Recognizing Epithelial Cells in Prostatic Glands Using Deep Learning
Abstract
1. Introduction
- For each size of patches, the accuracy of the trained GlandNet was investigated to determine the effect of patch size on prediction performance, and we identified the one with superior performance.
- GlandNet development was continued using the best-performing patch size (Patch-256) and a semi-supervised approach conducted using the training and validation datasets.
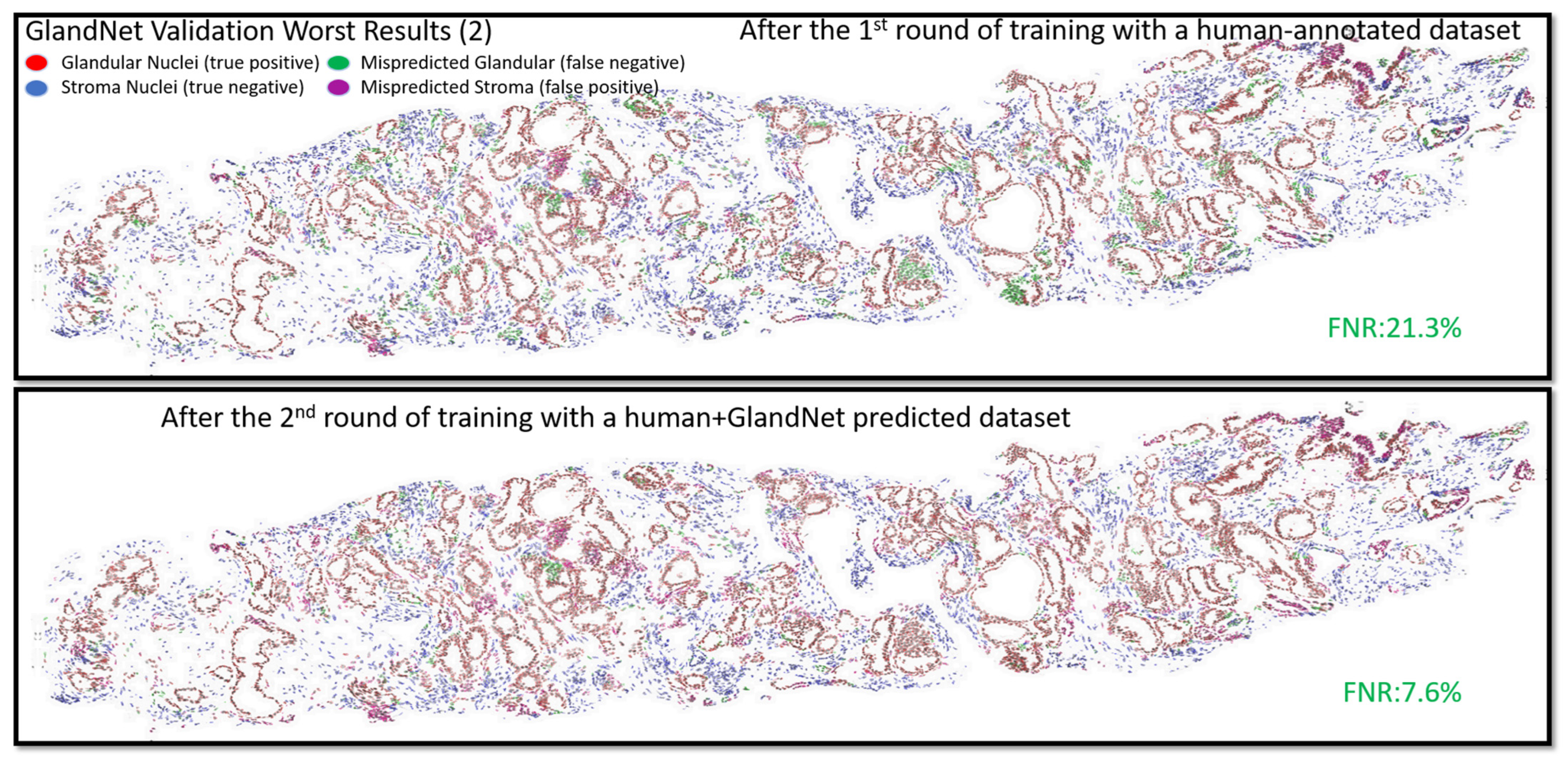
- In the second-round training, GlandNet integrated both human and machine predictions to boost its accuracy, and then it was applied to the test cohort.
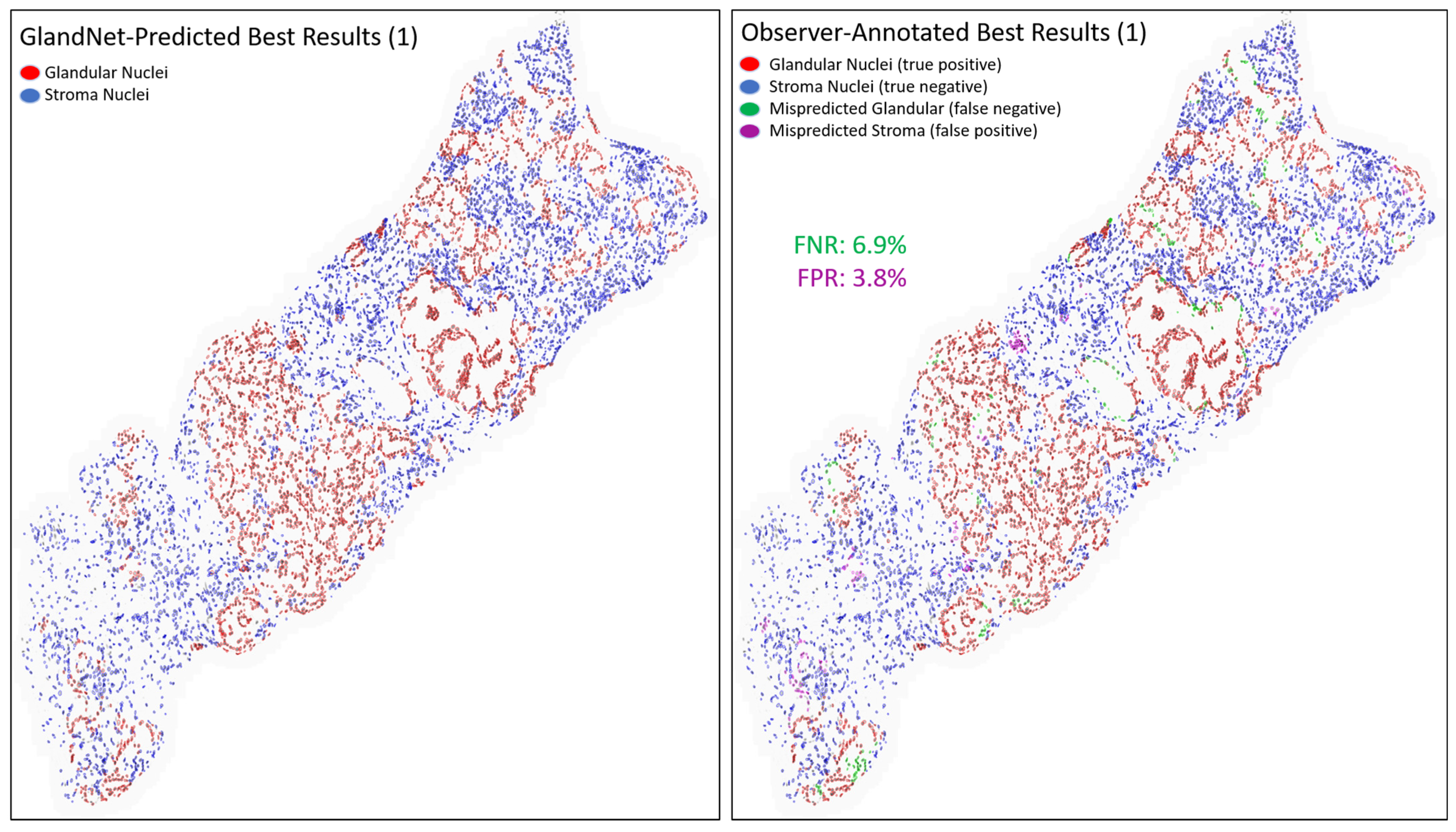
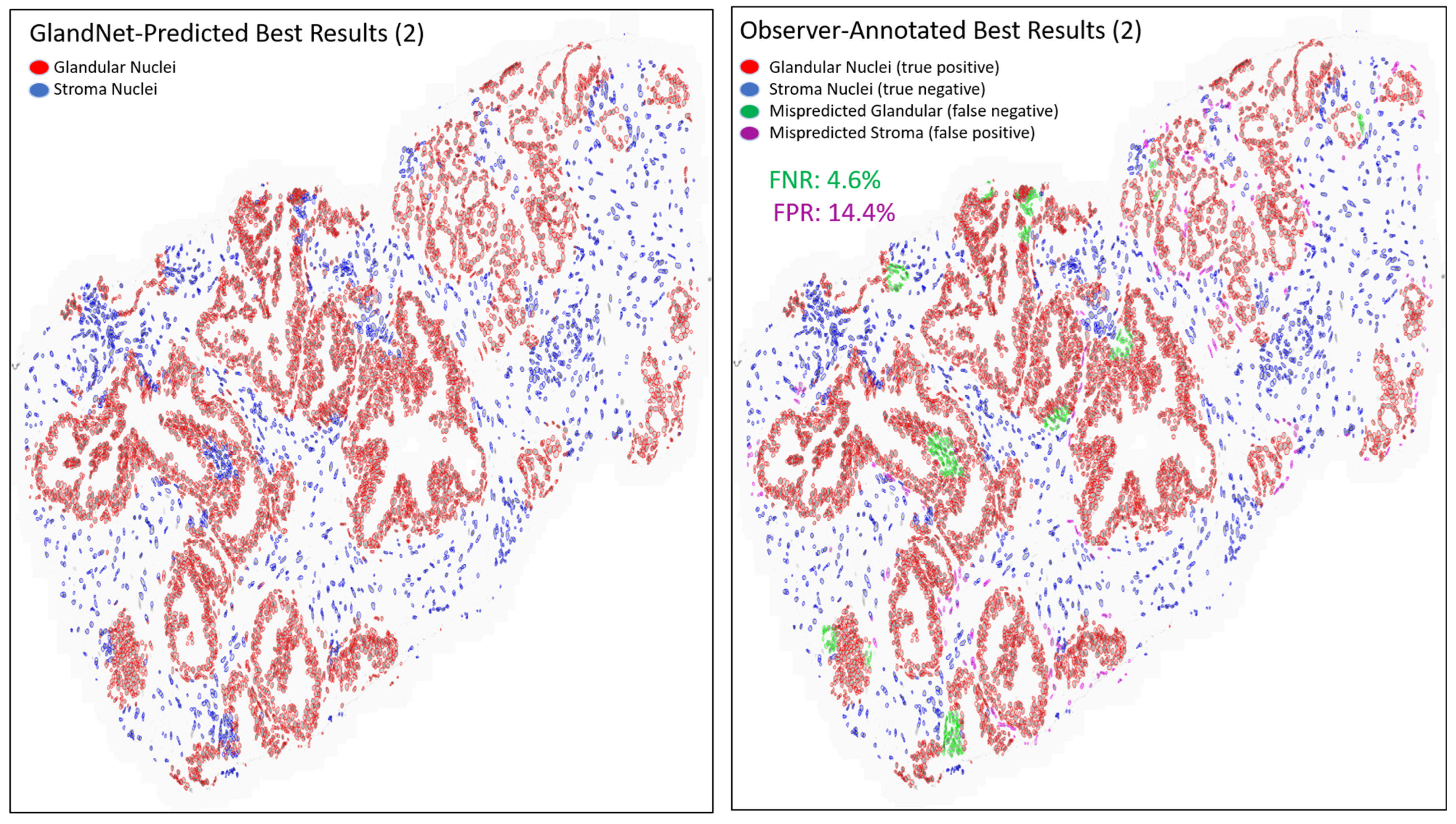
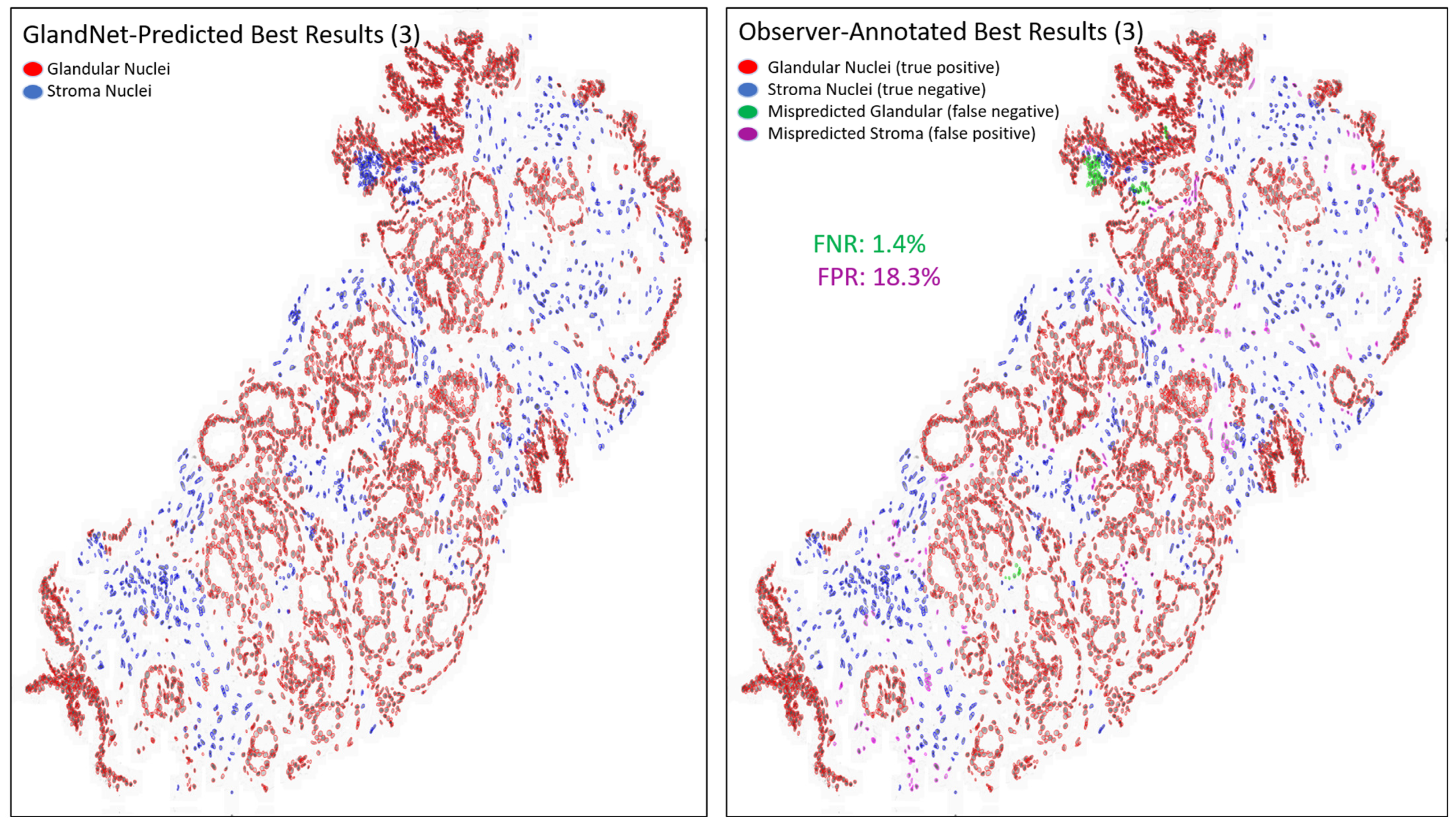
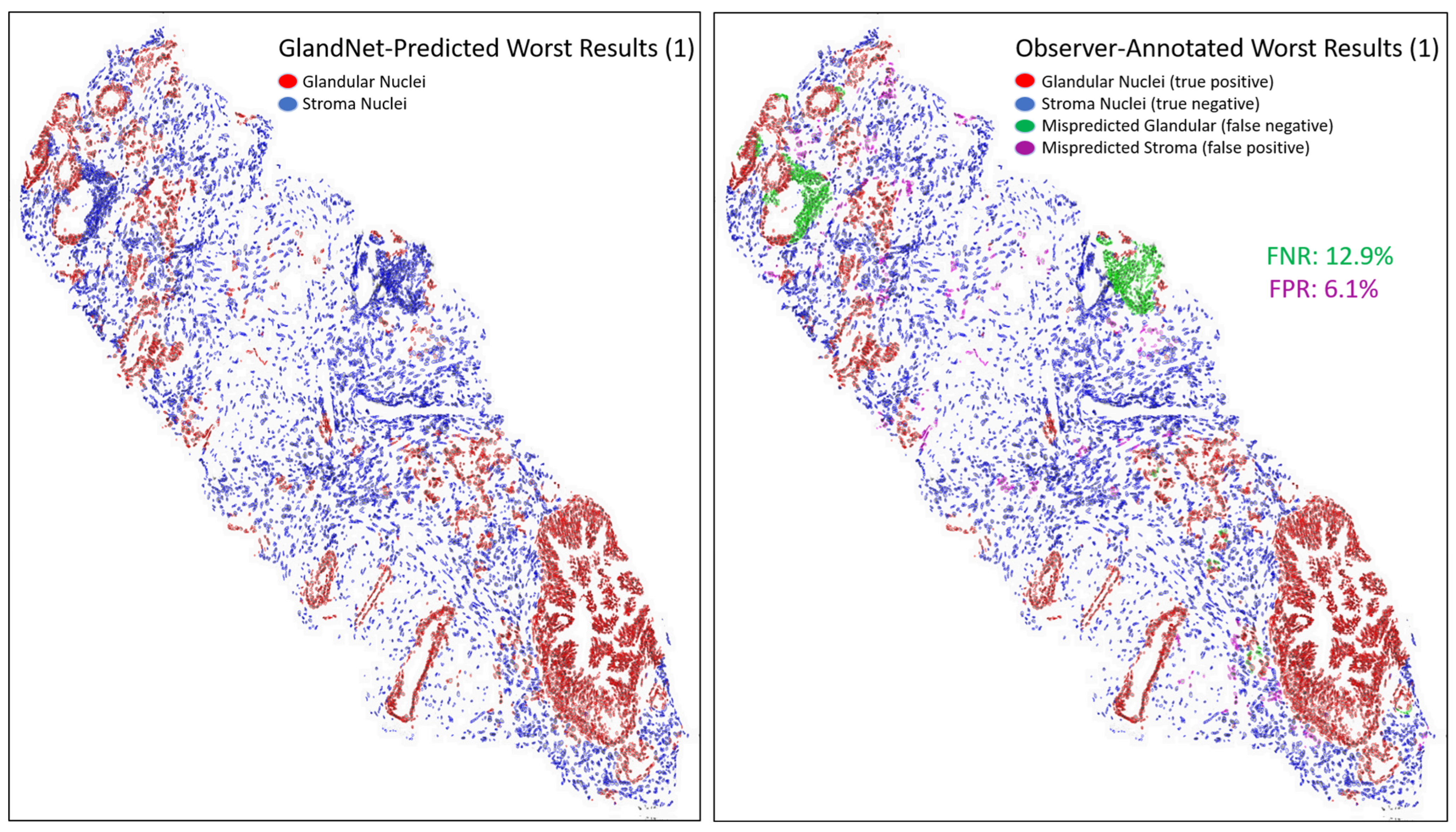
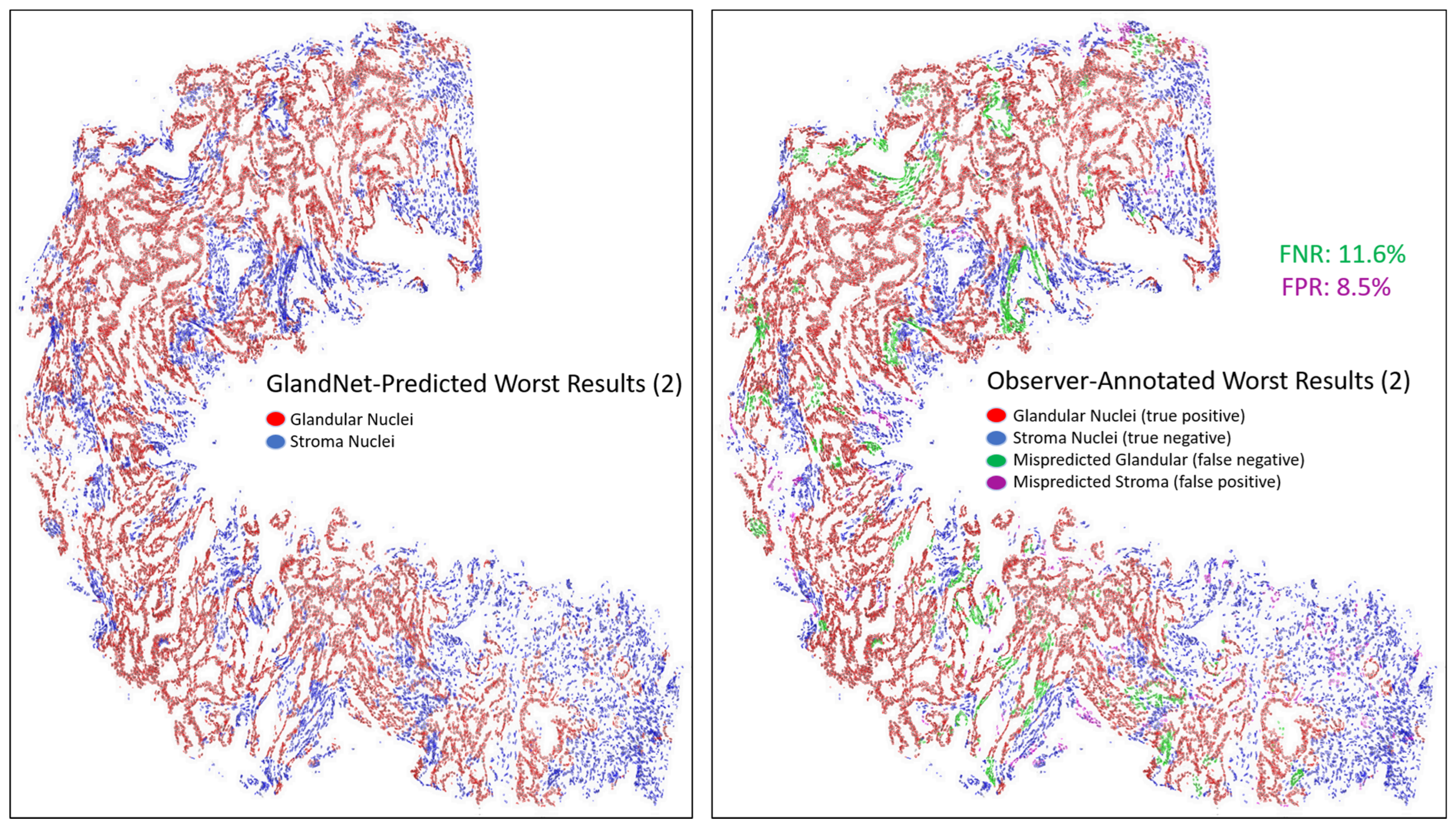
