The Long Non-Coding RNA Gene AC027288.3 Plays a Role in Human Endometrial Stromal Fibroblast Decidualization
Abstract
1. Introduction
2. Methods and Materials
2.1. Cell Isolation and Culture
2.2. Poly A+ RNA-Seq Library Preparation and Sequencing
2.3. RNA-Seq Analysis
2.4. Reverse Transcription-Quantitative PCR (RT-qPCR)
2.5. Cell Fractionation
2.6. Protein Extraction and Western Blot Analysis
2.7. ChIP-Seq Data Reanalysis
2.8. FOXO1 ChIP-PCR
2.9. Lentivirus Preparation and AC027288.3 Knockdown
2.10. Statistical analysis
3. Results
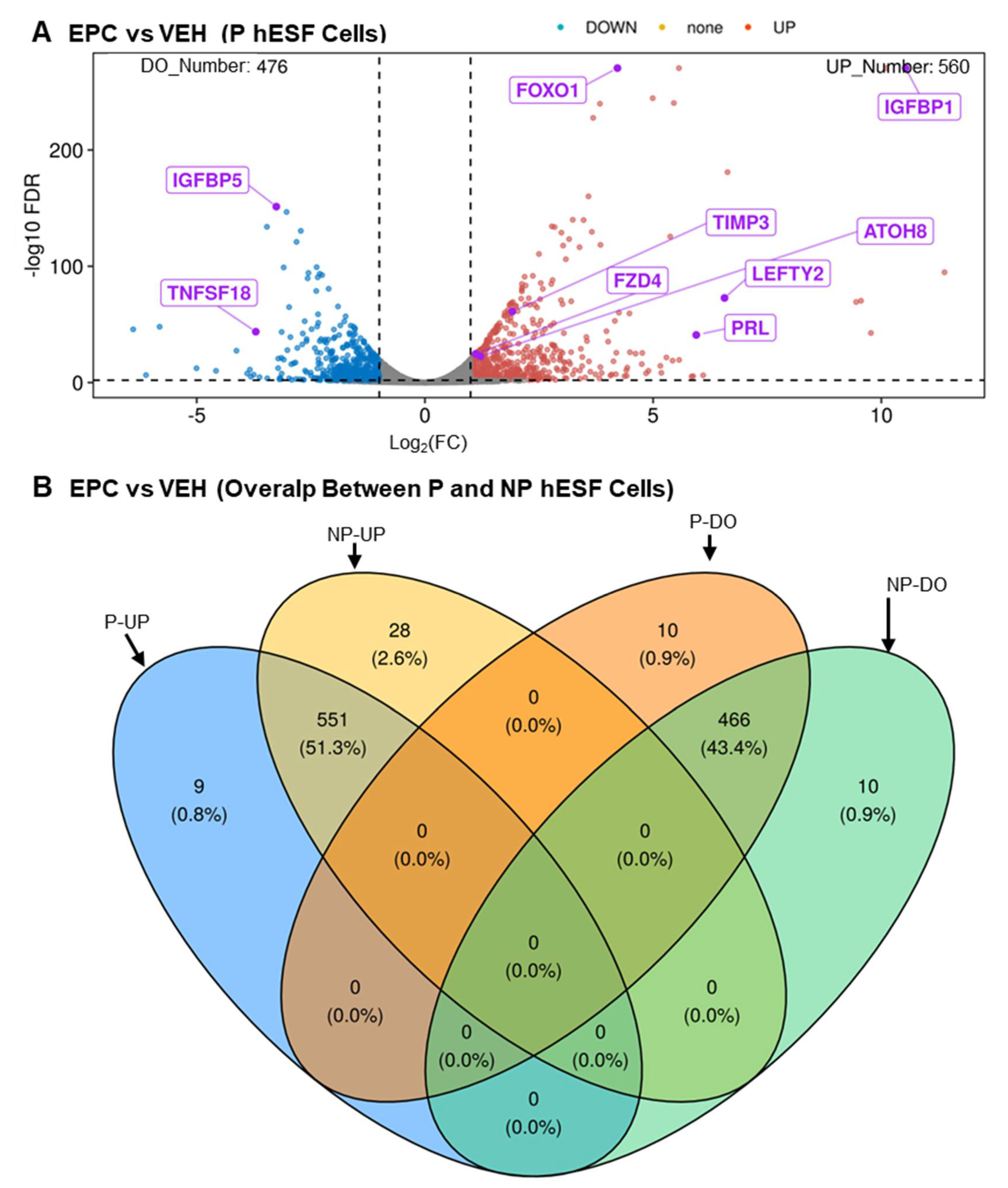
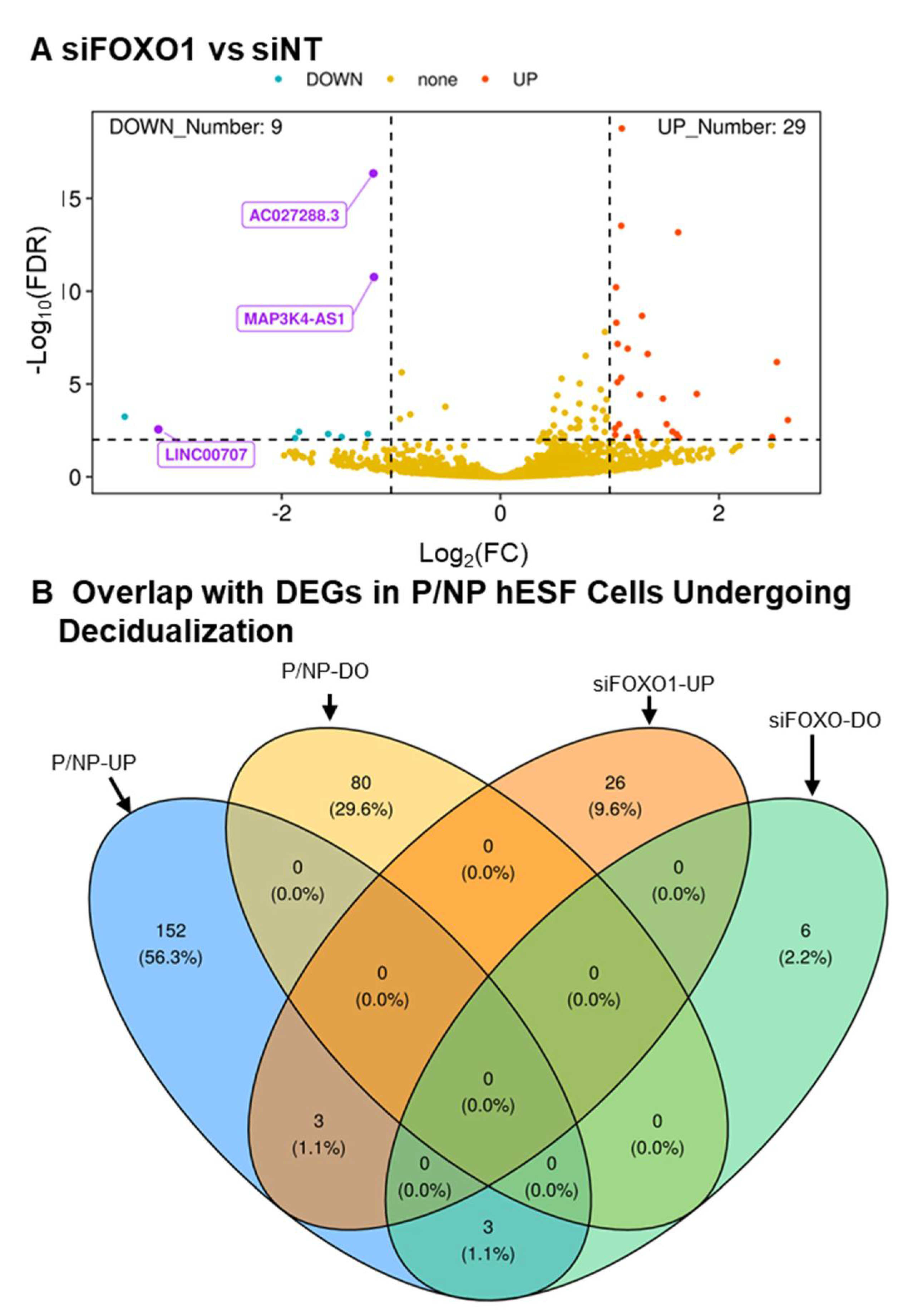
3.1. RNA-Seq Analysis
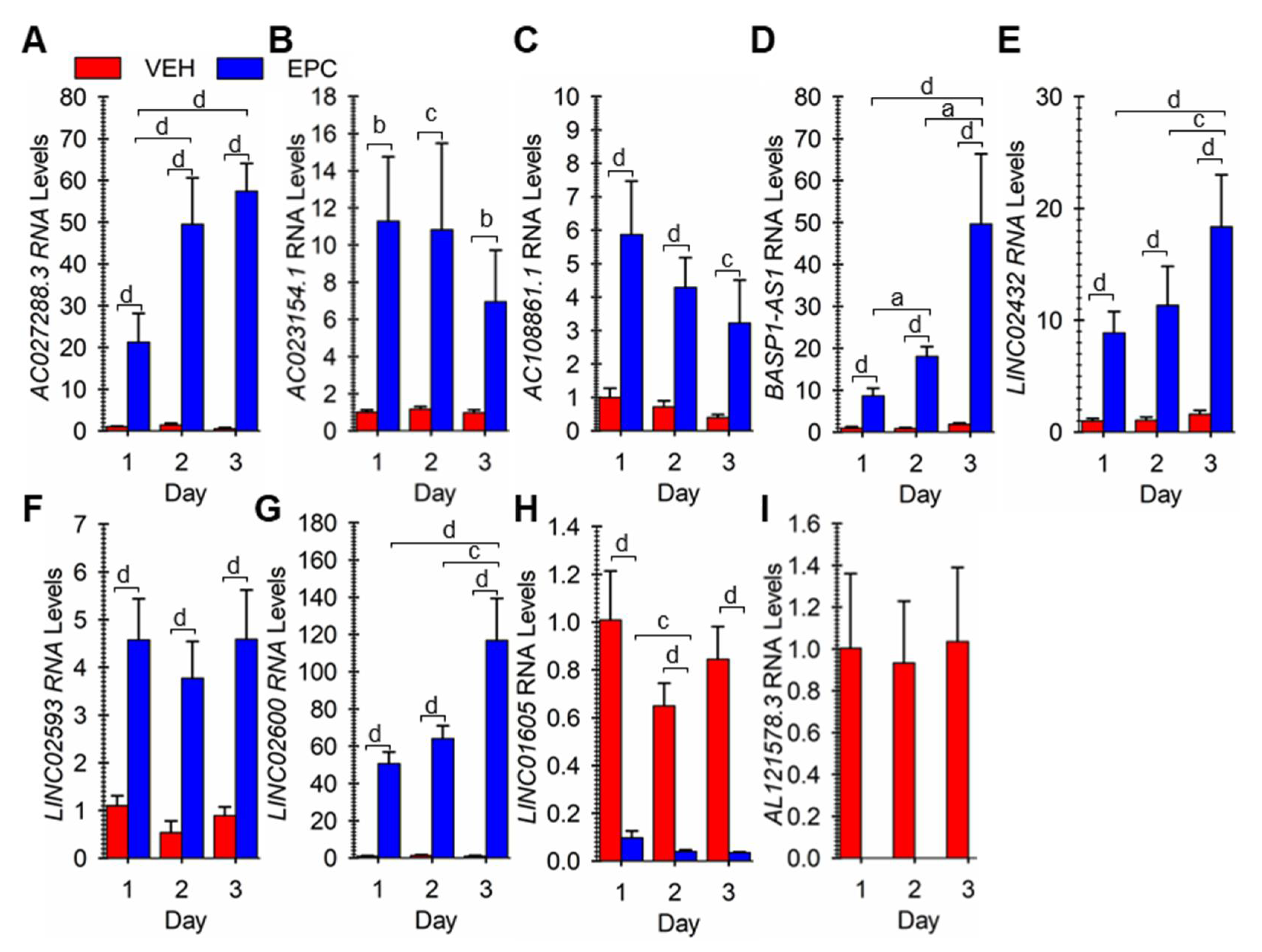
3.2. Verifying Differential Expression of 9 Novel LncRNA during hESF Decidualization
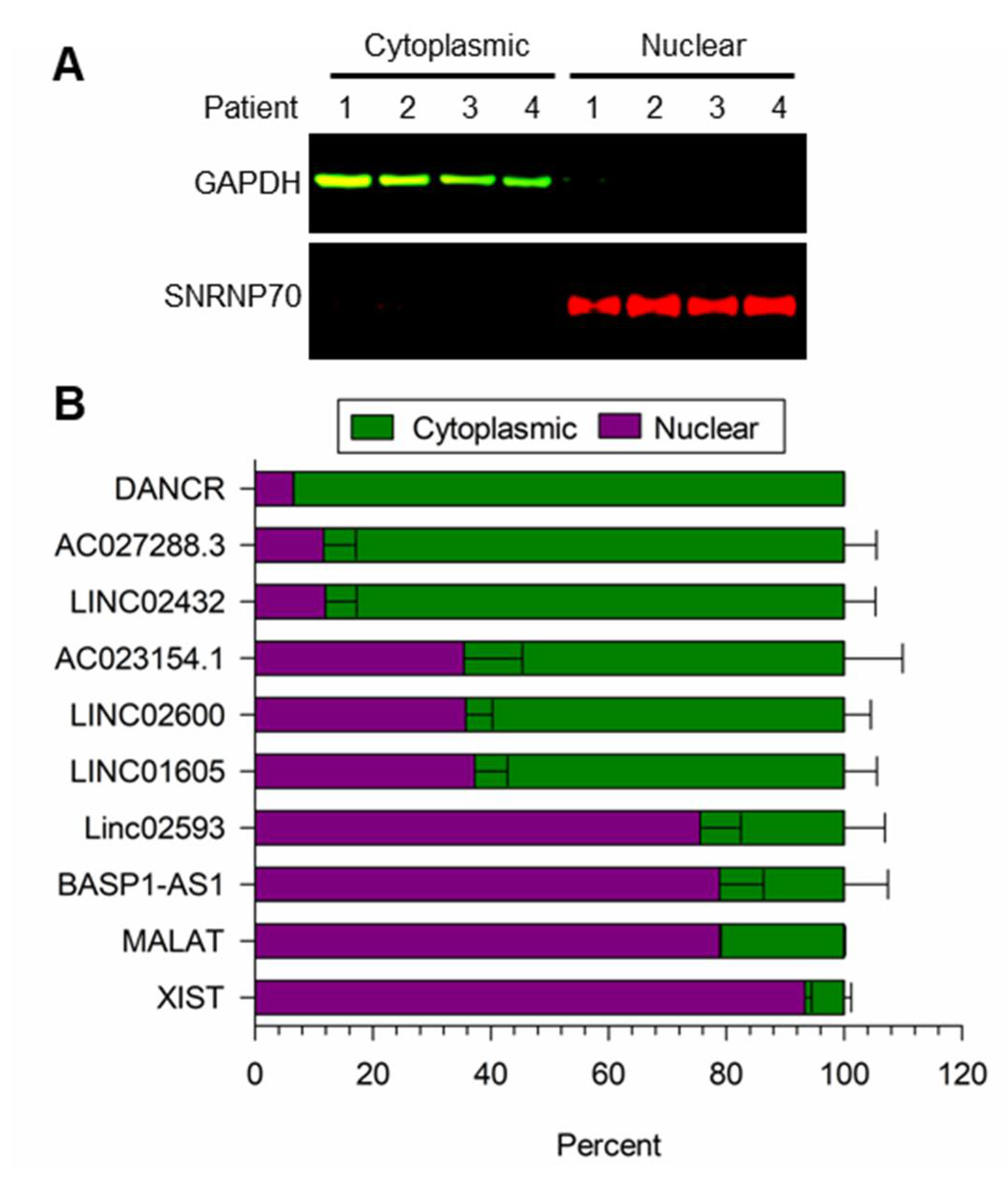
3.3. Localization of LncRNA Expression
3.4. Steroid and cAMP Control of LncRNA Expression
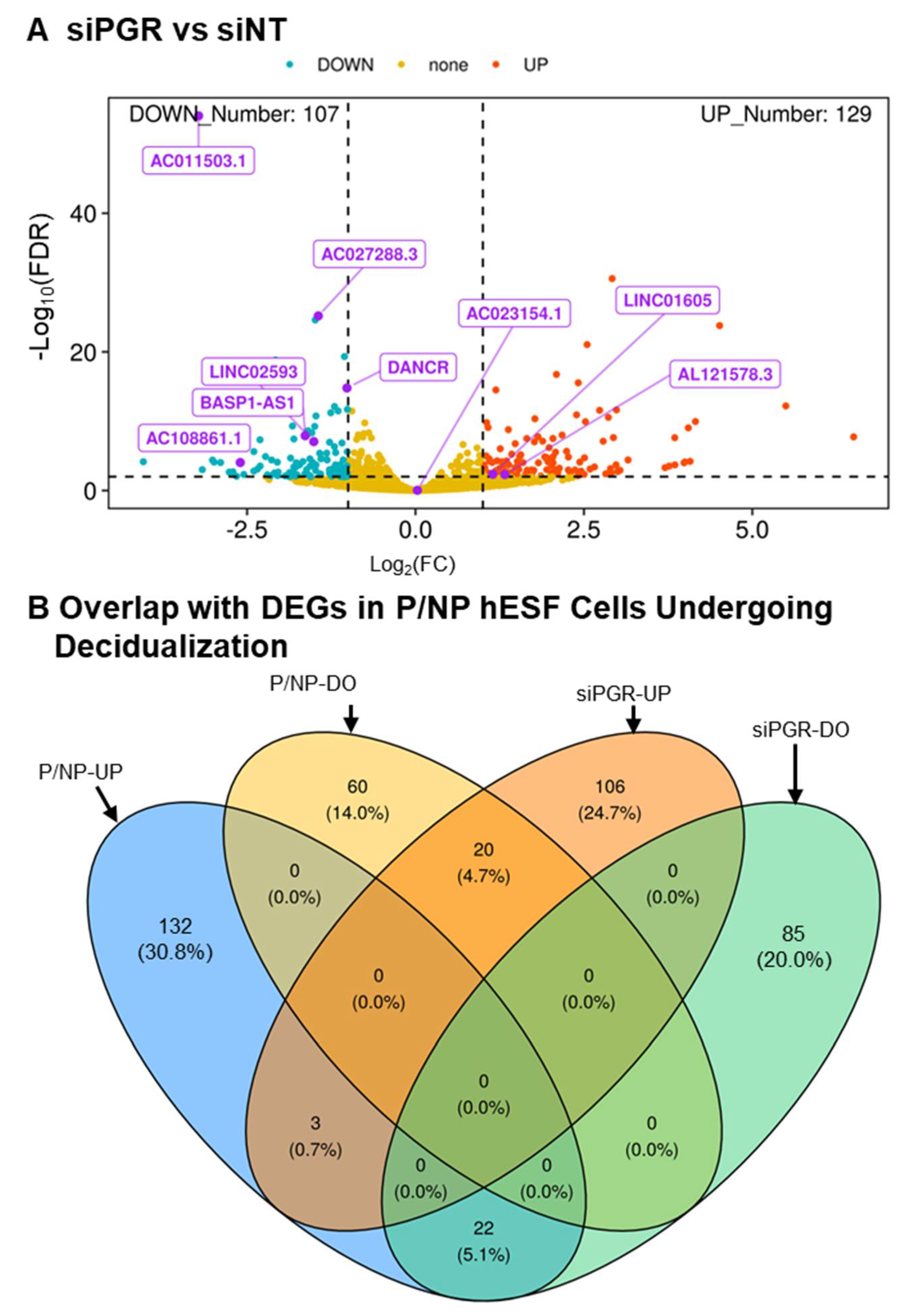
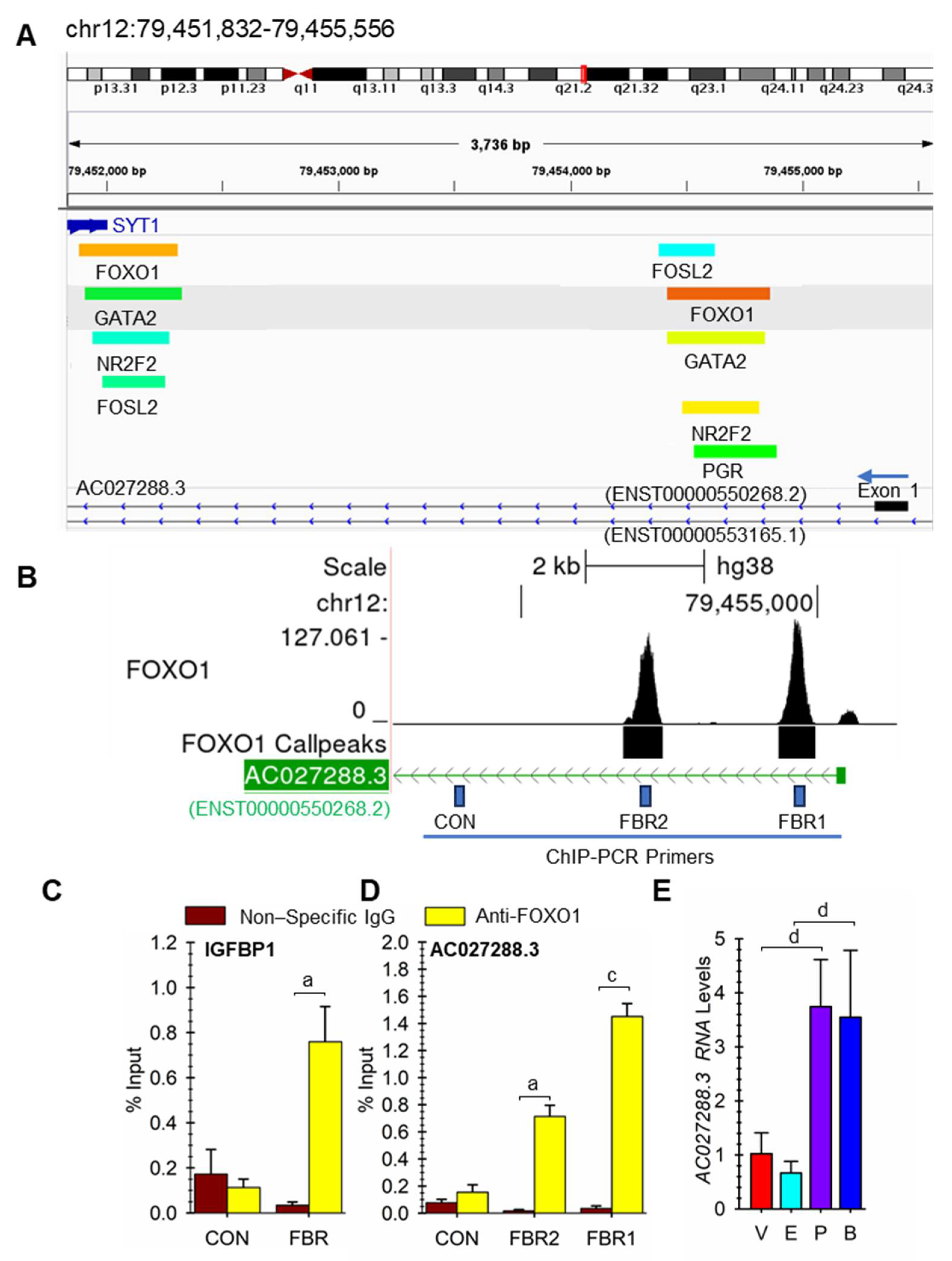
3.5. PGR and FOXO1 as Regulators of LncRNA Expression
3.6. Potential Roles of Specific Transcription Factors on AC027288.3 Expression
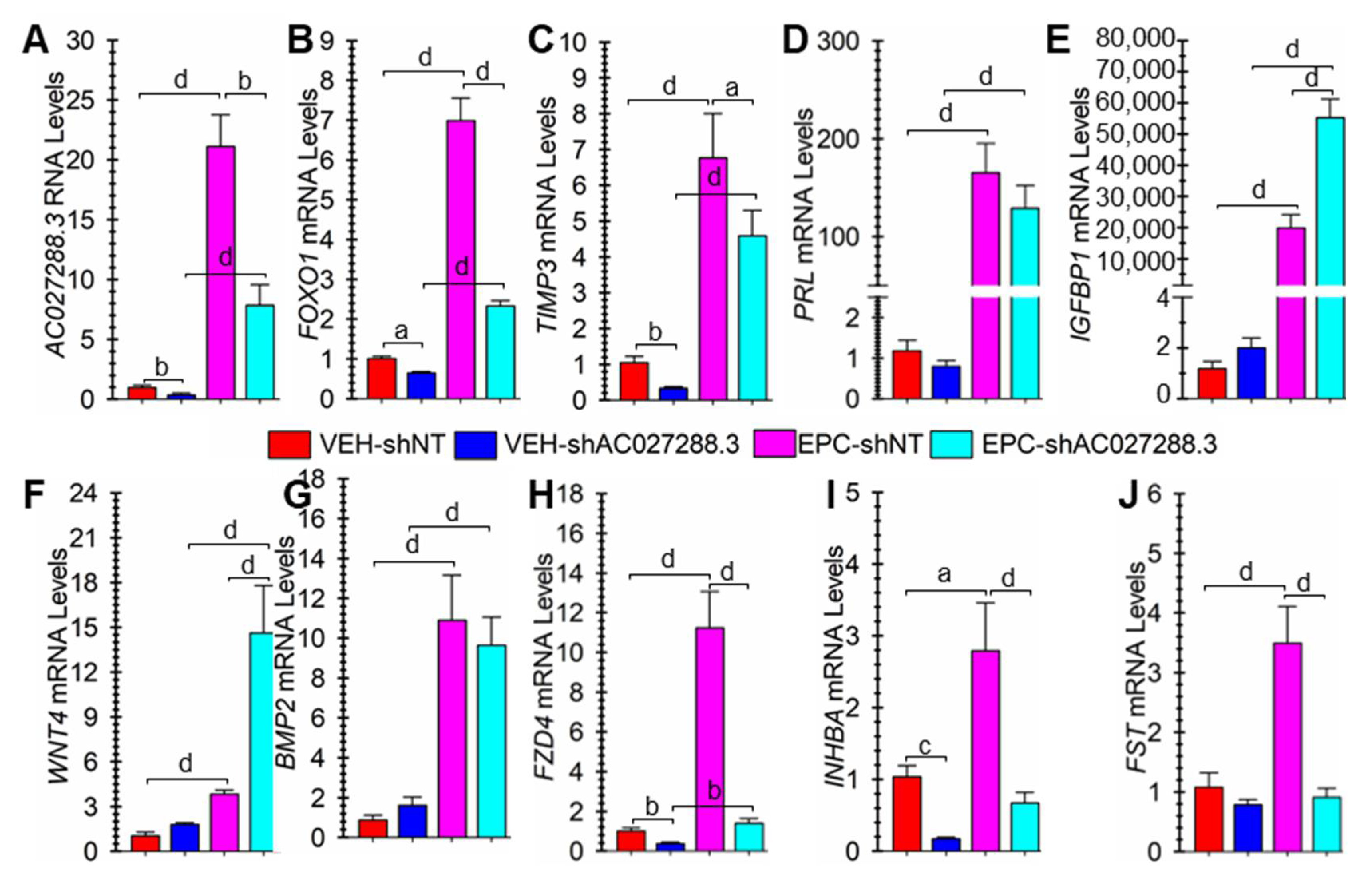
3.7. Targeting AC027288.3 Expression Affects hESF Decidualization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gellersen, B.; Brosens, J.J. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr. Rev. 2014, 35, 851–905. