Variation of Structure and Cellular Functions of Type IA Topoisomerases across the Tree of Life
Abstract
1. Introduction
2. Conserved N-Terminal Domains
2.1. Organization of N-Terminal Domains
2.2. Formation of Catalytic Site and the Presence of Mg2+ Ion
2.3. Interdomain Movements Observed from Crystal Structures
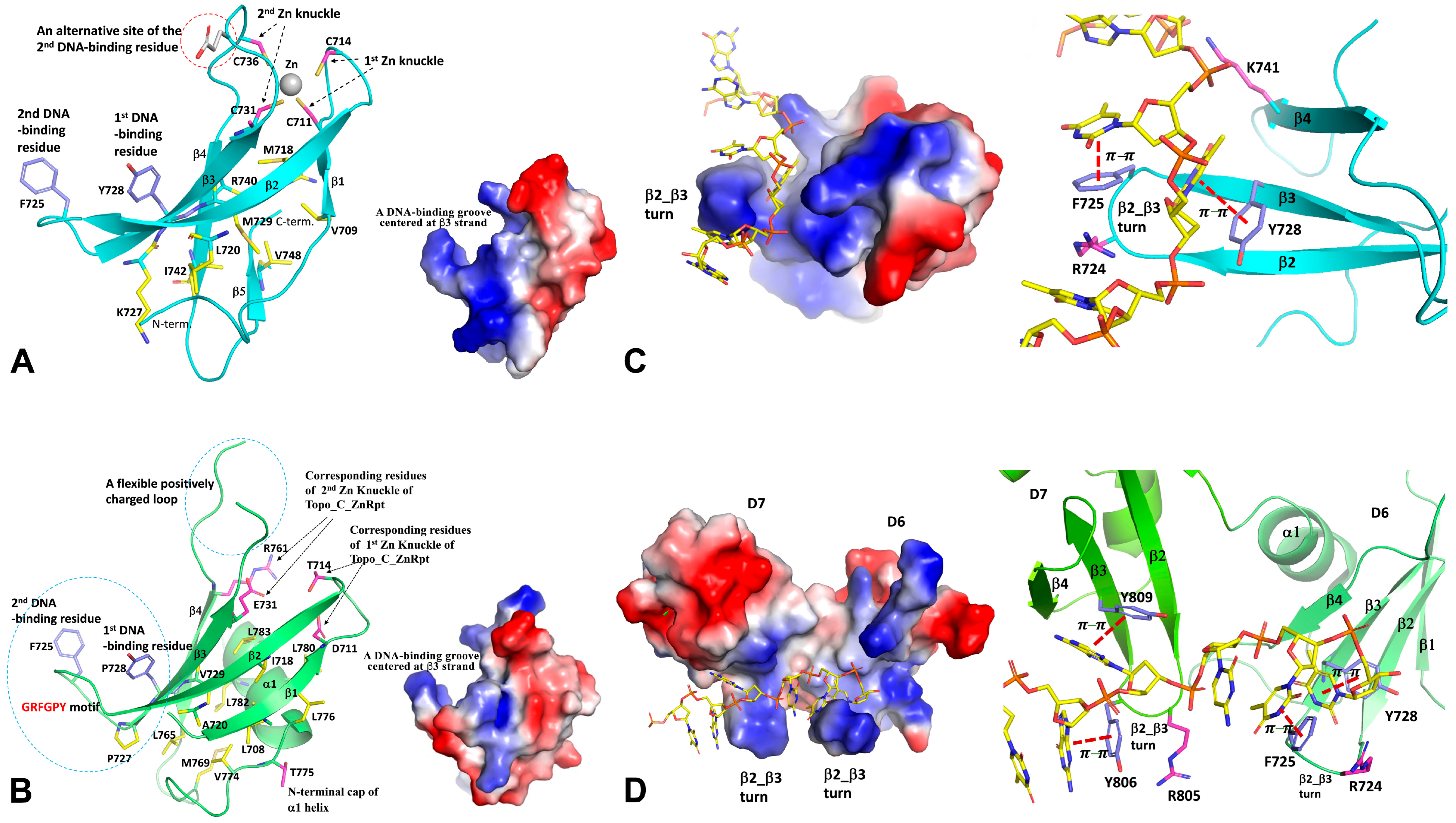
3. Variable C-Terminal Domains
3.1. C-Terminal Structural Motifs
3.2. Interaction between C-Terminal Domains and Nucleic Acid Substrates
4. Physiological Functions of Type IA Topoisomerases
4.1. Importance of Protein–Protein Interaction Partners
4.2. Resolution of Replication, Recombination, and Repair Intermediates in Bacteria and Archaea
4.3. DNA Supercoiling, Transcription and Regulation of R-Loops by Bacterial Topo I
4.4. Function of Reverse Gyrase
4.5. Function of TOP3A in Eukaryotes
4.6. Function of TOP3B in Eukaryotes
4.7. Cellular RNA Substrates for RNA Topoisomerase Activity
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, J.C. Interaction between DNA and an Escherichia coli protein omega. J. Mol. Biol. 1971, 55, 523–533. [Google Scholar] [CrossRef]
- Wang, J.C. A journey in the world of DNA rings and beyond. Annu. Rev. Biochem. 2009, 78, 31–54. [Google Scholar] [CrossRef]
- Gellert, M.; Mizuuchi, K.; O’Dea, M.H.; Nash, H.A. DNA gyrase: An enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. USA 1976, 73, 3872–3876. [Google Scholar] [CrossRef] [PubMed]
- Champoux, J.J.; McConaughy, B.L. Purification and characterization of the DNA untwisting enzyme from rat liver. Biochemistry 1976, 15, 4638–4642. [Google Scholar] [CrossRef] [PubMed]
- Champoux, J.J. DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef]
- McKie, S.J.; Neuman, K.C.; Maxwell, A. DNA topoisomerases: Advances in understanding of cellular roles and multi-protein complexes via structure-function analysis. Bioessays 2021, 43, e2000286. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Nussenzweig, A.; Takeda, S.; Austin, C. Human topoisomerases and their roles in genome stability and organization. Nat. Rev. Mol. Cell Biol. 2022, 23, 407–427. [Google Scholar] [CrossRef] [PubMed]
- Corbett, K.D.; Berger, J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004, 33, 95–118. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Chan, N.L.; Hsieh, T.S. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 2013, 82, 139–170. [Google Scholar] [CrossRef] [PubMed]
- Depew, R.E.; Liu, L.F.; Wang, J.C. Interaction between DNA and Escherichia coli protein omega. Formation of a complex between single-stranded DNA and omega protein. J. Biol. Chem. 1978, 253, 511–518. [Google Scholar] [CrossRef]
- Dasgupta, T.; Ferdous, S.; Tse-Dinh, Y.C. Mechanism of Type IA Topoisomerases. Molecules 2020, 25, 4769. [Google Scholar] [CrossRef]
- Viard, T.; de la Tour, C.B. Type IA topoisomerases: A simple puzzle? Biochimie 2007, 89, 456–467. [Google Scholar] [CrossRef]
