Functional Dynamics of Arginine Mono- and Di-Methylation
Abstract
1. Introduction
2. Detection and Analysis of Arginine Methylation
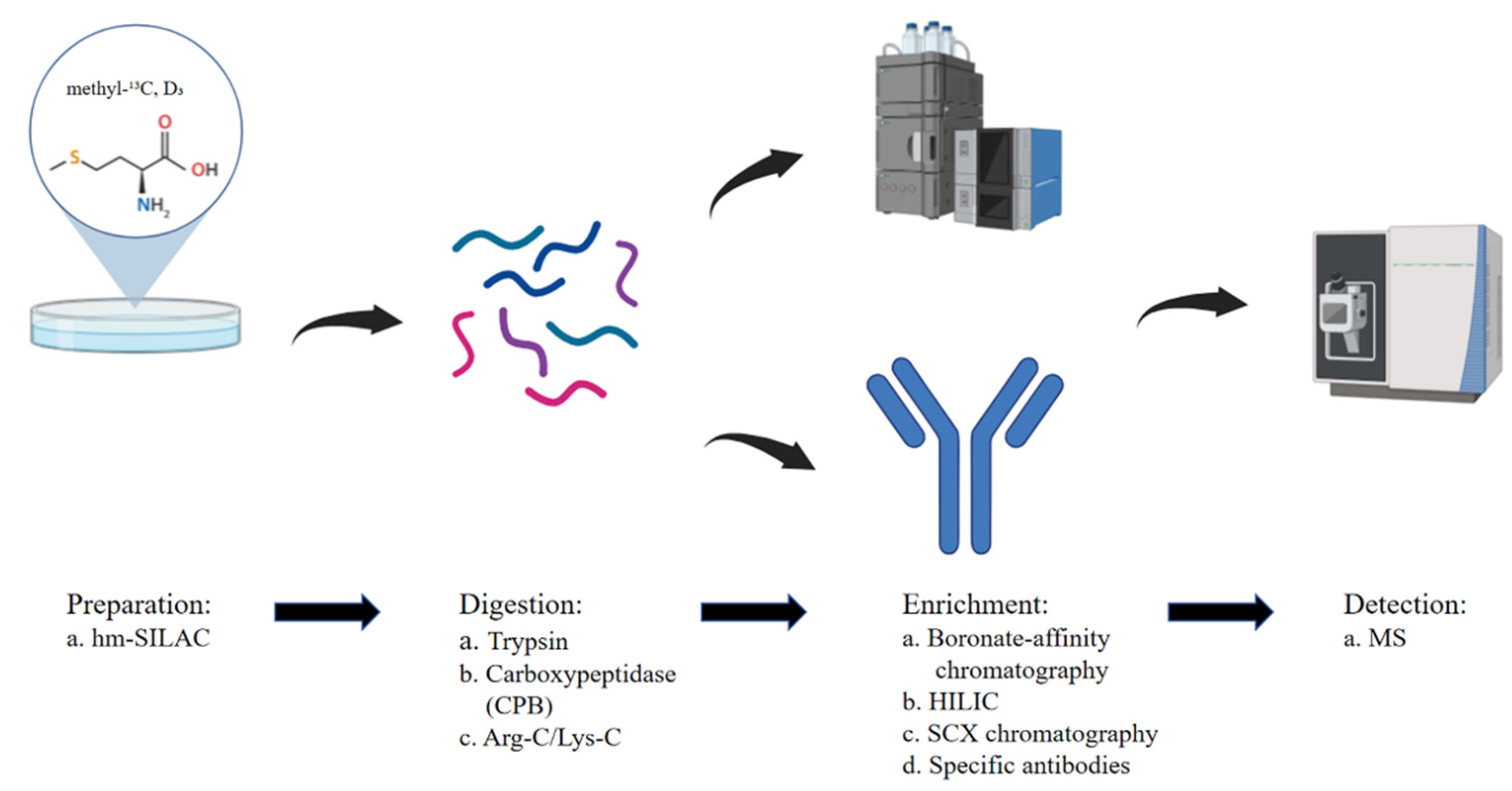
2.1. Advances in Proteomics Data Analysis
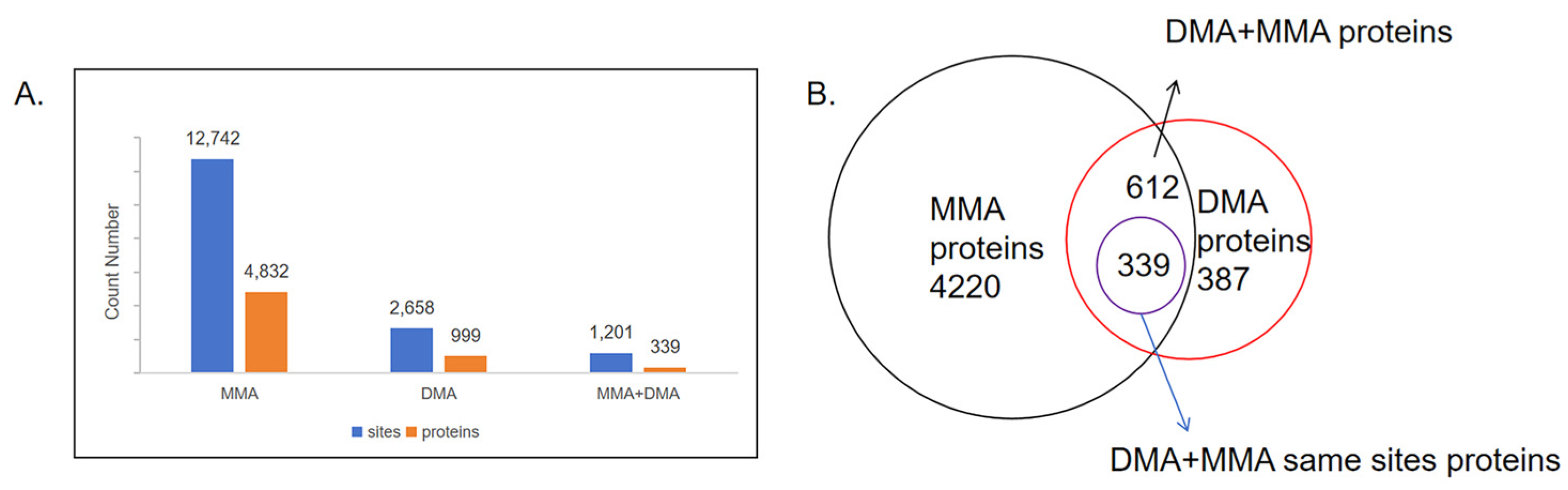
2.2. Quantification and Distribution of Methylation Sites
3. Functional Implications of Arginine Methylation
3.1. Impact on Protein Properties
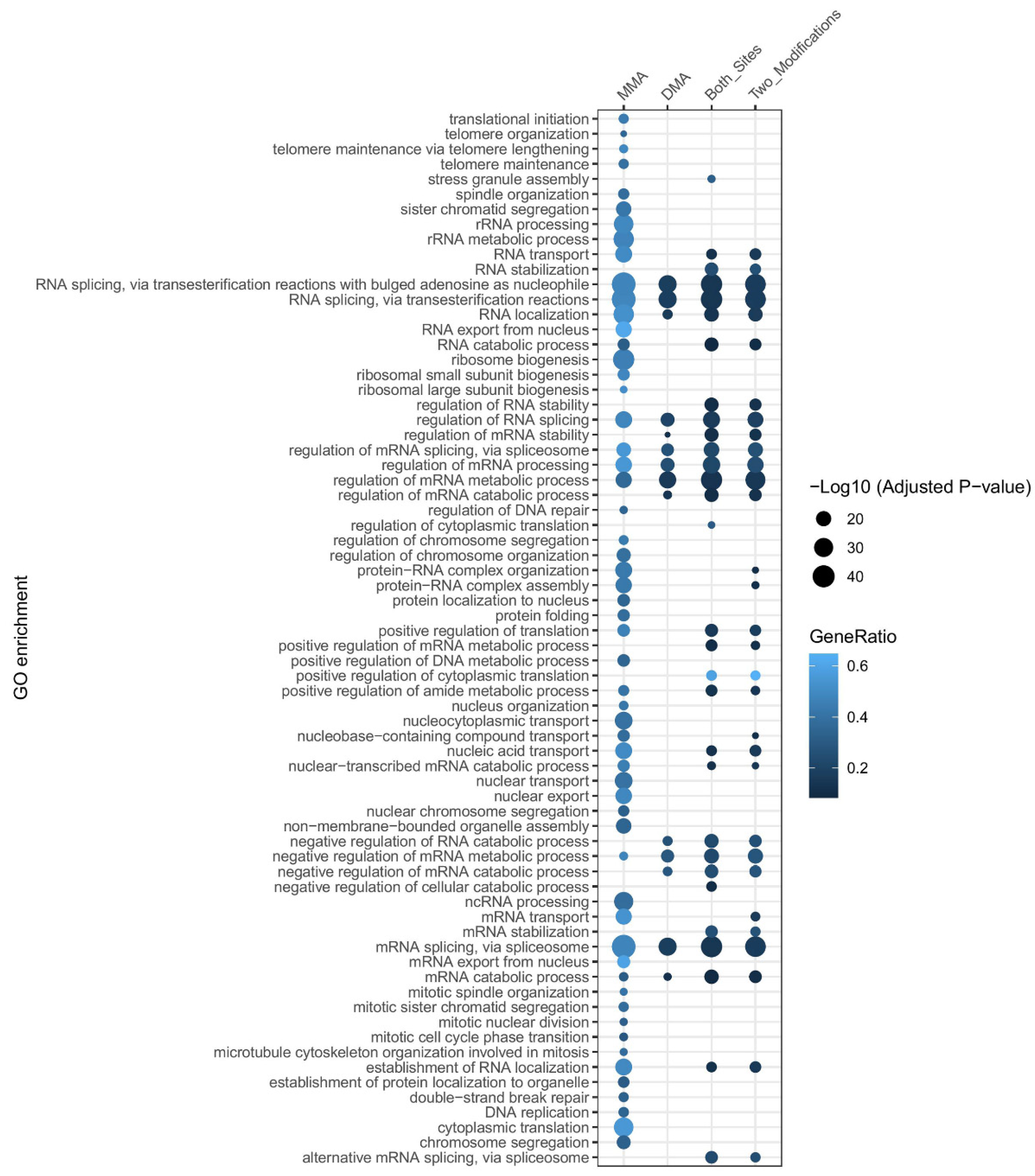
3.2. Diverse Functions of MMA and DMA
3.2.1. Overview
3.2.2. Tissue-Specific and Disease-Related Variability in Arginine Methylation
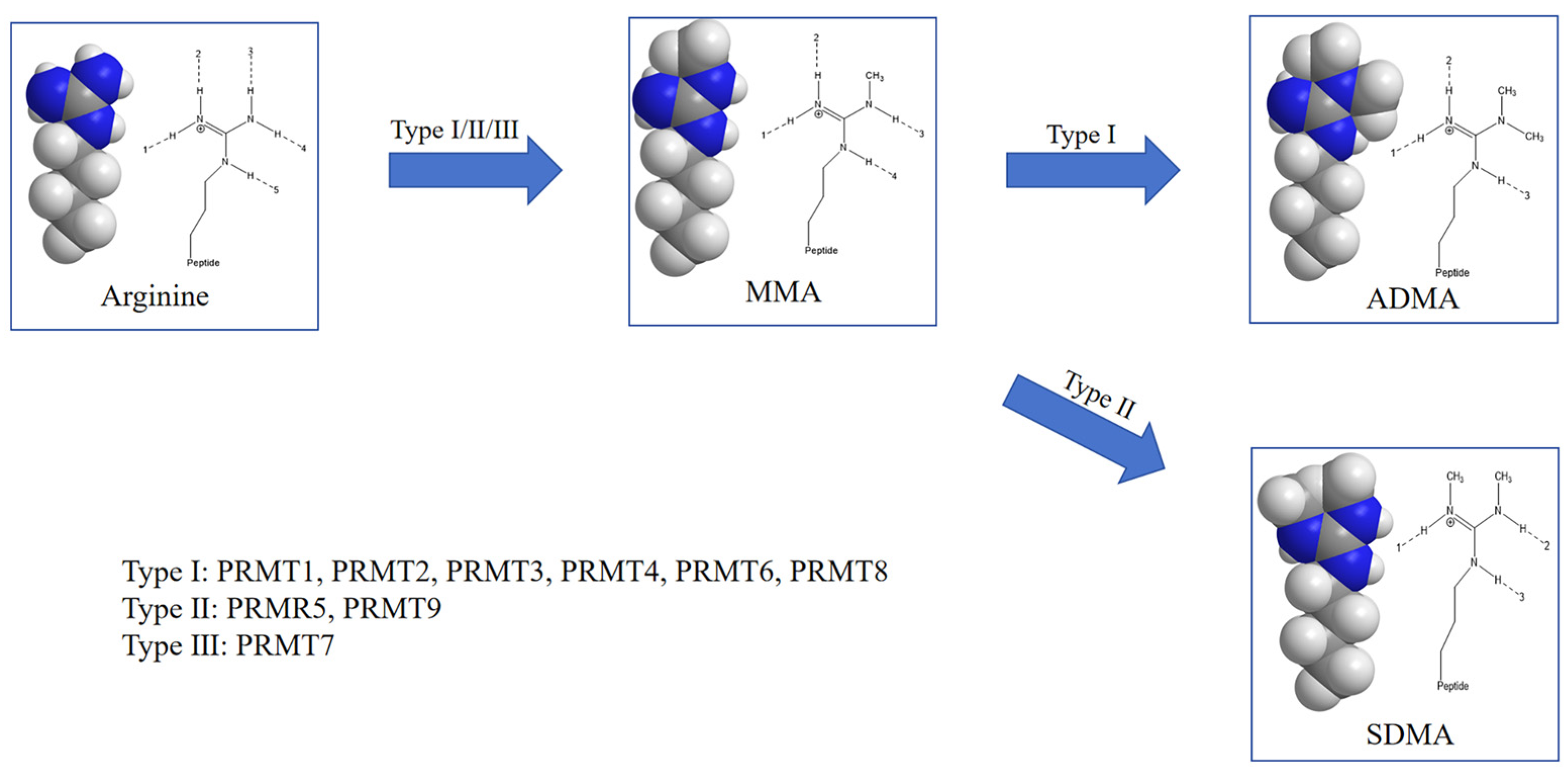
4. PRMTs and Their Role in Arginine Methylation
4.1. Functional Specificity of PRMTs
4.1.1. PRMT1 and PRMT5 in Arginine Methylation Regulation
4.1.2. Functional Redundancy and PRMT Interplay
4.1.3. Site-Specific Methylation and PRMT Crosstalk
4.1.4. Functional Specialization of PRMTs
4.2. Structural Basis of PRMT Activity
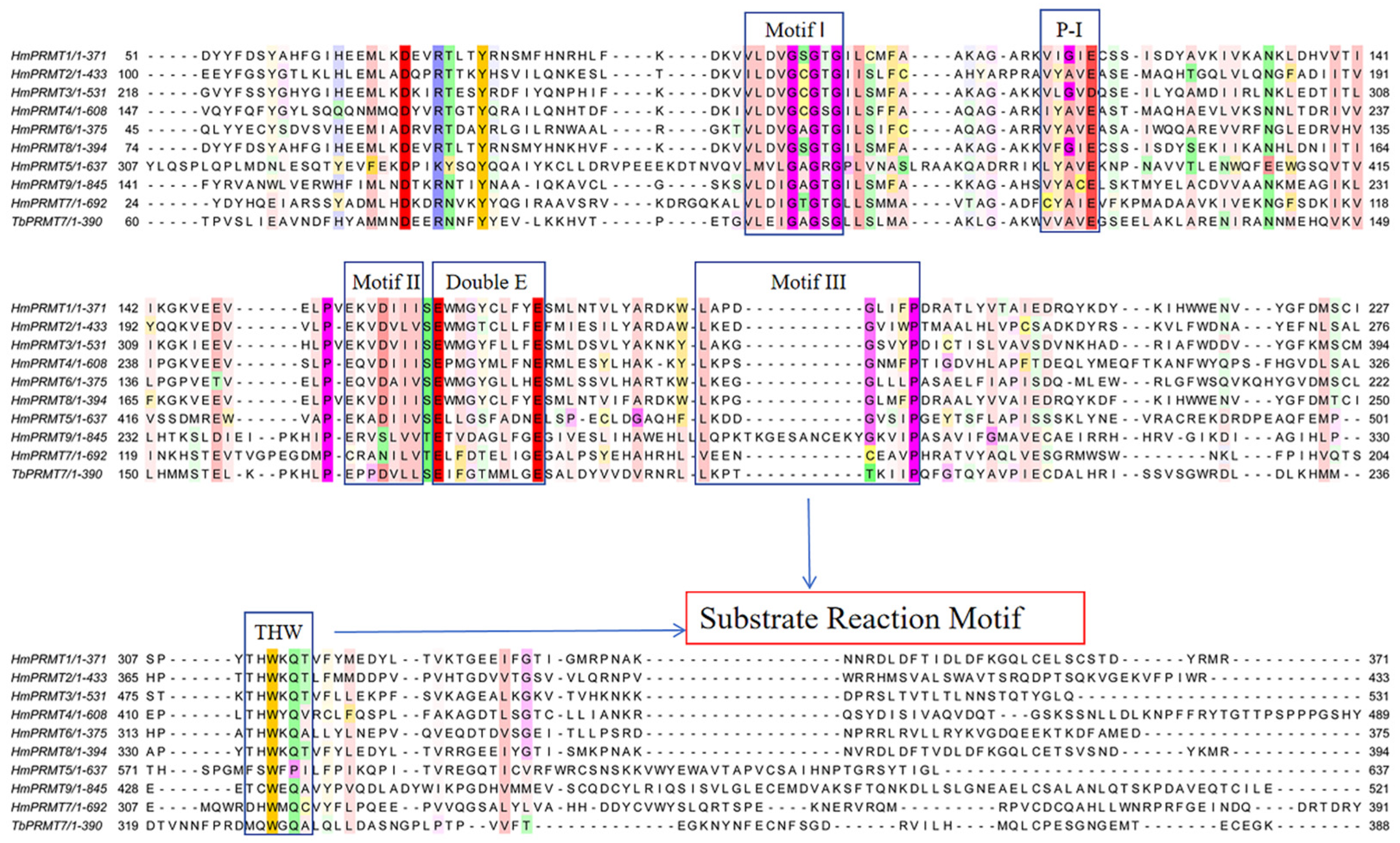
4.2.1. Catalytic Motifs and Substrate Recognition
4.2.2. Free-Energy Barriers and Methylation Specificity
4.2.3. Functional Implications of PRMT Mono- vs. Di-Methylation
5. Regulation and Dynamics of Arginine Methylation
5.1. Substrate Motifs and PRMT Specificity
5.2. Structural Basis of PRMT-Substrate Recognition
6. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deribe, Y.L.; Pawson, T.; Dikic, I. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 2010, 17, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Ambler, R.P.; Rees, M.W. Epsilon-N-Methyl-lysine in bacterial flagellar protein. Nature 1959, 184, 56–57. [Google Scholar] [CrossRef]
- Biggar, K.K.; Li, S.S. Non-histone protein methylation as a regulator of cellular signalling and function. Nat. Rev. Mol. Cell Biol. 2015, 16, 5–17. [Google Scholar] [CrossRef]
- Fulton, M.D.; Brown, T.; Zheng, Y.G. The Biological Axis of Protein Arginine Methylation and Asymmetric Dimethylarginine. Int. J. Mol. Sci. 2019, 20, 3322. [Google Scholar] [CrossRef]
- Guccione, E.; Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 642–657. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Schapira, M.; Arrowsmith, C.H.; Barsyte-Lovejoy, D. Protein arginine methylation: From enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 2021, 20, 509–530. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Richard, S. Cellular pathways influenced by protein arginine methylation: Implications for cancer. Mol. Cell 2021, 81, 4357–4368. [Google Scholar] [CrossRef]