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.W.; Norwitz, G.A.; Pavlicev, M.; Tilburgs, T.; Simon, C.; Norwitz, E.R. Endometrial Decidualization: The Primary Driver of Pregnancy Health. Int. J. Mol. Sci. 2020, 21, 4092. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.J.; Guan, H.Y.; Wang, L.; Zhang, W. Research progress on human endometrium decidualization cell models. Reprod. Dev. Med. 2021, 5, 119–127. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermuller, J.; Hofacker, I.L.; et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef] [PubMed]
- Frankish, A.; Carbonell-Sala, S.; Diekhans, M.; Jungreis, I.; Loveland, J.E.; Mudge, J.M.; Sisu, C.; Wright, J.C.; Arnan, C.; Barnes, I.; et al. GENCODE: Reference annotation for the human and mouse genomes in 2023. Nucleic Acids Res 2023, 51, D942–D949. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.J.; Amode, M.R.; Aneja, A.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Becker, A.; Bennett, R.; Berry, A.; Bhai, J.; et al. Ensembl 2023. Nucleic Acids Res. 2023, 51, D933–D941. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Fu, X.D. Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat. Rev. Genet. 2019, 20, 503–519. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Ghosal, S.; Das, S.; Balti, S.; Chakrabarti, J. Competing endogenous RNA: The key to posttranscriptional regulation. ScientificWorldJournal 2014, 2014, 896206. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Chang, C.P. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015, 12, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Szaflik, T.; Romanowicz, H.; Szyllo, K.; Smolarz, B. MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) in endometriosis—Review of literature. Ginekol. Pol. 2023, 94, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Abbaszadeh, M.; Karimi, M.; Rajaei, S. The landscape of non-coding RNAs in the immunopathogenesis of Endometriosis. Front. Immunol. 2023, 14, 1223828. [Google Scholar] [CrossRef] [PubMed]
- Panir, K.; Schjenken, J.E.; Robertson, S.A.; Hull, M.L. Non-coding RNAs in endometriosis: A narrative review. Hum. Reprod. Update 2018, 24, 497–515. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Yang, Z.; Liu, K.; Wang, D. Genome-Wide Microarray Analysis of Long Non-Coding RNAs in Eutopic Secretory Endometrium with Endometriosis. Cell Physiol. Biochem. 2015, 37, 2231–2245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, S. The possible role of long non-coding RNAs in recurrent miscarriage. Mol. Biol. Rep. 2022, 49, 9687–9697. [Google Scholar] [CrossRef] [PubMed]
- Moradi, M.T.; Rahimi, Z.; Vaisi-Raygani, A. New insight into the role of long non-coding RNAs in the pathogenesis of preeclampsia. Hypertens. Pregnancy 2019, 38, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Dong, H.; Guan, Y.; Fan, T.; Sun, W.; Bagchi, I.C.; Miao, C.; Li, Q. Regulation of HAND2 Expression by LncRNA HAND2-AS1 in Ovarian Endometriosis Involving DNA Methylation. J. Endocr. Soc. 2023, 7, bvad049. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cai, X.; Liu, J.; Zhou, J.; Shi, Q.; Jiang, Y.; Kang, N.; Zhen, X.; Wu, M.; Qiu, P.; et al. A novel lncRNA lncSAMD11-1: 1 interacts with PIP4K2A to promote endometrial decidualization by stabilizing FoxO1 nuclear localization. Int. J. Biochem. Cell Biol. 2022, 151, 106280. [Google Scholar] [CrossRef]
- Lv, H.; Tong, J.; Yang, J.; Lv, S.; Li, W.P.; Zhang, C.; Chen, Z.J. Dysregulated Pseudogene HK2P1 May Contribute to Preeclampsia as a Competing Endogenous RNA for Hexokinase 2 by Impairing Decidualization. Hypertension 2018, 71, 648–658. [Google Scholar] [CrossRef]
- Wang, Y.; Hu, S.; Yao, G.; Zhu, Q.; He, Y.; Lu, Y.; Qi, J.; Xu, R.; Ding, Y.; Li, J.; et al. A Novel Molecule in Human Cyclic Endometrium: LncRNA TUNAR Is Involved in Embryo Implantation. Front. Physiol. 2020, 11, 587448. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Ma, L.; Chang, Z.; Yu, T. Increased expression of prokineticin 2 and its receptor in endometrium of recurrent implantation failure patients decreased the expression of MMP9 important for decidualization. Reprod. Biol. Endocrinol. 2022, 20, 76. [Google Scholar] [CrossRef]
- Liang, X.H.; Deng, W.B.; Liu, Y.F.; Liang, Y.X.; Fan, Z.M.; Gu, X.W.; Liu, J.L.; Sha, A.G.; Diao, H.L.; Yang, Z.M. Non-coding RNA LINC00473 mediates decidualization of human endometrial stromal cells in response to cAMP signaling. Sci. Rep. 2016, 6, 22744. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Gao, R.; Chen, X.; Geng, Y.; Yin, X.; Peng, C.; Mu, X.; Su, Y.; Zhang, Y.; Li, F.; et al. lincRNA RP24-315D19.10 promotes endometrial decidualization via upregulation of hnRNPA2B1. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1869, 166762. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Cai, R.; Yu, T.; Zhang, R.; Liu, S.; Guo, X.; Shang, C.; Wang, A.; Jin, Y.; Lin, P. Progesterone-induced RNA Hand2os1 regulates decidualization in mice uteri. Reproduction 2020, 159, 303–314. [Google Scholar] [CrossRef]
- Deng, W.B.; Liang, X.H.; Liu, J.L.; Yang, Z.M. Regulation and function of deiodinases during decidualization in female mice. Endocrinology 2014, 155, 2704–2717. [Google Scholar] [CrossRef]
- Thapa, R.; Druessel, L.; Ma, L.; Torry, D.S.; Bany, B.M. ATOH8 Expression Is Regulated by BMP2 and Plays a Key Role in Human Endometrial Stromal Cell Decidualization. Endocrinology 2023, 165, bqad188. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Gingeras, T.R. Optimizing RNA-Seq Mapping with STAR. Methods Mol. Biol. 2016, 1415, 245–262. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Chen, Y.; McCarthy, D.; Baldoni, P.; Ritchie, M.; Robinson, M.; Smyth, G. edgeR: Differential Analysis of Sequence Read Count Data. Available online: https://www.bioconductor.org/packages/devel/bioc/vignettes/edgeR/inst/doc/edgeRUsersGuide.pdf (accessed on 10 January 2024).