- Bhat, A.G.; Leelaram, M.N.; Hegde, S.M.; Nagaraja, V. Deciphering the distinct role for the metal coordination motif in the catalytic activity of Mycobacterium smegmatis topoisomerase I. J. Mol. Biol. 2009, 393, 788–802. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Xue, Y.; Lee, S.K.; Martindale, J.L.; Shen, W.; Li, W.; Zou, S.; Ciaramella, M.; Debat, H.; Nadal, M.; et al. RNA topoisomerase is prevalent in all domains of life and associates with polyribosomes in animals. Nucleic Acids Res. 2016, 44, 6335–6349. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Xu, D.; Wang, W. Type IA topoisomerases can be “magicians” for both DNA and RNA in all domains of life. RNA Biol. 2017, 14, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Garnier, F.; Debat, H.; Nadal, M. Type IA DNA Topoisomerases: A Universal Core and Multiple Activities. In DNA Topoisomerases: Methods and Protocols; Humana Press: New York, NY, USA, 2018; Volume 1703, pp. 1–20. [Google Scholar] [CrossRef]
- Baker, N.M.; Rajan, R.; Mondragón, A. Structural studies of type I topoisomerases. Nucleic Acids Res. 2009, 37, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Lima, C.D.; Wang, J.C.; Mondragon, A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature 1994, 367, 138–146. [Google Scholar] [CrossRef]
- Tse, Y.C.; Kirkegaard, K.; Wang, J.C. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J. Biol. Chem. 1980, 255, 5560–5565. [Google Scholar] [CrossRef]
- Aravind, L.; Leipe, D.D.; Koonin, E.V. Toprim—A conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998, 26, 4205–4213. [Google Scholar] [CrossRef]
- Zhu, C.X.; Tse-Dinh, Y.C. The acidic triad conserved in type IA DNA topoisomerases is required for binding of Mg(II) and subsequent conformational change. J. Biol. Chem. 2000, 275, 5318–5322. [Google Scholar] [CrossRef]
- Cao, N.; Tan, K.; Annamalai, T.; Joachimiak, A.; Tse-Dinh, Y.C. Investigating mycobacterial topoisomerase I mechanism from the analysis of metal and DNA substrate interactions at the active site. Nucleic Acids Res. 2018, 46, 7296–7308. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.H.; Burgin, A.B.; Deweese, J.E.; Osheroff, N.; Berger, J.M. A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases. Nature 2010, 465, 641–644. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Saha, S.; Yang, W.; Neuman, K.C.; Pommier, Y. Structural and biochemical basis for DNA and RNA catalysis by human Topoisomerase 3β. Nat. Commun. 2022, 13, 4656. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Garnier, F.; Debat, H.; Strick, T.R.; Nadal, M. Direct observation of helicase-topoisomerase coupling within reverse gyrase. Proc. Natl. Acad. Sci. USA 2020, 117, 10856–10864. [Google Scholar] [CrossRef]
- Rudolph, M.G.; del Toro Duany, Y.; Jungblut, S.P.; Ganguly, A.; Klostermeier, D. Crystal structures of Thermotoga maritima reverse gyrase: Inferences for the mechanism of positive DNA supercoiling. Nucleic Acids Res. 2013, 41, 1058–1070. [Google Scholar] [CrossRef]
- Rodriguez, A.C.; Stock, D. Crystal structure of reverse gyrase: Insights into the positive supercoiling of DNA. EMBO J. 2002, 21, 418–426. [Google Scholar] [CrossRef]
- Forterre, P. A hot story from comparative genomics: Reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002, 18, 236–237. [Google Scholar] [CrossRef]
- Garnier, F.; Couturier, M.; Débat, H.; Nadal, M. Archaea: A Gold Mine for Topoisomerase Diversity. Front. Microbiol. 2021, 12, 661411. [Google Scholar] [CrossRef]