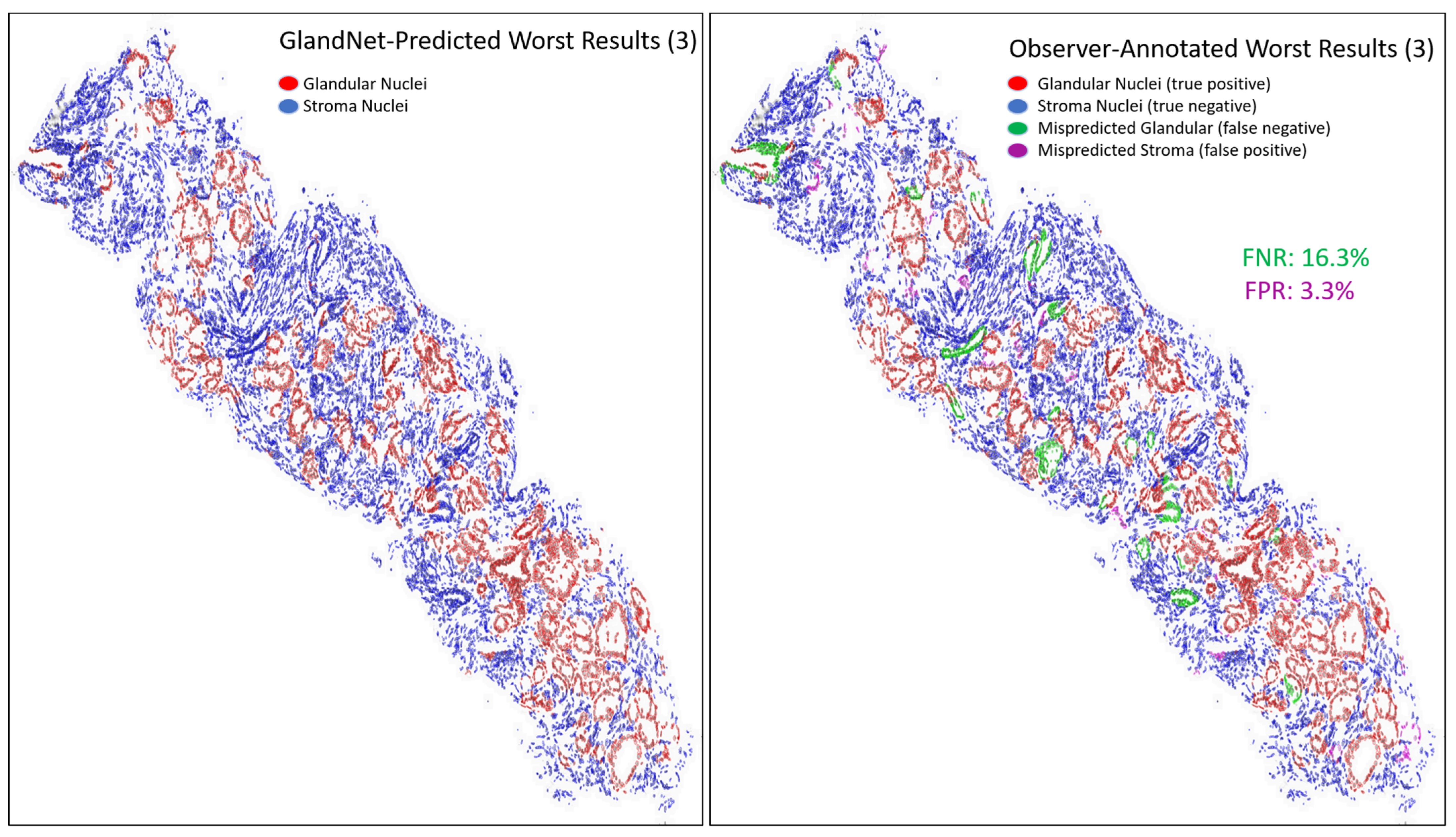
- Prostate glandular cell recognition in the test cohort was evaluated through a majority vote process among three observers to visually identify the three biopsies with the best predictions and the three biopsies with the worst predictions. For these six biopsies, all the 77,952 cells were hand-annotated (C.M.) as within glands or not within glands to estimate the accuracy range of GlandNet.