- Vasquez, Y.M.; Mazur, E.C.; Li, X.; Kommagani, R.; Jiang, L.; Chen, R.; Lanz, R.B.; Kovanci, E.; Gibbons, W.E.; DeMayo, F.J. FOXO1 is required for binding of PR on IRF4, novel transcriptional regulator of endometrial stromal decidualization. Mol. Endocrinol. 2015, 29, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Mazur, E.C.; Vasquez, Y.M.; Li, X.; Kommagani, R.; Jiang, L.; Chen, R.; Lanz, R.B.; Kovanci, E.; Gibbons, W.E.; DeMayo, F.J. Progesterone receptor transcriptome and cistrome in decidualized human endometrial stromal cells. Endocrinology 2015, 156, 2239–2253. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, Z. GraphBio: A shiny web app to easily perform popular visualization analysis for omics data. Front. Genet. 2022, 13, 957317. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, F.; Ryan, D.P.; Gruning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Dundar, F.; Manke, T. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Buzzio, O.L.; Lu, Z.; Miller, C.D.; Unterman, T.G.; Kim, J.J. FOXO1A differentially regulates genes of decidualization. Endocrinology 2006, 147, 3870–3876. [Google Scholar] [CrossRef]
- Herington, J.L.; Bi, J.; Martin, J.D.; Bany, B.M. Beta-catenin (CTNNB1) in the mouse uterus during decidualization and the potential role of two pathways in regulating its degradation. J. Histochem. Cytochem. 2007, 55, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Cech, M.; Chilton, J.; Clements, D.; Coraor, N.; Gruning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Feng, J.; Liu, T.; Qin, B.; Zhang, Y.; Liu, X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012, 7, 1728–1740. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Ohta, T.; Miura, F.; Oki, S. ChIP-Atlas 2021 update: A data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res. 2022, 50, W175–W182. [Google Scholar] [CrossRef]
- Oki, S.; Ohta, T.; Shioi, G.; Hatanaka, H.; Ogasawara, O.; Okuda, Y.; Kawaji, H.; Nakaki, R.; Sese, J.; Meno, C. ChIP-Atlas: A data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018, 19, e46255. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdottir, H.; Turner, D.; Mesirov, J.P. igv.js: An embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). Bioinformatics 2023, 39, btac830. [Google Scholar] [CrossRef] [PubMed]
- Kommagani, R.; Szwarc, M.M.; Vasquez, Y.M.; Peavey, M.C.; Mazur, E.C.; Gibbons, W.E.; Lanz, R.B.; DeMayo, F.J.; Lydon, J.P. The Promyelocytic Leukemia Zinc Finger Transcription Factor Is Critical for Human Endometrial Stromal Cell Decidualization. PLoS Genet. 2016, 12, e1005937. [Google Scholar] [CrossRef]
- Li, X.; Large, M.J.; Creighton, C.J.; Lanz, R.B.; Jeong, J.W.; Young, S.L.; Lessey, B.A.; Palomino, W.A.; Tsai, S.Y.; Demayo, F.J. COUP-TFII regulates human endometrial stromal genes involved in inflammation. Mol. Endocrinol. 2013, 27, 2041–2054. [Google Scholar] [CrossRef] [PubMed]
- Mika, K.M.; Li, X.; DeMayo, F.J.; Lynch, V.J. An Ancient Fecundability-Associated Polymorphism Creates a GATA2 Binding Site in a Distal Enhancer of HLA-F. Am. J. Hum. Genet. 2018, 103, 509–521. [Google Scholar] [CrossRef]
- Szwarc, M.M.; Hai, L.; Gibbons, W.E.; White, L.D.; Mo, Q.; Kommagani, R.; Lanz, R.B.; DeMayo, F.J.; O’Malley, B.W.; Lydon, J.P. Retinoid signaling controlled by SRC-2 in decidualization revealed by transcriptomics. Reproduction 2018, 156, 387–395. [Google Scholar] [CrossRef]
- Sullivan, A.E.; Santos, S.D.M. An Optimized Protocol for ChIP-Seq from Human Embryonic Stem Cell Cultures. STAR Protoc. 2020, 1, 100062. [Google Scholar] [CrossRef]
- Bany, B.M.; Torry, D.S. Regulation of maternal blood pressure by the conceptus during early pregnancy. Biol. Reprod. 2012, 86, 61. [Google Scholar] [CrossRef] [PubMed]
- Ganeff, C.; Chatel, G.; Munaut, C.; Frankenne, F.; Foidart, J.M.; Winkler, R. The IGF system in in-vitro human decidualization. Mol. Hum. Reprod. 2009, 15, 27–38. [Google Scholar] [CrossRef]
- Arun, G.; Aggarwal, D.; Spector, D.L. MALAT1 Long Non-Coding RNA: Functional Implications. Noncoding RNA 2020, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S.; et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef] [PubMed]
- Grinius, L.; Kessler, C.; Schroeder, J.; Handwerger, S. Forkhead transcription factor FOXO1A is critical for induction of human decidualization. J. Endocrinol. 2006, 189, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Kannan, A.; Das, A.; Demayo, F.J.; Hornsby, P.J.; Young, S.L.; Taylor, R.N.; Bagchi, M.K.; Bagchi, I.C. WNT4 acts downstream of BMP2 and functions via beta-catenin signaling pathway to regulate human endometrial stromal cell differentiation. Endocrinology 2013, 154, 446–457. [Google Scholar] [CrossRef]
- Jones, R.L.; Salamonsen, L.A.; Findlay, J.K. Activin A promotes human endometrial stromal cell decidualization in vitro. J. Clin. Endocrinol. Metab. 2002, 87, 4001–4004. [Google Scholar] [CrossRef] [PubMed]
- Thompson, T.B.; Lerch, T.F.; Cook, R.W.; Woodruff, T.K.; Jardetzky, T.S. The structure of the Follistatin:Activin complex reveals antagonism of both type I and type II receptor binding. Dev. Cell 2005, 9, 535–543. [Google Scholar] [CrossRef]
- Huyen, D.V.; Bany, B.M. Evidence for a conserved function of heart and neural crest derivatives expressed transcript 2 in mouse and human decidualization. Reproduction 2011, 142, 353–368. [Google Scholar] [CrossRef]