- Diaz, B.; Mederos, C.; Tan, K.; Tse-Dinh, Y.C. Microbial Type IA Topoisomerase C-Terminal Domain Sequence Motifs, Distribution and Combination. Int. J. Mol. Sci. 2022, 23, 8709. [Google Scholar] [CrossRef]
- Cao, N.; Tan, K.; Zuo, X.; Annamalai, T.; Tse-Dinh, Y.C. Mechanistic insights from structure of Mycobacterium smegmatis topoisomerase I with ssDNA bound to both N- and C-terminal domains. Nucleic Acids Res. 2020, 48, 4448–4462. [Google Scholar] [CrossRef]
- Strzalka, A.; Szafran, M.J.; Strick, T.; Jakimowicz, D. C-terminal lysine repeats in Streptomyces topoisomerase I stabilize the enzyme-DNA complex and confer high enzyme processivity. Nucleic Acids Res. 2017, 45, 11908–11924. [Google Scholar] [CrossRef] [PubMed]
- Dorn, A.; Röhrig, S.; Papp, K.; Schröpfer, S.; Hartung, F.; Knoll, A.; Puchta, H. The topoisomerase 3α zinc-finger domain T1 of Arabidopsis thaliana is required for targeting the enzyme activity to Holliday junction-like DNA repair intermediates. PLoS Genet. 2018, 14, e1007674. [Google Scholar] [CrossRef]
- Tan, K.; Zhou, Q.; Cheng, B.; Zhang, Z.; Joachimiak, A.; Tse-Dinh, Y.C. Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I. Nucleic Acids Res. 2015, 43, 11031–11046. [Google Scholar] [CrossRef]
- Suerbaum, S.; Brauer-Steppkes, T.; Labigne, A.; Cameron, B.; Drlica, K. Topoisomerase I of Helicobacter pylori: Juxtaposition with a flagellin gene (flaB) and functional requirement of a fourth zinc finger motif. Gene 1998, 210, 151–161. [Google Scholar] [CrossRef]
- Cheng, B.; Zhu, C.X.; Ji, C.; Ahumada, A.; Tse-Dinh, Y.C. Direct interaction between Escherichia coli RNA polymerase and the zinc ribbon domains of DNA topoisomerase I. J. Biol. Chem. 2003, 278, 30705–30710. [Google Scholar] [CrossRef] [PubMed]
- Allemand, F.; Mathy, N.; Brechemier-Baey, D.; Condon, C. The 5S rRNA maturase, ribonuclease M5, is a Toprim domain family member. Nucleic Acids Res. 2005, 33, 4368–4376. [Google Scholar] [CrossRef]
- Podobnik, M.; McInerney, P.; O’Donnell, M.; Kuriyan, J. A TOPRIM domain in the crystal structure of the catalytic core of Escherichia coli primase confirms a structural link to DNA topoisomerases. J. Mol. Biol. 2000, 300, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.A.; Hevener, K.E. Crystal structure of the 65-kilodalton amino-terminal fragment of DNA topoisomerase I from the gram-positive model organism Streptococcus mutans. Biochem. Biophys. Res. Commun. 2019, 516, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cheng, B.; Tse-Dinh, Y.C. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc. Natl. Acad. Sci. USA 2011, 108, 6939–6944. [Google Scholar] [CrossRef]
- Tan, K.; Cao, N.; Cheng, B.; Joachimiak, A.; Tse-Dinh, Y.C. Insights from the Structure of Mycobacterium tuberculosis Topoisomerase I with a Novel Protein Fold. J. Mol. Biol. 2016, 428, 182–193. [Google Scholar] [CrossRef]
