wang, P.; He, X.; Hong, M.; Liu, F. A Comparative Study of Endoderm Differentiation Between Activin A and Small Molecules. Exp. Clin. Endocrinol. Diabetes 2023, 131, 667–675. [Google Scholar] [CrossRef]
- Siller, R.; Naumovska, E.; Mathapati, S.; Lycke, M.; Greenhough, S.; Sullivan, G.J. Development of a rapid screen for the endodermal differentiation potential of human pluripotent stem cell lines. Sci. Rep. 2016, 22, 37178. [Google Scholar] [CrossRef]
- Mathapati, S.; Siller, R.; Impellizzeri, A.A.; Lycke, M.; Vegheim, K.; Almaas, R.; Sullivan, G.J. Small-Molecule-Directed Hepatocyte-Like Cell Differentiation of Human Pluripotent Stem Cells. Curr. Protoc. Stem Cell Biol. 2016, 38, 1G.6.1–1G.6.18. [Google Scholar] [CrossRef]
- Asumda, F.Z.; Hatzistergos, K.E.; Dykxhoorn, D.M.; Jakubski, S.; Edwards, J.; Thomas, E.; Schiff, E.R. Differentiation of hepatocyte-like cells from human pluripotent stem cells using small molecules. Differentiation 2018, 101, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Carpentier, A.; Cheng, X.; Block, P.D.; Zhao, Y.; Zhang, Z.; Protzer, U.; Liang, T.J. Human stem cell-derived hepatocytes as a model for hepatitis B virus infection, spreading and virus-host interactions. J. Hepatol. 2017, 66, 494–503. [Google Scholar] [CrossRef] [PubMed]
- Glass, J.; Robinson, R.; Edupuganti, N.; Altman, J.; Greenway, G.; Lee, T.J.; Zhi, W.; Sharma, A.; Sharma, S. Proteomic Alterations in Retinal Müller Glial Cells Lacking Interleukin-6 Receptor: A Comprehensive Analysis. Investig. Ophthalmol. Vis. Sci. 2024, 65, 33. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.G.; Araújo, T.G.; Carvalho, B.M.; Rocha, G.Z.; Santos, A.; Saad, M.J.A. The Role of Hepatocyte Growth Factor (HGF) in Insulin Resistance and Diabetes. Front. Endocrinol. 2018, 9, 503. [Google Scholar] [CrossRef]
- Kitade, M.; Factor, V.M.; Andersen, J.B.; Tomokuni, A.; Kaji, K.; Akita, H.; Holczbauer, A.; Seo, D.; Marquardt, J.U.; Conner, E.A.; et al. Specific fate decisions in adult hepatic progenitor cells driven by MET and EGFR signaling. Genes. Dev. 2013, 27, 1706–1717. [Google Scholar] [CrossRef]
- Han, H.S.; Kang, G.; Kim, J.S.; Choi, B.H.; Koo, S.H. Regulation of glucose metabolism from a liver-centric perspective. Exp. Mol. Med. 2016, 48, e218. [Google Scholar] [CrossRef]
- Jellali, R.; Lereau, B.M.; Tauran, Y.; Gilard, F.; Danoy, M.; Kido, T.; Miyajima, A.; Sakai, Y.; Leclerc, E. Metabolomic profiling during the differentiation of human induced pluripotent stem cells into hepatocyte-like cells. Differentiation 2020, 112, 17–26. [Google Scholar] [CrossRef]
- Moreno-Torres, M.; Kumar, M.; García-Llorens, G.; Quintás, G.; Tricot, T.; Boon, R.; Tolosa, L.; Toprakhisar, B.; Chesnais, F.; Verfaillie, C.; et al. A Novel UPLC-MS Metabolomic Analysis-Based Strategy to Monitor the Course and Extent of iPSC Differentiation to Hepatocytes. J. Proteome Res. 2022, 21, 702–712. [Google Scholar] [CrossRef]