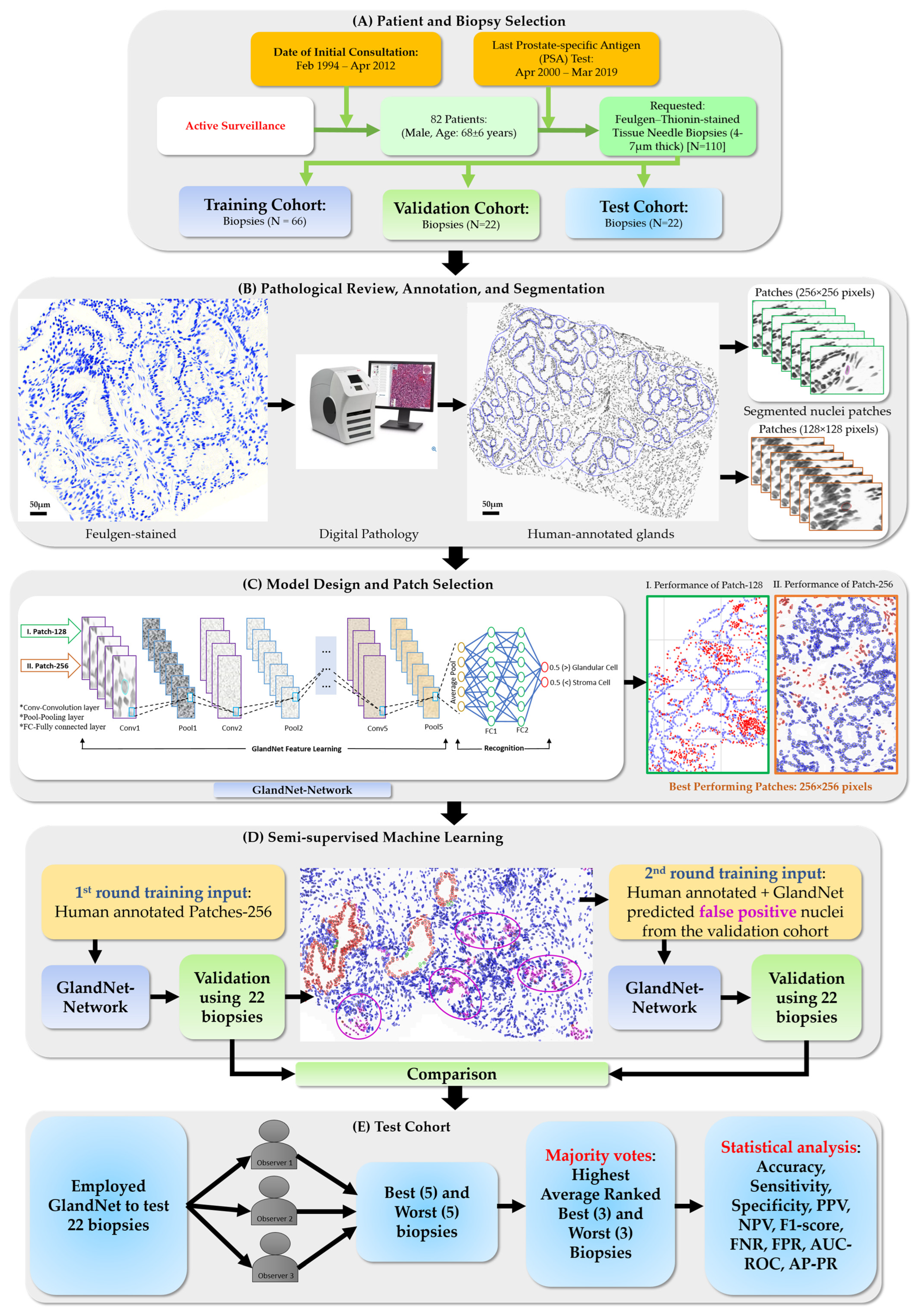
2. Materials and Methods
2.1. Sample Preparation
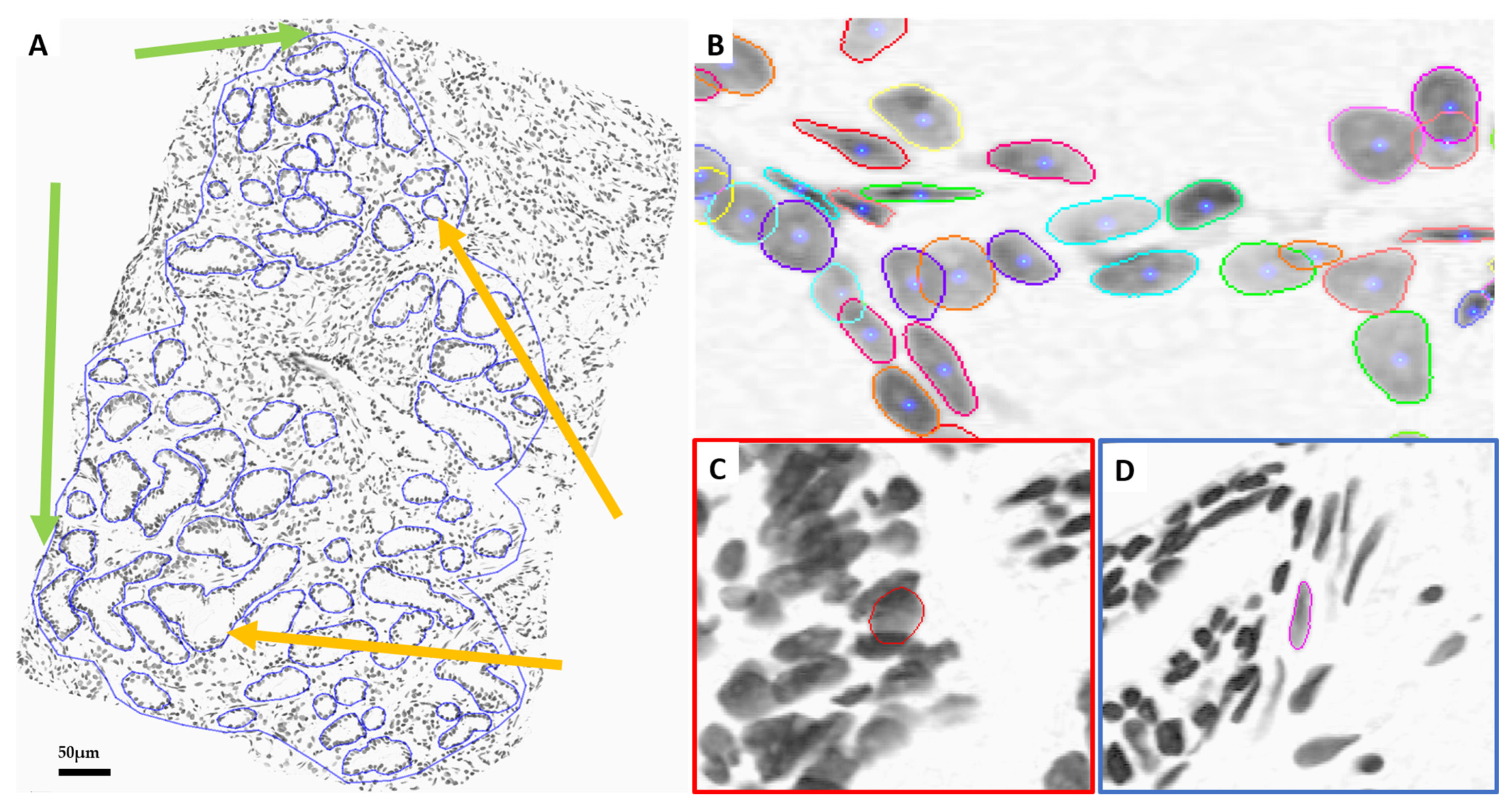
2.2. Pathological Review and Annotation
2.3. Nuclei Segmentation
2.4. GlandNet Algorithm Design
2.5. GlandNet Implementation with Semi-Supervised Learning Technique
3. Results
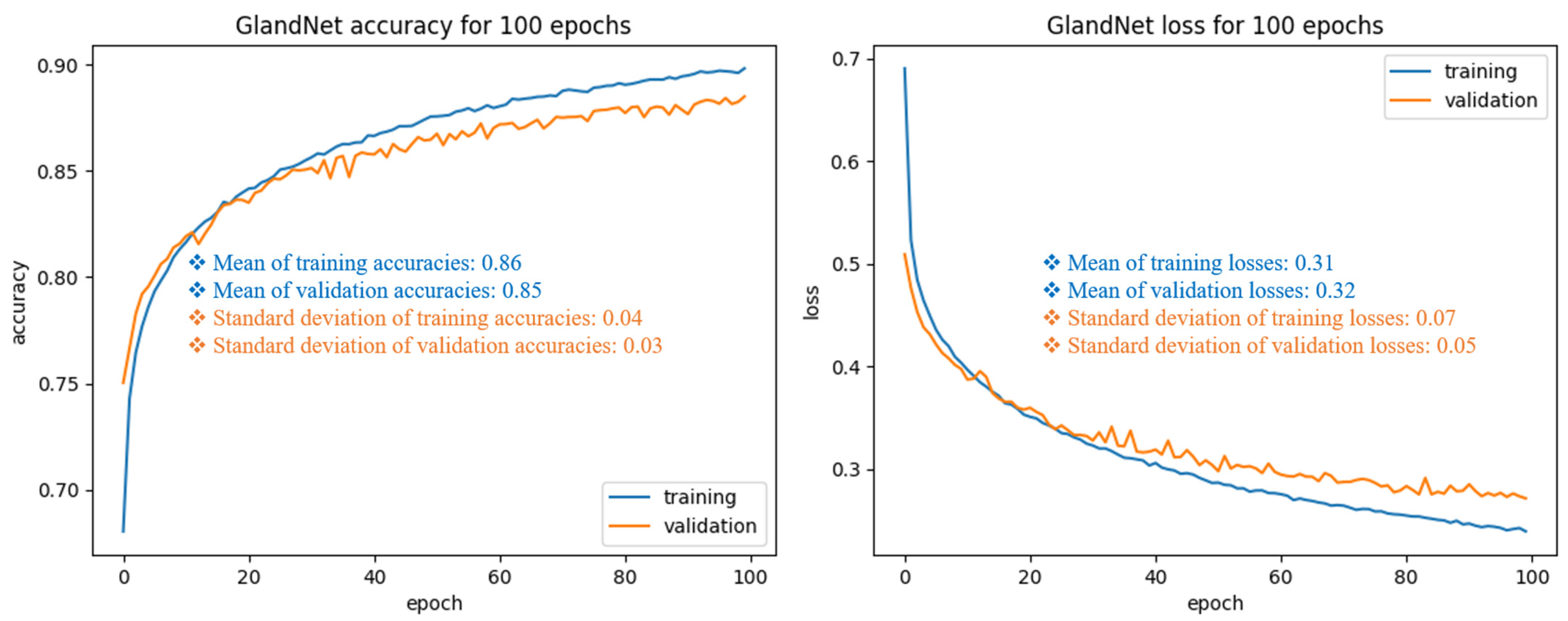
3.1. Training Cohort: Patch Size Selection and Tuning the Process
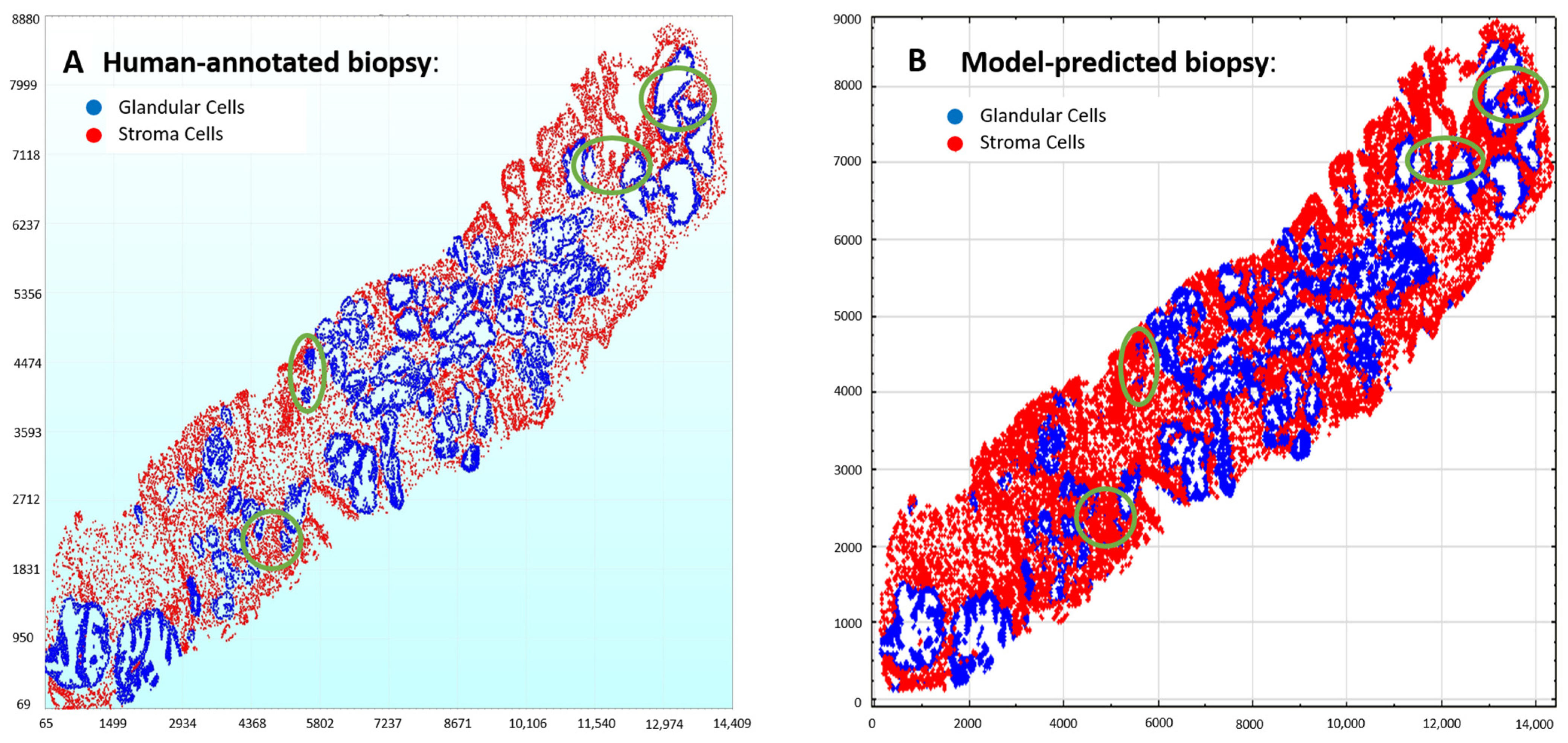
3.2. Validation Cohort: Semi-Supervised Learning
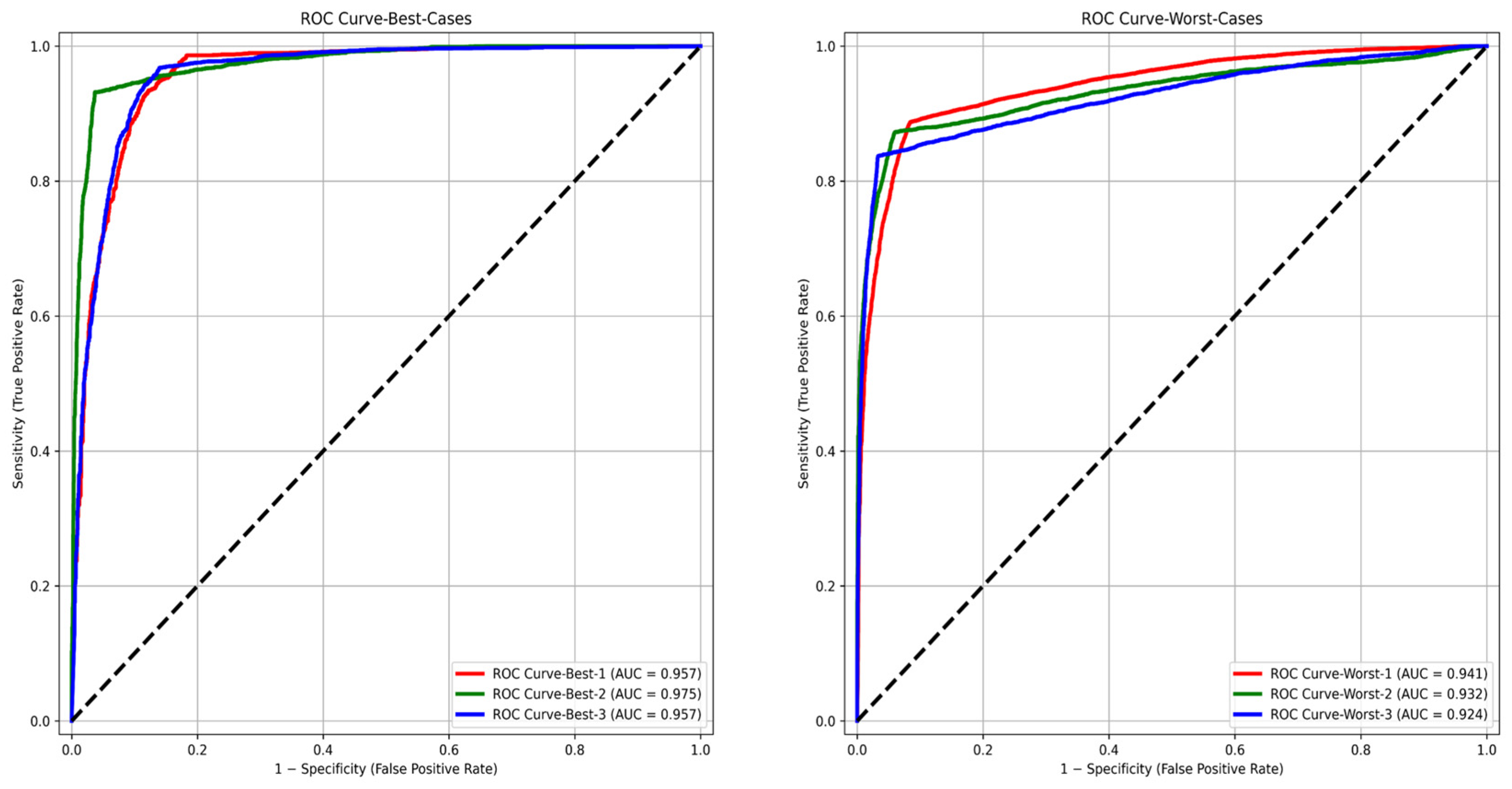
3.3. Test Cohort: Majority Votes Across Three Observers
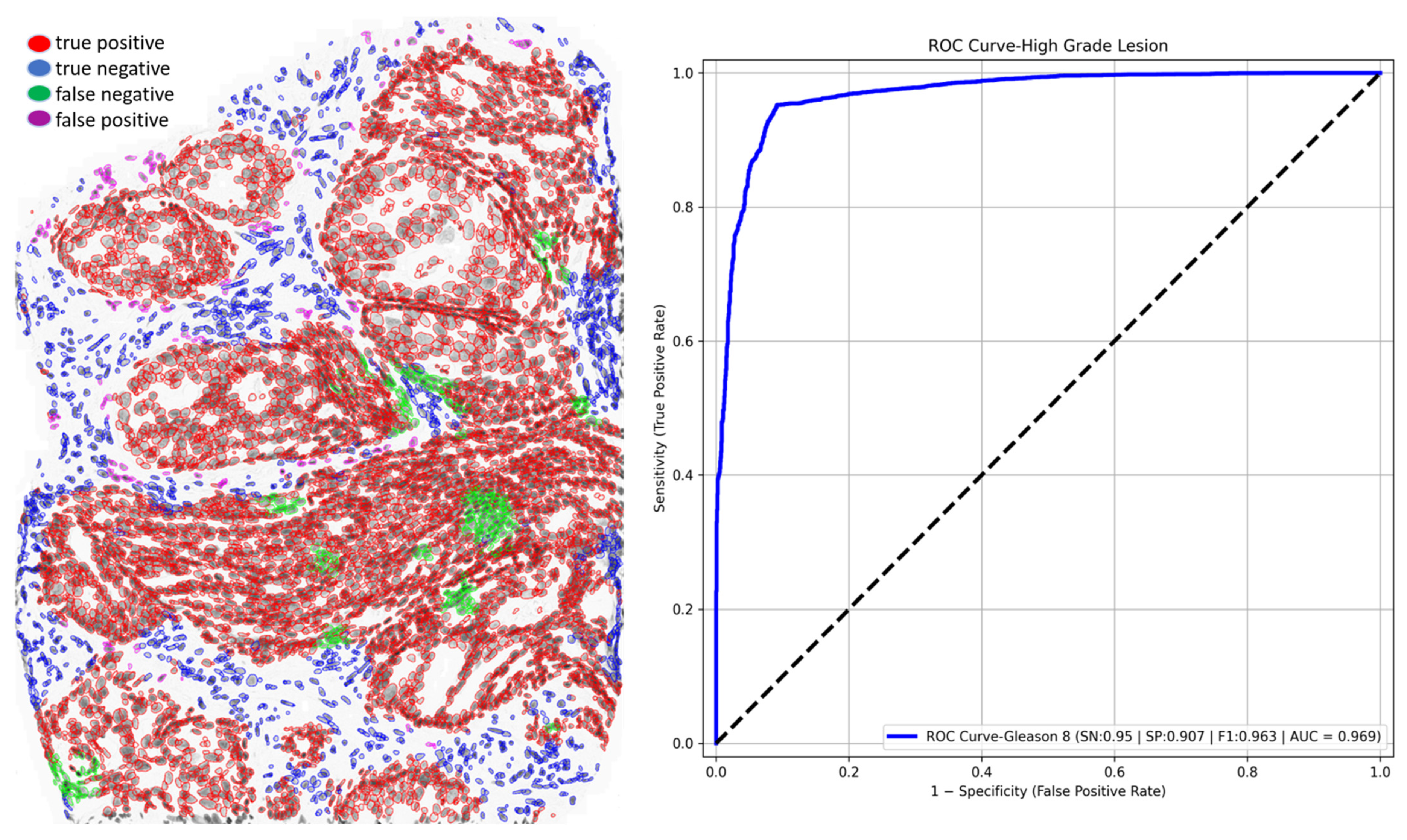
3.4. Out-of-Scope Validation: GlandNet Performance on High-Grade Lesion
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| GlandNet | Glandular cell recognition network |
| DNA | Deoxyribonucleic Acid |
| PCa | Prostate Cancer |
| TME | Tissue Microenvironment |
| H&E | Hematoxylin and Eosin |
| MIDI | Musical Instrument Digital Interface |
| CCD | Charge Coupled Device |
| ROI | Region of Interest |
| CMG | Compressed Media Group |
| CNN | Convolutional Neural Network |
| VGG16 | Deep Learning Architecture |
| PPV | Positive Prediction Value |
| NPV | Negative Prediction Value |
| FPR | False Positive Rate |
| FNR | False Negative Rate |
| AUC-ROC | Area Under the Curve-Receiver Operating Characteristic |
| AP-PR | Average precision (AP) under the precision-recall (PR) curve |