- Berger, J.M.; Fass, D.; Wang, J.C.; Harrison, S.C. Structural similarities between topoisomerases that cleave one or both DNA strands. Proc. Natl. Acad. Sci. USA 1998, 95, 7876–7881. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, S.; Dasgupta, T.; Annamalai, T.; Tan, K.; Tse-Dinh, Y.C. The interaction between transport-segment DNA and topoisomerase IA-crystal structure of MtbTOP1 in complex with both G- and T-segments. Nucleic Acids Res. 2023, 51, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Castillo, R.M.; Mizuguchi, K.; Dhanaraj, V.; Albert, A.; Blundell, T.L.; Murzin, A.G. A six-stranded double-psi beta barrel is shared by several protein superfamilies. Structure 1999, 7, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Bianco, P.R. OB-fold Families of Genome Guardians: A Universal Theme Constructed from the Small β-barrel Building Block. Front. Mol. Biosci. 2022, 9, 784451. [Google Scholar] [CrossRef] [PubMed]
- Arcus, V. OB-fold domains: A snapshot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 2002, 12, 794–801. [Google Scholar] [CrossRef]
- Theobald, D.L.; Mitton-Fry, R.M.; Wuttke, D.S. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 115–133. [Google Scholar] [CrossRef]
- Tse-Dinh, Y.C.; McCarron, B.G.; Arentzen, R.; Chowdhry, V. Mechanistic study of E. coli DNA topoisomerase I: Cleavage of oligonucleotides. Nucleic Acids Res. 1983, 11, 8691–8701. [Google Scholar] [CrossRef]
- Feinberg, H.; Lima, C.D.; Mondragón, A. Conformational changes in E. coli DNA topoisomerase I. Nat. Struct. Biol. 1999, 6, 918–922. [Google Scholar] [CrossRef]
- Changela, A.; DiGate, R.J.; Mondragon, A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature 2001, 411, 1077–1081. [Google Scholar] [CrossRef]
- Perry, K.; Mondragon, A. Biochemical characterization of an invariant histidine involved in Escherichia coli DNA topoisomerase I catalysis. J. Biol. Chem. 2002, 277, 13237–13245. [Google Scholar] [CrossRef]
- Tse-Dinh, Y.C.; Beran-Steed, R.K. Escherichia coli DNA topoisomerase I is a zinc metalloprotein with three repetitive zinc-binding domains. J. Biol. Chem. 1988, 263, 15857–15859. [Google Scholar] [CrossRef] [PubMed]
- Grishin, N.V. C-terminal domains of Escherichia coli topoisomerase I belong to the zinc-ribbon superfamily. J. Mol. Biol. 2000, 299, 1165–1177. [Google Scholar] [CrossRef] [PubMed]
- Krishna, S.S.; Majumdar, I.; Grishin, N.V. Structural classification of zinc fingers: Survey and summary. Nucleic Acids Res. 2003, 31, 532–550. [Google Scholar] [CrossRef] [PubMed]
- Murzin, A.G.; Brenner, S.E.; Hubbard, T.; Chothia, C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995, 247, 536–540. [Google Scholar] [CrossRef]
- Yu, L.; Zhu, C.X.; Tse-Dinh, Y.C.; Fesik, S.W. Solution structure of the C-terminal single-stranded DNA-binding domain of Escherichia coli topoisomerase I. Biochemistry 1995, 34, 7622–7628. [Google Scholar] [CrossRef] [PubMed]
- Moreira, F.; Arenas, M.; Videira, A.; Pereira, F. Molecular Evolution of DNA Topoisomerase III Beta (TOP3B) in Metazoa. J. Mol. Evol. 2021, 89, 384–395. [Google Scholar] [CrossRef]
- Dello Stritto, M.R.; Vojtassakova, N.; Velkova, M.; Hamminger, P.; Ulm, P.; Jantsch, V. The topoisomerase 3 zinc finger domain cooperates with the RMI1 scaffold to promote stable association of the BTR complex to recombination intermediates in the Caenorhabditis elegans germline. Nucleic Acids Res. 2022, 50, 5652–5671. [Google Scholar] [CrossRef] [PubMed]