- Shindoh, H.; Okada, H.; Tsuzuki, T.; Nishigaki, A.; Kanzaki, H. Requirement of heart and neural crest derivatives-expressed transcript 2 during decidualization of human endometrial stromal cells in vitro. Fertil. Steril. 2014, 101, 1781–1790. [Google Scholar] [CrossRef]
- Wang, M.; Gu, J.; Zhang, X.; Yang, J.; Zhang, X.; Fang, X. Long Non-coding RNA DANCR in Cancer: Roles, Mechanisms, and Implications. Front. Cell Dev. Biol. 2021, 9, 753706. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shu, L.; Hu, M.; Zhou, X.; Yang, F.; Zhou, X.H. Emerging role of lncRNA DANCR in progenitor cells: Beyond cancer. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Buzzio, O.L.; Li, S.; Lu, Z. Role of FOXO1A in the regulation of insulin-like growth factor-binding protein-1 in human endometrial cells: Interaction with progesterone receptor. Biol. Reprod. 2005, 73, 833–839. [Google Scholar] [CrossRef] [PubMed]
- Lynch, V.J.; Brayer, K.; Gellersen, B.; Wagner, G.P. HoxA-11 and FOXO1A cooperate to regulate decidual prolactin expression: Towards inferring the core transcriptional regulators of decidual genes. PLoS ONE 2009, 4, e6845. [Google Scholar] [CrossRef]
- Park, Y.; Nnamani, M.C.; Maziarz, J.; Wagner, G.P. Cis-Regulatory Evolution of Forkhead Box O1 (FOXO1), a Terminal Selector Gene for Decidual Stromal Cell Identity. Mol. Biol. Evol. 2016, 33, 3161–3169. [Google Scholar] [CrossRef] [PubMed]
- Popovici, R.M.; Kao, L.C.; Giudice, L.C. Discovery of new inducible genes in in vitro decidualized human endometrial stromal cells using microarray technology. Endocrinology 2000, 141, 3510–3513. [Google Scholar] [CrossRef] [PubMed]
- Brar, A.K.; Handwerger, S.; Kessler, C.A.; Aronow, B.J. Gene induction and categorical reprogramming during in vitro human endometrial fibroblast decidualization. Physiol. Genom. 2001, 7, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.L.; Salamonsen, L.A.; Critchley, H.O.; Rogers, P.A.; Affandi, B.; Findlay, J.K. Inhibin and activin subunits are differentially expressed in endometrial cells and leukocytes during the menstrual cycle, in early pregnancy and in women using progestin-only contraception. Mol. Hum. Reprod. 2000, 6, 1107–1117. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.Q.; Lin, H.Y.; Zhu, C.; Yang, X.; Wang, H. Maternal Smad3 deficiency compromises decidualization in mice. J. Cell Biochem. 2012, 113, 3266–3275. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dong, X.Y.; Yang, P.W.; Yang, S.L.; Hu, D.; Zhang, H.W.; Sui, C. Activation of Uterine Smad3 Pathway Is Crucial for Embryo Implantation. Curr. Med. Sci. 2019, 39, 997–1002. [Google Scholar] [CrossRef]
- Menkhorst, E.; Salamonsen, L.A.; Zhang, J.; Harrison, C.A.; Gu, J.; Dimitriadis, E. Interleukin 11 and activin A synergise to regulate progesterone-induced but not cAMP-induced decidualization. J. Reprod. Immunol. 2010, 84, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Brar, A.K.; Frank, G.R.; Kessler, C.A.; Cedars, M.I.; Handwerger, S. Progesterone-dependent decidualization of the human endometrium is mediated by cAMP. Endocrine 1997, 6, 301–307. [Google Scholar] [CrossRef]
- Prajapati, B.; Fatima, M.; Fatma, M.; Maddhesiya, P.; Arora, H.; Naskar, T.; Devasenapathy, S.; Seth, P.; Sinha, S. Temporal transcriptome analysis of neuronal commitment reveals the preeminent role of the divergent lncRNA biotype and a critical candidate gene during differentiation. Cell Death Discov. 2020, 6, 28. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.K.; Kessler, C.A.; Handwerger, S. Critical role for TWIST1 in the induction of human uterine decidualization. Endocrinology 2011, 152, 4368–4376. [Google Scholar] [CrossRef] [PubMed]
- Kommagani, R.; Szwarc, M.M.; Kovanci, E.; Creighton, C.J.; O’Malley, B.W.; Demayo, F.J.; Lydon, J.P. A murine uterine transcriptome, responsive to steroid receptor coactivator-2, reveals transcription factor 23 as essential for decidualization of human endometrial stromal cells. Biol. Reprod. 2014, 90, 75. [Google Scholar] [CrossRef]
- Deepak, V.; Ravikumar, N.; Badell, M.L.; Sidell, N.; Rajakumar, A. Transcription factor ID1 is involved in decidualization of stromal cells: Implications in preeclampsia. Pregnancy Hypertens. 2020, 21, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Wang, Y.; Chang, H.M.; Zhu, H.; Yang, J.; Leung, P.C.K. ID3 mediates BMP2-induced downregulation of ICAM1 expression in human endometiral stromal cells and decidual cells. Front. Cell Dev. Biol. 2023, 11, 1090593. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.C.; Chen, S.T.; Hong, Z.K.; Li, S.Y.; Yang, Z.S.; Quan, S.; Yang, Z.M. Tryptophan and kynurenine stimulate human decidualization via activating Aryl hydrocarbon receptor: Short title: Kynurenine action on human decidualization. Reprod. Toxicol. 2020, 96, 282–292. [Google Scholar] [CrossRef]