| panCK | pan Cytokeratin |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Kumar, N.; Patil, A.; Kurian, N.C.; Rane, S.; Graham, S.; Vu, Q.D.; Zwager, M.; Raza, S.E.; Rajpoot, N.; et al. MoNuSAC2020: A multi-organ nuclei segmentation and classification challenge. IEEE Trans. Med. Imaging 2021, 40, 3413–3423. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Zitvogel, L.; Sautès–Fridman, C.; Kroemer, G. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 2017, 14, 717–734. [Google Scholar] [CrossRef] [PubMed]
- Gleason, D.F. Classification of prostatic carcinomas. Cancer Chemother. Rep. 1966, 50, 125–128. [Google Scholar]
- MacAulay, C.; Keyes, M.; Hayes, M.; Lo, A.; Wang, G.; Guillaud, M.; Gleave, M.; Fazli, L.; Korbelik, J.; Collins, C.; et al. Quantification of large scale DNA organization for predicting prostate cancer recurrence. Cytom. Part A 2017, 91, 1164–1174. [Google Scholar] [CrossRef]
- Zarei, N.; Bakhtiari, P.; Gallagher, P.; Keys, M.; MacAulay, C. Automated prostate glandular and nuclei detection using hyperspectral imaging. In Proceedings of the 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia, 18–21 April 2017. [Google Scholar]
- Donovan, M.; Khan, F.; Powell, D.; Fernandez, G.; Feliz, A.; Hansen, T.; Bernardino, L.; Capodieci, P.; Bayer-Zubek, V.; Liu, Q.; et al. 1293 Previously developed systems-based biopsy model (prostate PX1) identifies favorable-risk prostate cancer for men enrolled in an active surveillance program. J. Urol. 2011, 185, e517–e518. [Google Scholar] [CrossRef]