- Fuhrmann, J.; Clancy, K.W.; Thompson, P.R. Chemical biology of protein arginine modifications in epigenetic regulation. Chem. Rev. 2015, 115, 5413–5461. [Google Scholar] [CrossRef]
- Fischer, N.; Neumann, P.; Konevega, A.L.; Bock, L.V.; Ficner, R.; Rodnina, M.V.; Stark, H. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature 2015, 520, 567–570. [Google Scholar] [CrossRef]
- Pellegrino, S.; Dent, K.C.; Spikes, T.; Warren, A.J. Cryo-EM reconstruction of the human 40S ribosomal subunit at 2.15 A resolution. Nucleic Acids Res. 2023, 51, 4043–4054. [Google Scholar] [CrossRef]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef]
- Bremang, M.; Cuomo, A.; Agresta, A.M.; Stugiewicz, M.; Spadotto, V.; Bonaldi, T. Mass spectrometry-based identification and characterisation of lysine and arginine methylation in the human proteome. Mol. Biosyst. 2013, 9, 2231–2247. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, V.; Guo, A.; Trudgian, D.; Thomas, B.; Acuto, O. Comprehensive identification of arginine methylation in primary T cells reveals regulatory roles in cell signalling. Nat. Commun. 2015, 6, 6758. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Gu, H.; Zhou, J.; Mulhern, D.; Wang, Y.; Lee, K.A.; Yang, V.; Aguiar, M.; Kornhauser, J.; Jia, X.; et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol. Cell. Proteom. 2014, 13, 372–387. [Google Scholar] [CrossRef] [PubMed]
- Sylvestersen, K.B.; Horn, H.; Jungmichel, S.; Jensen, L.J.; Nielsen, M.L. Proteomic analysis of arginine methylation sites in human cells reveals dynamic regulation during transcriptional arrest. Mol. Cell. Proteom. 2014, 13, 2072–2088. [Google Scholar] [CrossRef]
- Ong, S.E.; Mittler, G.; Mann, M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat. Methods 2004, 1, 119–126. [Google Scholar] [CrossRef]
- Uhlmann, T.; Geoghegan, V.L.; Thomas, B.; Ridlova, G.; Trudgian, D.C.; Acuto, O. A method for large-scale identification of protein arginine methylation. Mol. Cell. Proteom. 2012, 11, 1489–1499. [Google Scholar] [CrossRef]
- Larsen, S.C.; Sylvestersen, K.B.; Mund, A.; Lyon, D.; Mullari, M.; Madsen, M.V.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Proteome-wide analysis of arginine monomethylation reveals widespread occurrence in human cells. Sci. Signal. 2016, 9, rs9. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Q.; Wang, Y.; Wang, K.; Liu, Z.; Zhang, W.; Ye, M. An efficient approach based on basic strong cation exchange chromatography for enriching methylated peptides with high specificity for methylproteomics analysis. Anal. Chim. Acta 2021, 1161, 338467. [Google Scholar] [CrossRef]