- Lv, S.; Wang, N.; Ma, J.; Li, W.P.; Chen, Z.J.; Zhang, C. Impaired decidualization caused by downregulation of circadian clock gene BMAL1 contributes to human recurrent miscarriagedagger. Biol. Reprod. 2019, 101, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Meng, N.; Bao, H.; Jiang, Y.; Yang, N.; Wu, K.; Wu, J.; Wang, H.; Kong, S.; Zhang, Y. Circadian gene PER1 senses progesterone signal during human endometrial decidualization. J. Endocrinol. 2019, 243, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.M.; Zhang, T.T.; He, Y.Y.; Luo, H.N.; Hong, Y.Q.; Yang, Z.M. Human Chorionic Gonadotropin-Stimulated Interleukin-4-Induced-1 (IL4I1) Promotes Human Decidualization via Aryl Hydrocarbon Receptor. Int. J. Mol. Sci. 2023, 24, 3163. [Google Scholar] [CrossRef]
- Maurya, V.K.; Szwarc, M.M.; Lonard, D.M.; Gibbons, W.E.; Wu, S.P.; O’Malley, B.W.; DeMayo, F.J.; Lydon, J.P. Decidualization of human endometrial stromal cells requires steroid receptor coactivator-3. Front. Reprod. Health 2022, 4, 1033581. [Google Scholar] [CrossRef] [PubMed]
- Kommagani, R.; Szwarc, M.M.; Kovanci, E.; Gibbons, W.E.; Putluri, N.; Maity, S.; Creighton, C.J.; Sreekumar, A.; DeMayo, F.J.; Lydon, J.P.; et al. Acceleration of the Glycolytic Flux by Steroid Receptor Coactivator-2 Is Essential for Endometrial Decidualization. PLoS Genet. 2013, 9, e1003900. [Google Scholar]
- Shang, Q.; Yang, Y.; Li, H. LINC01605 knockdown induces apoptosis in human Tenon’s capsule fibroblasts by inhibiting autophagy. Exp. Ther. Med. 2022, 23, 343. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; He, X.; Wang, Y.; Liu, H.; Zhang, F.; Wu, Z.; Mo, S.; Chen, D. LINC01605 promotes aerobic glycolysis through lactate dehydrogenase A in triple-negative breast cancer. Cancer Sci. 2022, 113, 2484–2495. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.S.; Fu, L.; Han, S.Y.; Li, X.L.; Zhang, L.D. LINC01605 regulates proliferation, migration and invasion of colorectal cancer cells via targeting miR-3960/SOX11. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 1322–1329. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Li, M.; Liu, Z.; Li, T.; Zhao, L.; Fu, W. Glycolysis-Related LINC02432/Hsa-miR-98-5p/HK2 Axis Inhibits Ferroptosis and Predicts Immune Infiltration, Tumor Mutation Burden, and Drug Sensitivity in Pancreatic Adenocarcinoma. Front. Pharmacol. 2022, 13, 937413. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Chen, C.; Qian, J.; Cui, L.; Wang, S. Energy metabolism and maternal-fetal tolerance working in decidualization. Front. Immunol. 2023, 14, 1203719. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Lin, S.; Huang, P.; Qiu, L.; Jiang, Y.; Zhang, Y.; Meng, N.; Meng, M.; Wang, L.; Deng, W.; et al. Maternal anxiety affects embryo implantation via impairing adrenergic receptor signaling in decidual cells. Commun. Biol. 2022, 5, 840. [Google Scholar] [CrossRef] [PubMed]


| Gene | Upstream | Downstream |
|---|---|---|
| AC023154.1 | AAGCCTTCTGGAGGAGAAGCA | TGGTGGCTCAGCAACATCTA |
| AC027288.3 | GCAGCTTGGCAATGAAGTCA | CTCTGTCAGCCTCCCTCTTC |
| BMP2 | CAGACCACCGGTTGGAGA | CCACTCGTTTCTGGTAGTTCTTC |
| AC108861.1 | AGAAGCGTGCGTGCTACA | TTTGAAGCTGTCTGTGCAGTTG |
| AL121578.3 | ACCTTCACATGTCCGAATGC | TCCTGTCCCCACCTCTAAGA |
| BASP1-AS1 | AGCACCGGGACACAGAATAG | TTTGCGGGAAGGTAAAATTG |
| DANCR | CTGCATTCCTGAACCGTTATCT | GGGTGTAATCCACGTTTCTCAT |
| FOXO1 | CTGGCTCTCACAGCAATGAT | CACCATAGAATGCACATCCC |
| FZD4 | AACTTTCACACCGCTCATCC | TGCACATTGGCACATAAACA |
| IGFBP1 | GCTCTCCATGTCACCAACAT | TCTCCTGATGTCTCCTGTGC |
| H36B4 | GACACCCTCCAGGAAGCGA | GTGTTCGACAATGGCAGCAT |
| LEFTY2 | CCTGAGAGGGTGCTAAGAG | GGTAGGTAGGGGCTGTCT |
| LINC01605 | TTCCCGTTACAAACAGCCGA | ACTGCCTCTGTCTCCTGTCA |
| LINC02432 | ATGCTGTGGAGGCTCTGTTG | AAAAGCAGTGTGGACCCGAA |
| LINC02593 | TCCAAGTCCAGTCTGTCCAC | AGCATCCCTGACTACACACC |
| LINC02600 | CAGGAGGAGCTGAGGATGAG | GGACGTCACTCTCCAAAGGA |
| MALAT1 | GAATTGCGTCATTTAAAGCCTAGTT | GTTTCATCCTACCACTCCCAATTAAT |
| PGR | TCGCCTTAGAAAGTGCTGTC | GCTTGGCTTTCATTTGGAACG |
| PRL | AGCCAGGTTCATCCTGAAA | AGCAGAAAGGCGAGACTCTT |
| TIMP3 | TGACAGGTCGCGTCTATGAT | CAACCCAGGTGATACCGATAG |
| WNT4 | AGCCCTCATGAACCTCCAC | CACCCGCATGTGTGTCAG |
| XIST | ACGCTGCATGTGTCCTTAG | GAGCCTCTTATAGCTGTTTG |
| Near Gene (Site) | Upstream | Downstream |
|---|---|---|
| AC027288.3(FBR1) | GCCTGACTCTGGTGGTAAGC | TGCTGGCATTCAGTGCAGTT |
| AC027288.3(FBR2) | CAGTTACCTTCCACCCCGAC | GGTAGAGATTTGTGTGCAGCC |
| AC027288.3(CON) | AGGACGCTTTATAGTCGGGC | TGGCATCGTTCTTGCCAATC |
| IGFBP (CON) | AGATAGGGATTGGTTCGCGTAT | GGTCTGTGCTAACAATGCCAC |
| IGFBP1 (FBR) | TTCAGAGCATGGATTGCCCA | CTTCCCAATCCTGCCTTCGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thapa, R.; Marmo, K.; Ma, L.; Torry, D.S.; Bany, B.M. The Long Non-Coding RNA Gene AC027288.3 Plays a Role in Human Endometrial Stromal Fibroblast Decidualization. Cells 2024, 13, 778. https://doi.org/10.3390/cells13090778
Thapa R, Marmo K, Ma L, Torry DS, Bany BM. The Long Non-Coding RNA Gene AC027288.3 Plays a Role in Human Endometrial Stromal Fibroblast Decidualization. Cells. 2024; 13(9):778. https://doi.org/10.3390/cells13090778
Chicago/Turabian StyleThapa, Rupak, Kevin Marmo, Liang Ma, Donald S. Torry, and Brent M. Bany. 2024. "The Long Non-Coding RNA Gene AC027288.3 Plays a Role in Human Endometrial Stromal Fibroblast Decidualization" Cells 13, no. 9: 778. https://doi.org/10.3390/cells13090778
APA StyleThapa, R., Marmo, K., Ma, L., Torry, D. S., & Bany, B. M. (2024). The Long Non-Coding RNA Gene AC027288.3 Plays a Role in Human Endometrial Stromal Fibroblast Decidualization. Cells, 13(9), 778. https://doi.org/10.3390/cells13090778