- Donovan, M.J.; Khan, F.M.; Fernandez, G.; Mesa-Tejada, R.; Sapir, M.; Zubek, V.B.; Powell, D.; Fogarasi, S.; Vengrenyuk, Y.; Teverovskiy, M.; et al. Personalized prediction of tumor response and cancer progression on prostate needle biopsy. J. Urol. 2009, 182, 125–132. [Google Scholar] [CrossRef]
- Donovan, M.J.; Hamann, S.; Clayton, M.; Khan, F.M.; Sapir, M.; Bayer-Zubek, V.; Fernandez, G.; Mesa-Tejada, R.; Teverovskiy, M.; Reuter, V.E.; et al. Systems pathology approach for the prediction of prostate cancer progression after radical prostatectomy. J. Clin. Oncol. 2008, 26, 3923–3929. [Google Scholar] [CrossRef]
- Ryu, H.S.; Jin, M.S.; Park, J.H.; Lee, S.; Cho, J.; Oh, S.; Kwak, T.Y.; Woo, J.I.; Mun, Y.; Kim, S.W.; et al. Automated Gleason scoring and tumor quantification in prostate core needle biopsy images using deep neural networks and its comparison with pathologist-based assessment. Cancers 2019, 11, 1860. [Google Scholar] [CrossRef]
- Singhal, N.; Soni, S.; Bonthu, S.; Chattopadhyay, N.; Samanta, P.; Joshi, U.; Jojera, A. A deep learning system for prostate cancer diagnosis and grading in whole slide images of core needle biopsies. Sci. Rep. 2022, 12, 3383. [Google Scholar] [CrossRef]
- Raciti, P.; Sue, J.; Ceballos, R.; Godrich, R.; Kunz, J.D.; Kapur, S.; Reuter, V.; Grady, L.; Kanan, C.; Klimstra, D.S.; et al. Novel artificial intelligence system increases the detection of prostate cancer in whole slide images of core needle biopsies. Mod. Pathol. 2020, 33, 2058–2066. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, K.; Foote, D.; Liu, Y.; Chen, P.H.; Wulczyn, E.; Tan, F.; Olson, N.; Smith, J.L.; Mohtashamian, A.; Wren, J.H.; et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit. Med. 2019, 2, 48. [Google Scholar] [CrossRef]