- Ma, M.; Zhao, X.; Chen, S.; Zhao, Y.; Yang, L.; Feng, Y.; Qin, W.; Li, L.; Jia, C. Strategy Based on Deglycosylation, Multiprotease, and Hydrophilic Interaction Chromatography for Large-Scale Profiling of Protein Methylation. Anal. Chem. 2017, 89, 12909–12917. [Google Scholar] [CrossRef]
- Wang, K.; Dong, M.; Mao, J.; Wang, Y.; Jin, Y.; Ye, M.; Zou, H. Antibody-Free Approach for the Global Analysis of Protein Methylation. Anal. Chem. 2016, 88, 11319–11327. [Google Scholar] [CrossRef]
- Hartel, N.G.; Chew, B.; Qin, J.; Xu, J.; Graham, N.A. Deep Protein Methylation Profiling by Combined Chemical and Immunoaffinity Approaches Reveals Novel PRMT1 Targets. Mol. Cell. Proteom. 2019, 18, 2149–2164. [Google Scholar] [CrossRef]
- Lu, L.; Ye, Z.; Zhang, R.; Olsen, J.V.; Yuan, Y.; Mao, Y. ETD-Based Proteomic Profiling Improves Arginine Methylation Identification and Reveals Novel PRMT5 Substrates. J. Proteome Res. 2024, 23, 1014–1027. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Z.; Zhou, J.; Wang, Y.; Wang, K.; Qin, H.; Ye, M. Chemical Depletion of Histidine-Containing Peptides Allows Identification of More Low-Abundance Methylation Sites from Proteome Samples. J. Proteome Res. 2021, 20, 2497–2505. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Z.; Zhang, S.; Li, Y.; Wang, Y.; Fang, Z.; Ma, Y.; Liu, Z.; Zhang, W.; Li, D.; et al. Global profiling of arginine dimethylation in regulating protein phase separation by a steric effect-based chemical-enrichment method. Proc. Natl. Acad. Sci. USA 2022, 119, e2205255119. [Google Scholar] [CrossRef]
- Liu, Z.; Fang, Z.; Wang, K.; Ye, M. Hydrophobic Derivatization Strategy Facilitates Comprehensive Profiling of Protein Methylation. J. Proteome Res. 2023, 22, 3275–3281. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yan, X.; Fu, B.; Xu, Y.; Li, L.; Chang, C.; Jia, C. mNeuCode Empowers Targeted Proteome Analysis of Arginine Dimethylation. Anal. Chem. 2023, 95, 3684–3693. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, Q.; Zhou, J.; Wang, Y.; Liu, Z.; Wang, K.; Ye, M. A Chemoenzymatic Method Enables Global Enrichment and Characterization of Protein Arginine Methylation. Anal. Chem. 2024, 96, 14612–14620. [Google Scholar] [CrossRef]
- Bedford, M.T.; Richard, S. Arginine methylation an emerging regulator of protein function. Mol. Cell 2005, 18, 263–272. [Google Scholar] [CrossRef]