- Wallace, B.D.; Berman, Z.; Mueller, G.A.; Lin, Y.; Chang, T.; Andres, S.N.; Wojtaszek, J.L.; DeRose, E.F.; Appel, C.D.; London, R.E.; et al. APE2 Zf-GRF facilitates 3′-5′ resection of DNA damage following oxidative stress. Proc. Natl. Acad. Sci. USA 2017, 114, 304–309. [Google Scholar] [CrossRef]
- Klebanov-Akopyan, O.; Mishra, A.; Glousker, G.; Tzfati, Y.; Shlomai, J. Trypanosoma brucei UMSBP2 is a single-stranded telomeric DNA binding protein essential for chromosome end protection. Nucleic Acids Res. 2018, 46, 7757–7771. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, Y.; Pang, Y.; Yu, H.; Zhang, W.; Zhao, X.; Yu, J. The distinct roles of zinc finger CCHC-type (ZCCHC) superfamily proteins in the regulation of RNA metabolism. RNA Biol. 2021, 18, 2107–2126. [Google Scholar] [CrossRef]
- Aceituno-Valenzuela, U.; Micol-Ponce, R.; Ponce, M.R. Genome-wide analysis of CCHC-type zinc finger (ZCCHC) proteins in yeast, Arabidopsis, and humans. Cell Mol. Life Sci. 2020, 77, 3991–4014. [Google Scholar] [CrossRef]
- Szafran, M.J.; Strzalka, A.; Jakimowicz, D. A highly processive actinobacterial topoisomerase I—Thoughts on Streptomyces’ demand for an enzyme with a unique C-terminal domain. Microbiology 2020, 166, 120–128. [Google Scholar] [CrossRef]
- Xu, D.; Shen, W.; Guo, R.; Xue, Y.; Peng, W.; Sima, J.; Yang, J.; Sharov, A.; Srikantan, S.; Yang, J.; et al. Top3beta is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013, 16, 1238–1247. [Google Scholar] [CrossRef]
- Yang, J.; Annamalai, T.; Cheng, B.; Banda, S.; Tyagi, R.; Tse-Dinh, Y.C. Antimicrobial Susceptibility and SOS-Dependent Increase in Mutation Frequency Are Impacted by Escherichia coli Topoisomerase I C-Terminal Point Mutation. Antimicrob. Agents Chemother. 2015, 59, 6195–6202. [Google Scholar] [CrossRef] [PubMed]
- Sutormin, D.; Galivondzhyan, A.; Musharova, O.; Travin, D.; Rusanova, A.; Obraztsova, K.; Borukhov, S.; Severinov, K. Interaction between transcribing RNA polymerase and topoisomerase I prevents R-loop formation in E. coli. Nat. Commun. 2022, 13, 4524. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.B.; Chapagain, P.P.; Banda, S.; Darici, Y.; Uren, A.; Tse-Dinh, Y.C. Characterization of molecular interactions between Escherichia coli RNA polymerase and topoisomerase I by molecular simulations. FEBS Lett. 2016, 590, 2844–2851. [Google Scholar] [CrossRef]
- Lee, C.M.; Wang, G.; Pertsinidis, A.; Marians, K.J. Topoisomerase III Acts at the Replication Fork To Remove Precatenanes. J. Bacteriol. 2019, 201. [Google Scholar] [CrossRef] [PubMed]
- Harami, G.M.; Pálinkás, J.; Seol, Y.; Kovács, Z.J.; Gyimesi, M.; Harami-Papp, H.; Neuman, K.C.; Kovács, M. The toposiomerase IIIalpha-RMI1-RMI2 complex orients human Bloom’s syndrome helicase for efficient disruption of D-loops. Nat. Commun. 2022, 13, 654. [Google Scholar] [CrossRef] [PubMed]
- Bachrati, C.Z.; Hickson, I.D. Dissolution of double Holliday junctions by the concerted action of BLM and topoisomerase IIIalpha. Methods Mol. Biol. 2009, 582, 91–102. [Google Scholar] [CrossRef]
- Bizard, A.H.; Hickson, I.D. The many lives of type IA topoisomerases. J. Biol. Chem. 2020, 295, 7138–7153. [Google Scholar] [CrossRef] [PubMed]
- Bakx, J.A.M.; Biebricher, A.S.; King, G.A.; Christodoulis, P.; Sarlós, K.; Bizard, A.H.; Hickson, I.D.; Wuite, G.J.L.; Peterman, E.J.G. Duplex DNA and BLM regulate gate opening by the human TopoIIIα-RMI1-RMI2 complex. Nat. Commun. 2022, 13, 584. [Google Scholar] [CrossRef]