- Mun, Y.; Paik, I.; Shin, S.J.; Kwak, T.Y.; Chang, H. Yet another automated Gleason grading system (YAAGGS) by weakly supervised deep learning. NPJ Digit. Med. 2021, 4, 99. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, A.; Ghelichkhan, E.; Manoochehri, H.; Ho, M.M.; Albertson, D.J.; Brintz, B.J.; Tasdizen, T.; Whitaker, R.T.; Knudsen, B.S. HistoEM: A Pathologist-guided and explainable workflow using histogram embedding for gland classification. Mod. Pathol. 2024, 37, 100447. [Google Scholar] [CrossRef]
- Oner, M.U.; Ng, M.Y.; Giron, D.M.; Xi, C.E.; Xiang, L.A.; Singh, M.; Yu, W.; Sung, W.K.; Wong, C.F.; Lee, H.K. An AI-assisted tool for efficient prostate cancer diagnosis in low-grade and low-volume cases. Patterns 2022, 3, 100642. [Google Scholar] [CrossRef]
- Inamdar, M.A.; Raghavendra, U.; Gudigar, A.; Bhandary, S.; Salvi, M.; Deo, R.C.; Barua, P.D.; Ciaccio, E.J.; Molinari, F.; Acharya, U.R. A Novel Attention based model for Semantic Segmentation of Prostate Glands using Histopathological Images. IEEE Access 2023, 11, 108982–108994. [Google Scholar] [CrossRef]
- Spatial Biology Incorporating Single Cell Measurements of Large Scale DNA Alterations and Structural DNA Changes as Biomarkers to Predict Future Behavior of Cancers. BC Cancer Research Institute, Canada, Gordon and Leslie Diamond Family Theatre, Presentation on 6 February 2023. Available online: https://tube.bccrc.ca/videos/watch/d1173dfc-9a77-4f95-9a5d-39b57a9e766b (accessed on 1 May 2025).