- Gui, S.; Wooderchak-Donahue, W.L.; Zang, T.; Chen, D.; Daly, M.P.; Zhou, Z.S.; Hevel, J.M. Substrate-induced control of product formation by protein arginine methyltransferase 1. Biochemistry 2013, 52, 199–209. [Google Scholar] [CrossRef]
- Evich, M.; Stroeva, E.; Zheng, Y.G.; Germann, M.W. Effect of methylation on the side-chain pKa value of arginine. Protein Sci. 2016, 25, 479–486. [Google Scholar] [CrossRef]
- Santos, M. Conformational analysis of arginine methylated and di methylated and serine phosphorylated in silico. Preprint 2024. [Google Scholar] [CrossRef]
- Ryan, V.H.; Dignon, G.L.; Zerze, G.H.; Chabata, C.V.; Silva, R.; Conicella, A.E.; Amaya, J.; Burke, K.A.; Mittal, J.; Fawzi, N.L. Mechanistic View of hnRNPA2 Low-Complexity Domain Structure, Interactions, and Phase Separation Altered by Mutation and Arginine Methylation. Mol. Cell 2018, 69, 465–479.e467. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.P.; Zhou, W.; Wang, J.; Huang, X.; Zuo, Y.; Wang, T.S.; Gao, X.; Xu, Y.Y.; Zou, S.W.; Liu, Y.B.; et al. Arginine Methylation of MDH1 by CARM1 Inhibits Glutamine Metabolism and Suppresses Pancreatic Cancer. Mol. Cell 2016, 64, 673–687. [Google Scholar] [CrossRef]
- Wang, K.; Luo, L.; Fu, S.; Wang, M.; Wang, Z.; Dong, L.; Wu, X.; Dai, L.; Peng, Y.; Shen, G.; et al. PHGDH arginine methylation by PRMT1 promotes serine synthesis and represents a therapeutic vulnerability in hepatocellular carcinoma. Nat. Commun. 2023, 14, 1011. [Google Scholar] [CrossRef]
- Lei, Y.; Han, P.; Chen, Y.; Wang, H.; Wang, S.; Wang, M.; Liu, J.; Yan, W.; Tian, D.; Liu, M. Protein arginine methyltransferase 3 promotes glycolysis and hepatocellular carcinoma growth by enhancing arginine methylation of lactate dehydrogenase A. Clin. Transl. Med. 2022, 12, e686. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.; Lee, J.Y.; Ha, S.J.; Yu, S.; Shin, J.K.; Kim, H.C. Proteome-wide identification of arginine methylation in colorectal cancer tissues from patients. Proteome Sci. 2020, 18, 6. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Liu, F.; Miles, H.N.; Kim, E.J.; Fields, L.; Xu, W.; Li, L. Proteome-wide Profiling of Asymmetric Dimethylated Arginine in Human Breast Tumors. J. Am. Soc. Mass Spectrom. 2023, 34, 1692–1700. [Google Scholar] [CrossRef]
- Onwuli, D.O.; Rigau-Roca, L.; Cawthorne, C.; Beltran-Alvarez, P. Mapping arginine methylation in the human body and cardiac disease. Proteom. Clin. Appl. 2017, 11, 1600106. [Google Scholar] [CrossRef]