- Yang, Y.; McBride, K.M.; Hensley, S.; Lu, Y.; Chedin, F.; Bedford, M.T. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 2014, 53, 484–497. [Google Scholar] [CrossRef]
- Goto-Ito, S.; Yamagata, A.; Takahashi, T.S.; Sato, Y.; Fukai, S. Structural basis of the interaction between Topoisomerase IIIbeta and the TDRD3 auxiliary factor. Sci. Rep. 2017, 7, 42123. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Xue, Y.; Sharov, A.; Zhang, Y.; Lee, S.K.; Martindale, J.L.; Li, W.; Ku, W.L.; Zhao, K.; De, S.; et al. A dual-activity topoisomerase complex regulates mRNA translation and turnover. Nucleic Acids Res. 2022, 50, 7013–7033. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Xue, Y.; Lee, S.K.; Zhang, Y.; Fan, J.; De, S.; Sharov, A.; Wang, W. A dual-activity topoisomerase complex promotes both transcriptional activation and repression in response to starvation. Nucleic Acids Res. 2023, 51, 2415–2433. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.C. Cellular roles of DNA topoisomerases: A molecular perspective. Nat. Rev. Mol. Cell Biol. 2002, 3, 430–440. [Google Scholar] [CrossRef] [PubMed]
- Bizard, A.H.; Yang, X.; Debat, H.; Fogg, J.M.; Zechiedrich, L.; Strick, T.R.; Garnier, F.; Nadal, M. TopA, the Sulfolobus solfataricus topoisomerase III, is a decatenase. Nucleic Acids Res. 2018, 46, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Guo, L.; Deng, L.; Feng, D.; Ren, Y.; Chu, Y.; She, Q.; Huang, L. Deletion of the topoisomerase III gene in the hyperthermophilic archaeon Sulfolobus islandicus results in slow growth and defects in cell cycle control. J. Genet. Genom. 2011, 38, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Terekhova, K.; Marko, J.F.; Mondragon, A. Single-molecule analysis uncovers the difference between the kinetics of DNA decatenation by bacterial topoisomerases I and III. Nucleic Acids Res. 2014, 42, 11657–11667. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P.; Levine, C.; Hassing, H.; Marians, K.J. Topoisomerase III can serve as the cellular decatenase in Escherichia coli. J. Biol. Chem. 2003, 278, 8653–8660. [Google Scholar] [CrossRef]
- Conin, B.; Billault-Chaumartin, I.; El Sayyed, H.; Quenech’Du, N.; Cockram, C.; Koszul, R.; Espéli, O. Extended sister-chromosome catenation leads to massive reorganization of the E. coli genome. Nucleic Acids Res. 2022, 50, 2635–2650. [Google Scholar] [CrossRef]
- Pruss, G.J.; Manes, S.H.; Drlica, K. Escherichia coli DNA topoisomerase I mutants: Increased supercoiling is corrected by mutations near gyrase genes. Cell 1982, 31, 35–42. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, S.; Voelkel, K.A.; Sternglanz, R.; Reynolds, A.E.; Wright, A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell 1982, 31, 43–51. [Google Scholar] [CrossRef]
- Menzel, R.; Gellert, M. Regulation of the genes for E. coli DNA gyrase: Homeostatic control of DNA supercoiling. Cell 1983, 34, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Tse-Dinh, Y.C. Regulation of the Escherichia coli DNA topoisomerase I gene by DNA supercoiling. Nucleic Acids Res. 1985, 13, 4751–4763. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Menon, S.; Karthik, P.V.; Nagaraja, V. Autoregulation of topoisomerase I expression by supercoiling sensitive transcription. Nucleic Acids Res. 2016, 44, 1541–1552. [Google Scholar] [CrossRef] [PubMed]