- MacAulay, C.; Guillad, M.; Keyes, M. 106 Degree of Immune Infiltration in Active Surveillance Prostate Patients Detected by Quantitative DNA Staining Helps Predict Progression. J. ImmunoTherapy Cancer 2023, 11, A120. [Google Scholar]
- MacAulay, C.; Gallagher, P. Sequential Convolutional Neural Networks for Nuclei Segmentation. US Continuation Patent US 18/339M 193, 21 June 2023. [Google Scholar]
- Albalate, A.; Minker, W. Semi-Supervised and Unsupervised Machine Learning: Novel Strategies; John Wiley & Sons: Hoboken, NJ, USA, 2013. [Google Scholar]
- Eckardt, J.N.; Bornhäuser, M.; Wendt, K.; Middeke, J.M. Semi-supervised learning in cancer diagnostics. Front. Oncol. 2022, 12, 960984. [Google Scholar] [CrossRef] [PubMed]
- Mohanasundaram, R.; Malhotra, A.S.; Arun, R.; Periasamy, P.S. Deep learning and semi-supervised and transfer learning algorithms for medical imaging. In Deep Learning and Parallel Computing Environment for Bioengineering Systems; Academic Press: Cambridge, MA, USA, 2019; pp. 139–151. [Google Scholar]
- Peikari, M.; Salama, S.; Nofech-Mozes, S.; Martel, A.L. A cluster-then-label semi-supervised learning approach for pathology image classification. Sci. Rep. 2018, 8, 7193. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Li, F.-F. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. 6.2.2.3 Softmax Units for Multinoulli Output Distributions. In Deep Learning; MIT Press: Cambridge, MA, USA, 2016; pp. 180–184. ISBN 978-0-26203561-3. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference on Learning Representations (ICLR), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Devnath, L.; Janzen, I.; Lam, S.; Yuan, R.; MacAulay, C. Predicting future lung cancer risk in low-dose CT screening patients with AI tools. In Proceedings of the Medical Imaging 2025: Computer-Aided Diagnosis, San Diego, CA, USA, 16–21 February 2025; Volume 13407, pp. 634–639. [Google Scholar]
- Devnath, L. Black Lung Detection on Chest X-ray Radiographs Using Deep Learning. Ph.D. Thesis, The University of Newcastle, Newcastle, Australia, 2021. [Google Scholar]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem. In Proceedings of the 2018 International Interdisciplinary PhD Workshop (IIPhDW), Swinoujscie, Poland, 9–12 May 2018. [Google Scholar]
- Shorten, C.; Khoshgoftaar, T.M. A survey on image data augmentation for deep learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Shekar, B.H.; Dagnew, G. Grid search-based hyperparameter tuning and classification of microarray cancer data. In Proceedings of the 2019 Second International Conference on Advanced Computational and Communication Paradigms (ICACCP), Gangtok, India, 25–28 February 2019. [Google Scholar]
- Radzi, S.F.; Karim, M.K.; Saripan, M.I.; Rahman, M.A.; Isa, I.N.; Ibahim, M.J. Hyperparameter tuning and pipeline optimization via grid search method and tree-based autoML in breast cancer prediction. J. Pers. Med. 2021, 11, 978. [Google Scholar] [CrossRef] [PubMed]
- Belete, D.M.; Huchaiah, M.D. Grid search in hyperparameter optimization of machine learning models for prediction of HIV/AIDS test results. Int. J. Comput. Appl. 2022, 44, 875–886. [Google Scholar] [CrossRef]
- Fuadah, Y.N.; Pramudito, M.A.; Lim, K.M. An optimal approach for heart sound classification using grid search in hyperparameter optimization of machine learning. Bioengineering 2022, 10, 45. [Google Scholar] [CrossRef]

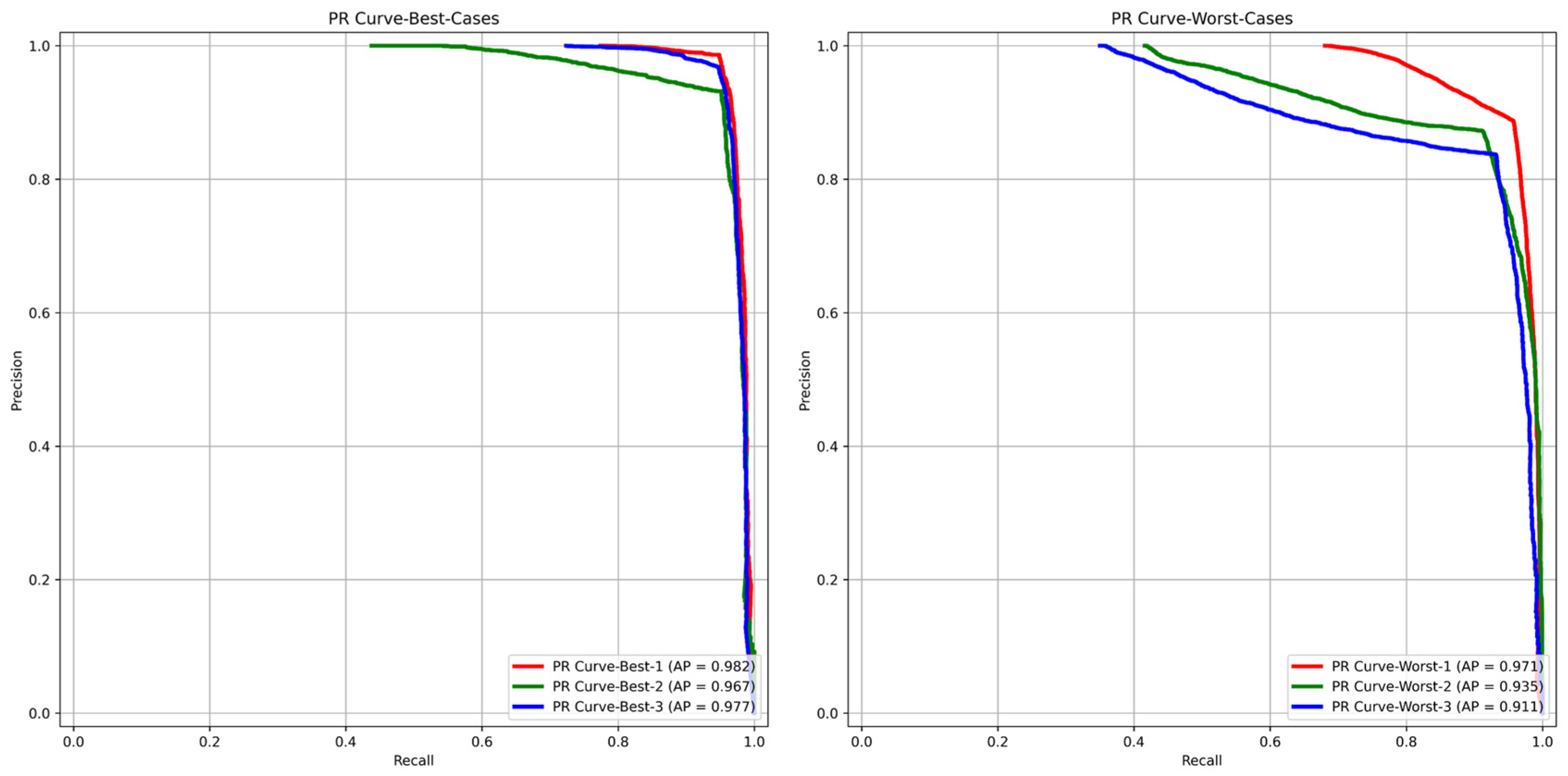
| Random Splits | Accuracy (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | F1-Score (%) |
|---|---|---|---|---|---|---|
| 1 | 87.6 | 91.1 | 84.6 | 83.3 | 91.8 | 87.0 |
| 2 | 88.7 | 70.3 | 95.1 | 83.1 | 90.3 | 76.2 |
| 3 | 90.3 | 15.5 | 98.3 | 49.9 | 91.6 | 23.6 |
| Average | 88.8 | 58.9 | 92.6 | 72.1 | 91.2 | 62.3 |
| Best 3 | Accuracy (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | F1-Score (%) |
|---|---|---|---|---|---|---|
| 1 | 94.9 | 93.1 | 96.2 | 95.1 | 94.7 | 94.1 |
| 2 | 92.6 | 95.3 | 85.6 | 94.5 | 87.5 | 94.9 |
| 3 | 94.8 | 98.6 | 81.8 | 94.9 | 94.4 | 96.7 |
| Average | 94.1 | 95.7 | 87.8 | 94.8 | 92.2 | 95.2 |
| Worst 3 | Accuracy (%) | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | F1-Score (%) |
|---|---|---|---|---|---|---|
| 1 | 91.1 | 87.1 | 93.9 | 91.0 | 91.1 | 89.0 |
| 2 | 89.4 | 88.4 | 91.5 | 95.7 | 78.8 | 91.9 |
| 3 | 92.1 | 83.7 | 96.7 | 93.1 | 91.7 | 88.1 |
| Average | 90.9 | 86.4 | 94.0 | 93.3 | 87.2 | 89.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Devnath, L.; Arora, P.; Carraro, A.; Korbelik, J.; Keyes, M.; Wang, G.; Guillaud, M.; MacAulay, C. Recognizing Epithelial Cells in Prostatic Glands Using Deep Learning. Cells 2025, 14, 737. https://doi.org/10.3390/cells14100737
Devnath L, Arora P, Carraro A, Korbelik J, Keyes M, Wang G, Guillaud M, MacAulay C. Recognizing Epithelial Cells in Prostatic Glands Using Deep Learning. Cells. 2025; 14(10):737. https://doi.org/10.3390/cells14100737
Chicago/Turabian StyleDevnath, Liton, Puneet Arora, Anita Carraro, Jagoda Korbelik, Mira Keyes, Gang Wang, Martial Guillaud, and Calum MacAulay. 2025. "Recognizing Epithelial Cells in Prostatic Glands Using Deep Learning" Cells 14, no. 10: 737. https://doi.org/10.3390/cells14100737
APA StyleDevnath, L., Arora, P., Carraro, A., Korbelik, J., Keyes, M., Wang, G., Guillaud, M., & MacAulay, C. (2025). Recognizing Epithelial Cells in Prostatic Glands Using Deep Learning. Cells, 14(10), 737. https://doi.org/10.3390/cells14100737