- Yang, C.Y.; Chiu, L.L.; Chang, C.C.; Chuang, H.C.; Tan, T.H. Induction of DUSP14 ubiquitination by PRMT5-mediated arginine methylation. FASEB J. 2018, 32, 6760–6770. [Google Scholar] [CrossRef]
- Aikio, M.; Odeh, H.M.; Wobst, H.J.; Lee, B.L.; Chan, U.; Mauna, J.C.; Mack, K.L.; Class, B.; Ollerhead, T.A.; Ford, A.F.; et al. Opposing roles of p38alpha-mediated phosphorylation and PRMT1-mediated arginine methylation in driving TDP-43 proteinopathy. Cell Rep. 2025, 44, 115205. [Google Scholar] [CrossRef]
- Tang, J.; Frankel, A.; Cook, R.J.; Kim, S.; Paik, W.K.; Williams, K.R.; Clarke, S.; Herschman, H.R. PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 2000, 275, 7723–7730. [Google Scholar] [CrossRef]
- Li, W.J.; He, Y.H.; Yang, J.J.; Hu, G.S.; Lin, Y.A.; Ran, T.; Peng, B.L.; Xie, B.L.; Huang, M.F.; Gao, X.; et al. Profiling PRMT methylome reveals roles of hnRNPA1 arginine methylation in RNA splicing and cell growth. Nat. Commun. 2021, 12, 1946. [Google Scholar] [CrossRef] [PubMed]
- Hamey, J.J.; Rakow, S.; Bouchard, C.; Senst, J.M.; Kolb, P.; Bauer, U.M.; Wilkins, M.R.; Hart-Smith, G. Systematic investigation of PRMT6 substrate recognition reveals broad specificity with a preference for an RG motif or basic and bulky residues. FEBS J. 2021, 288, 5668–5691. [Google Scholar] [CrossRef] [PubMed]
- Musiani, D.; Bok, J.; Massignani, E.; Wu, L.; Tabaglio, T.; Ippolito, M.R.; Cuomo, A.; Ozbek, U.; Zorgati, H.; Ghoshdastider, U.; et al. Proteomics profiling of arginine methylation defines PRMT5 substrate specificity. Sci. Signal. 2019, 12, eaat8388. [Google Scholar] [CrossRef]
- Radzisheuskaya, A.; Shliaha, P.V.; Grinev, V.; Lorenzini, E.; Kovalchuk, S.; Shlyueva, D.; Gorshkov, V.; Hendrickson, R.C.; Jensen, O.N.; Helin, K. PRMT5 methylome profiling uncovers a direct link to splicing regulation in acute myeloid leukemia. Nat. Struct. Mol. Biol. 2019, 26, 999–1012. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, L.; Cheng, X. Crystal structure of the conserved core of protein arginine methyltransferase PRMT3. EMBO J. 2000, 19, 3509–3519. [Google Scholar] [CrossRef]
- Tewary, S.K.; Zheng, Y.G.; Ho, M.-C. Protein arginine methyltransferases: Insights into the enzyme structure and mechanism at the atomic level. Cell. Mol. Life Sci. 2019, 76, 2917–2932. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, X. Structure of the predominant protein arginine methyltransferase PRMT1 and analysis of its binding to substrate peptides. Structure 2003, 11, 509–520. [Google Scholar] [CrossRef]