- García-López, M.; Megias, D.; Ferrándiz, M.J.; de la Campa, A.G. The balance between gyrase and topoisomerase I activities determines levels of supercoiling, nucleoid compaction, and viability in bacteria. Front. Microbiol. 2022, 13, 1094692. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Gao, L.; Swoboda, A.R.; Ouellette, S.P. Targeted repression of topA by CRISPRi reveals a critical function for balanced DNA topoisomerase I activity in the Chlamydia trachomatis developmental cycle. mBio 2024, 15, e0258423. [Google Scholar] [CrossRef]
- Liu, L.F.; Wang, J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. USA 1987, 84, 7024–7027. [Google Scholar] [CrossRef]
- Banda, S.; Cao, N.; Tse-Dinh, Y.C. Distinct Mechanism Evolved for Mycobacterial RNA Polymerase and Topoisomerase I Protein-Protein Interaction. J. Mol. Biol. 2017, 429, 2931–2942. [Google Scholar] [CrossRef]
- Ferrándiz, M.J.; Hernández, P.; de la Campa, A.G. Genome-wide proximity between RNA polymerase and DNA topoisomerase I supports transcription in Streptococcus pneumoniae. PLoS Genet. 2021, 17, e1009542. [Google Scholar] [CrossRef]
- Ahmed, W.; Sala, C.; Hegde, S.R.; Jha, R.K.; Cole, S.T.; Nagaraja, V. Transcription facilitated genome-wide recruitment of topoisomerase I and DNA gyrase. PLoS Genet. 2017, 13, e1006754. [Google Scholar] [CrossRef] [PubMed]
- Martel, M.; Balleydier, A.; Sauriol, A.; Drolet, M. Constitutive stable DNA replication in Escherichia coli cells lacking type 1A topoisomerase activity. DNA Repair. 2015, 35, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Leela, J.K.; Raghunathan, N.; Gowrishankar, J. Topoisomerase I Essentiality, DnaA-Independent Chromosomal Replication, and Transcription-Replication Conflict in Escherichia coli. J. Bacteriol. 2021, 203, e0019521. [Google Scholar] [CrossRef] [PubMed]
- Brochu, J.; Breton, E.V.; Drolet, M. Supercoiling, R-loops, Replication and the Functions of Bacterial Type 1A Topoisomerases. Genes 2020, 11, 249. [Google Scholar] [CrossRef]
- Usongo, V.; Martel, M.; Balleydier, A.; Drolet, M. Mutations reducing replication from R-loops suppress the defects of growth, chromosome segregation and DNA supercoiling in cells lacking topoisomerase I and RNase HI activity. DNA Repair. 2016, 40, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Raji, A.; Zabel, D.J.; Laufer, C.S.; Depew, R.E. Genetic analysis of mutations that compensate for loss of Escherichia coli DNA topoisomerase I. J. Bacteriol. 1985, 162, 1173–1179. [Google Scholar] [CrossRef]
- McNairn, E.; Ni Bhriain, N.; Dorman, C.J. Overexpression of the Shigella flexneri genes coding for DNA topoisomerase IV compensates for loss of DNA topoisomerase I: Effect on virulence gene expression. Mol. Microbiol. 1995, 15, 507–517. [Google Scholar] [CrossRef]
- Dorman, C.J.; Lynch, A.S.; Ni Bhriain, N.; Higgins, C.F. DNA supercoiling in Escherichia coli: topA mutations can be suppressed by DNA amplifications involving the tolC locus. Mol. Microbiol. 1989, 3, 531–540. [Google Scholar] [CrossRef]