- Lin, J.; Rao, D.; Zhang, M.; Gao, Q. Metabolic reprogramming in the tumor microenvironment of liver cancer. J. Hematol. Oncol. 2024, 17, 6. [Google Scholar] [CrossRef]
- Laubach, K.; Turan, T.; Mathew, R.; Wilsbacher, J.; Engelhardt, J.; Samayoa, J. Tumor-intrinsic metabolic reprogramming and how it drives resistance to anti-PD-1/PD-L1 treatment. Cancer Drug Resist. 2023, 6, 611–641. [Google Scholar] [CrossRef]

| Sequence Notes | Bases | Sequence |
|---|---|---|
| AFP, HOMO_SAPIENS | 25 | TCT GCA TGA ATT ATA CAT TGA CCA C |
| 20 | AGG AGA TGT GCT GGA TTG TC | |
| FOXA2, HOMO_SAPIENS | 19 | TGT TCA TGC CGT TCA TCC C |
| 19 | GGA GCG GTG AAG ATG GAA G | |
| SOX17, HOMO_SAPIENS | 19 | GGC CGG TAC TTG TAG TTG G |
| 17 | AAC GCC GAG TTG AGC AA | |
| HNF4A, HOMO_SAPIENS | 21 | GAT GTA GTC CTC CAA GCT CAC |
| 21 | GCC ATC ATC TTC TTT GAC CCA | |
| CXCR4, HOMO_SAPIENS | 21 | GTA CTT GTC CGT CAT GCT TCT |
| 20 | AAA TCT TCC TGC CCA CCA TC | |
| GATA4, HOMO_SAPIENS | 18 | TTG CTG GAG TTG CTG GAA |
| 19 | GGA AGC CCA AGA ACC TGA A | |
| GAPDH, HOMO_SAPIENS | 22 | TGT AGT TGA GGT CAA TGA AGG G |
| 19 | ACA TCG CTC AGA CAC CAT G | |
| HHEX, HOMO_SAPIENS | 21 | CAA ATC TTG CCT CTG ATC ACA |
| 19 | TCA GCG AGA GAC AGG TCA A | |
| CER1, HOMO_SAPIENS | 21 | GAT GTG TCC ATC TTC ATG CTC |
| 22 | CAC TGA ACT GCA CTG AAC TTT C | |
| SOX9, HOMO_SAPIENS | 18 | CTT CAG GTC AGC CTT GCC |
| 18 | CAT GAG CGA GGT GCA CTC | |
| FOXA2, HOMO_SAPIENS | 19 | TGT TCA TGC CGT TCA TCC C |
| 20 | AGC GGG CGA GTT AAA GTA TG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Asumda, F.Z.; Alzoubi, S.; Padarath, K.; John, N.; Jones, K.; Kolhe, R.; Mondal, A.K.; Lee, T.J.; Zhi, W.; Huebert, R.C.; et al. Efficient Generation of Induced Pluripotent Stem Cell-Derived Definitive Endoderm Cells with Growth Factors and Small Molecules. Cells 2025, 14, 815. https://doi.org/10.3390/cells14110815
Asumda FZ, Alzoubi S, Padarath K, John N, Jones K, Kolhe R, Mondal AK, Lee TJ, Zhi W, Huebert RC, et al. Efficient Generation of Induced Pluripotent Stem Cell-Derived Definitive Endoderm Cells with Growth Factors and Small Molecules. Cells. 2025; 14(11):815. https://doi.org/10.3390/cells14110815
Chicago/Turabian StyleAsumda, Faizal Z., Shadia Alzoubi, Kiyasha Padarath, Nina John, Kimya Jones, Ravindra Kolhe, Ashis Kumar Mondal, Tae Jin Lee, Wenbo Zhi, Robert C. Huebert, and et al. 2025. "Efficient Generation of Induced Pluripotent Stem Cell-Derived Definitive Endoderm Cells with Growth Factors and Small Molecules" Cells 14, no. 11: 815. https://doi.org/10.3390/cells14110815
APA StyleAsumda, F. Z., Alzoubi, S., Padarath, K., John, N., Jones, K., Kolhe, R., Mondal, A. K., Lee, T. J., Zhi, W., Huebert, R. C., Staff, N. P., & Kirkeby, L. A. (2025). Efficient Generation of Induced Pluripotent Stem Cell-Derived Definitive Endoderm Cells with Growth Factors and Small Molecules. Cells, 14(11), 815. https://doi.org/10.3390/cells14110815