- Antonysamy, S.; Bonday, Z.; Campbell, R.M.; Doyle, B.; Druzina, Z.; Gheyi, T.; Han, B.; Jungheim, L.N.; Qian, Y.; Rauch, C.; et al. Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl. Acad. Sci. USA 2012, 109, 17960–17965. [Google Scholar] [CrossRef]
- Cura, V.; Troffer-Charlier, N.; Wurtz, J.M.; Bonnefond, L.; Cavarelli, J. Structural insight into arginine methylation by the mouse protein arginine methyltransferase 7: A zinc finger freezes the mimic of the dimeric state into a single active site. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 2401–2412. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Toma-Fukai, S.; Kim, J.D.; Fukamizu, A.; Shimizu, T. Protein arginine methyltransferase 7 has a novel homodimer-like structure formed by tandem repeats. FEBS Lett. 2014, 588, 1942–1948. [Google Scholar] [CrossRef] [PubMed]
- Rossi, V.; Nielson, S.E.; Ortolano, A.; Lonardo, I.; Haroldsen, E.; Comer, D.; Price, O.M.; Wallace, N.; Hevel, J.M. Oligomerization of protein arginine methyltransferase 1 and its effect on methyltransferase activity and substrate specificity. Protein Sci. 2024, 33, e5118. [Google Scholar] [CrossRef] [PubMed]
- Price, O.M.; Thakur, A.; Ortolano, A.; Towne, A.; Velez, C.; Acevedo, O.; Hevel, J.M. Naturally occurring cancer-associated mutations disrupt oligomerization and activity of protein arginine methyltransferase 1 (PRMT1). J. Biol. Chem. 2021, 297, 101336. [Google Scholar] [CrossRef]
- Wang, C.; Zhu, Y.; Caceres Tamar, B.; Liu, L.; Peng, J.; Wang, J.; Chen, J.; Chen, X.; Zhang, Z.; Zuo, X.; et al. Structural Determinants for the Strict Monomethylation Activity by Trypanosoma brucei Protein Arginine Methyltransferase 7. Structure 2014, 22, 756–768. [Google Scholar] [CrossRef]
- Chu, Y.; Li, G.; Guo, H. QM/MM MD and free energy simulations of the methylation reactions catalyzed by protein arginine methyltransferase PRMT3. Can. J. Chem. 2013, 91, 605–612. [Google Scholar] [CrossRef]
- Yue, Y.; Chu, Y.; Guo, H. Computational Study of Symmetric Methylation on Histone Arginine Catalyzed by Protein Arginine Methyltransferase PRMT5 through QM/MM MD and Free Energy Simulations. Molecules 2015, 20, 10032–10046. [Google Scholar] [CrossRef]
- Ren, W.S.; Jiang, K.B.; Deng, H.; Lu, N.; Yu, T.; Guo, H.; Qian, P. Catalytic Mechanism and Product Specificity of Protein Arginine Methyltransferase PRMT7: A Study from QM/MM Molecular Dynamics and Free Energy Simulations. J. Chem. Theory Comput. 2020, 16, 5301–5312. [Google Scholar] [CrossRef]
- Debler, E.W.; Jain, K.; Warmack, R.A.; Feng, Y.; Clarke, S.G.; Blobel, G.; Stavropoulos, P. A glutamate/aspartate switch controls product specificity in a protein arginine methyltransferase. Proc. Natl. Acad. Sci. USA 2016, 113, 2068–2073. [Google Scholar] [CrossRef]
- Jain, K.; Warmack, R.A.; Debler, E.W.; Hadjikyriacou, A.; Stavropoulos, P.; Clarke, S.G. Protein Arginine Methyltransferase Product Specificity Is Mediated by Distinct Active-site Architectures. J. Biol. Chem. 2016, 291, 18299–18308. [Google Scholar] [CrossRef]
- Ren, W.S.; Rahman, A.; Jiang, K.B.; Deng, H.; Zhao, Y.Y.; Zhang, W.J.; Liu, K.; Qian, P.; Guo, H. Unraveling the Origins of Changing Product Specificity Properties of Arginine Methyltransferase PRMT7 by the E181D and E181D/Q329A Mutations through QM/MM MD and Free-Energy Simulations. J. Chem. Theory Comput. 2022, 18, 2631–2641. [Google Scholar] [CrossRef] [PubMed]
- Hamey, J.J.; Separovich, R.J.; Wilkins, M.R. MT-MAMS: Protein Methyltransferase Motif Analysis by Mass Spectrometry. J. Proteome Res. 2018, 17, 3485–3491. [Google Scholar] [CrossRef]
- Pahlich, S.; Zakaryan, R.P.; Gehring, H. Identification of proteins interacting with protein arginine methyltransferase 8: The Ewing sarcoma (EWS) protein binds independent of its methylation state. Proteins 2008, 72, 1125–1137. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Li, X.; Cai, X.; Zhou, Z.; Liao, Q.; Liu, X.; Wang, J.; Xiao, W. Asymmetric arginine dimethylation of cytosolic RNA and DNA sensors by PRMT3 attenuates antiviral innate immunity. Proc. Natl. Acad. Sci. USA 2023, 120, e2214956120. [Google Scholar] [CrossRef]
- Ren, R.; Mayer, B.J.; Cicchetti, P.; Baltimore, D. Identification of a ten-amino acid proline-rich SH3 binding site. Science 1993, 259, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Gayatri, S.; Cowles, M.W.; Vemulapalli, V.; Cheng, D.; Sun, Z.W.; Bedford, M.T. Using oriented peptide array libraries to evaluate methylarginine-specific antibodies and arginine methyltransferase substrate motifs. Sci. Rep. 2016, 6, 28718. [Google Scholar] [CrossRef]
- Feng, Y.; Maity, R.; Whitelegge, J.P.; Hadjikyriacou, A.; Li, Z.; Zurita-Lopez, C.; Al-Hadid, Q.; Clark, A.T.; Bedford, M.T.; Masson, J.Y.; et al. Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J. Biol. Chem. 2013, 288, 37010–37025. [Google Scholar] [CrossRef]
- Osborne, T.C.; Obianyo, O.; Zhang, X.; Cheng, X.; Thompson, P.R. Protein arginine methyltransferase 1: Positively charged residues in substrate peptides distal to the site of methylation are important for substrate binding and catalysis. Biochemistry 2007, 46, 13370–13381. [Google Scholar] [CrossRef]
- Feustel, K.; Falchook, G.S. Protein Arginine Methyltransferase 5 (PRMT5) Inhibitors in Oncology Clinical Trials: A review. J. Immunother. Precis. Oncol. 2022, 5, 58–67. [Google Scholar] [CrossRef]
- Wu, J.; Li, D.; Wang, L. Overview of PRMT1 modulators: Inhibitors and degraders. Eur. J. Med. Chem. 2024, 279, 116887. [Google Scholar] [CrossRef]
- Cao, M.; Nguyen, T.; Song, J.; Zheng, Y.G. Biomedical effects of protein arginine methyltransferase inhibitors. J. Biol. Chem. 2025, 301, 108201. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; He, X.; Zhang, L.; Chen, W.; Lin, Y.C.; Liu, S.B.; Wang, H.; Nguyen, L.X.T.; Li, M.; Zhu, Y.; et al. Targeting PRMT9-mediated arginine methylation suppresses cancer stem cell maintenance and elicits cGAS-mediated anticancer immunity. Nat. Cancer 2024, 5, 601–624. [Google Scholar] [CrossRef] [PubMed]

| Methylation Formation Type | PRMT and Mutant Sites | Angular Orientation (°) (Nη2)/(Nη1) | Spatial Distance (Å) (Nη2)/(Nη1) | Free Energy (kcal/mol) (Nη2)/(Nη1) |
|---|---|---|---|---|
| MMA | RnPRMT3 | 37.4/54.6 | 3.6/4.9 | 20.4/28.5 |
| HsPRMT5 | Approximately equal | Approximately equal | 29.4/20.4 | |
| TbPRMT7 | Approximately equal | 2.9/4.43 | 32.7/54 | |
| DMA | RnPRMT3 | 11.7/27 | 3.2/3.7 | 18.4/25.4 |
| HsPRMT5 | <30/>30 | Approximately equal | 20.1/31.3 | |
| TbPRMT7 | ≈30/>30 | 3.71/3.33 | 44.34/40.3 | |
| ADMA | TbPRMT7 E181D | Approximately equal | 3.23/3.44 | 36.14/37.4 |
| SDMA | TbPRMT7 E181D/Q329A | ≈30/<30 | 3.76/2.86 | 45.8/32.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Zhu, B.; Winn, R.; Lu, S.; Wang, H. Functional Dynamics of Arginine Mono- and Di-Methylation. Cells 2025, 14, 796. https://doi.org/10.3390/cells14110796
Wang X, Zhu B, Winn R, Lu S, Wang H. Functional Dynamics of Arginine Mono- and Di-Methylation. Cells. 2025; 14(11):796. https://doi.org/10.3390/cells14110796
Chicago/Turabian StyleWang, Xi’ang, Bin Zhu, Robert Winn, Shanfa Lu, and Hengbin Wang. 2025. "Functional Dynamics of Arginine Mono- and Di-Methylation" Cells 14, no. 11: 796. https://doi.org/10.3390/cells14110796
APA StyleWang, X., Zhu, B., Winn, R., Lu, S., & Wang, H. (2025). Functional Dynamics of Arginine Mono- and Di-Methylation. Cells, 14(11), 796. https://doi.org/10.3390/cells14110796