- Brochu, J.; Vlachos-Breton, É.; Sutherland, S.; Martel, M.; Drolet, M. Topoisomerases I and III inhibit R-loop formation to prevent unregulated replication in the chromosomal Ter region of Escherichia coli. PLoS Genet. 2018, 14, e1007668. [Google Scholar] [CrossRef]
- Reuß, D.R.; Faßhauer, P.; Mroch, P.J.; Ul-Haq, I.; Koo, B.M.; Pöhlein, A.; Gross, C.A.; Daniel, R.; Brantl, S.; Stülke, J. Topoisomerase IV can functionally replace all type 1A topoisomerases in Bacillus subtilis. Nucleic Acids Res. 2019, 47, 5231–5242. [Google Scholar] [CrossRef]
- Zechiedrich, E.L.; Khodursky, A.B.; Bachellier, S.; Schneider, R.; Chen, D.; Lilley, D.M.; Cozzarelli, N.R. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000, 275, 8103–8113. [Google Scholar] [CrossRef]
- Ravishankar, S.; Ambady, A.; Awasthy, D.; Mudugal, N.V.; Menasinakai, S.; Jatheendranath, S.; Guptha, S.; Sharma, S.; Balakrishnan, G.; Nandishaiah, R.; et al. Genetic and chemical validation identifies Mycobacterium tuberculosis topoisomerase I as an attractive anti-tubercular target. Tuberculosis 2015, 95, 589–598. [Google Scholar] [CrossRef]
- Ahmed, W.; Menon, S.; Godbole, A.A.; Karthik, P.V.; Nagaraja, V. Conditional silencing of topoisomerase I gene of Mycobacterium tuberculosis validates its essentiality for cell survival. FEMS Microbiol. Lett. 2014, 353, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Menon, S.; Karthik, P.V.; Nagaraja, V. Reduction in DNA topoisomerase I level affects growth, phenotype and nucleoid architecture of Mycobacterium smegmatis. Microbiology 2015, 161, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Rifat, D.; Chen, L.; Kreiswirth, B.N.; Nuermberger, E.L. Genome-wide essentiality analysis of Mycobacterium abscessus by saturated transposon mutagenesis and deep sequencing. Mbio 2021, 12, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Hu, S.; Ma, N.; Song, P.; Liang, Q.; Zhang, H.; Li, Y.; Shen, L.; Duan, K.; Chen, L. Regulatory Effect of DNA Topoisomerase I on T3SS Activity, Antibiotic Susceptibility and Quorum-Sensing-Independent Pyocyanin Synthesis in Pseudomonas aeruginosa. Int. J. Mol. Sci. 2019, 20, 1116. [Google Scholar] [CrossRef]
- García-López, M.; Hernández, P.; Megias, D.; Ferrándiz, M.J.; Campa, A.G. Physiologic and Transcriptomic Effects Triggered by Overexpression of Wild Type and Mutant DNA Topoisomerase I in Streptococcus pneumoniae. Int. J. Mol. Sci. 2023, 24, 15800. [Google Scholar] [CrossRef] [PubMed]
- Villain, P.; Catchpole, R.; Forterre, P.; Oberto, J.; da Cunha, V.; Basta, T. Expanded Dataset Reveals the Emergence and Evolution of DNA Gyrase in Archaea. Mol. Biol. Evol. 2022, 39, msac155. [Google Scholar] [CrossRef] [PubMed]
- McKie, S.J.; Desai, P.R.; Seol, Y.; Allen, A.M.; Maxwell, A.; Neuman, K.C. Topoisomerase VI is a chirally-selective, preferential DNA decatenase. Elife 2022, 11, e67021. [Google Scholar] [CrossRef]
- Stevens, K.M.; Warnecke, T. Histone variants in archaea—An undiscovered country. Semin. Cell Dev. Biol. 2023, 135, 50–58. [Google Scholar] [CrossRef]
- Duprey, A.; Groisman, E.A. The regulation of DNA supercoiling across evolution. Protein Sci. 2021, 30, 2042–2056. [Google Scholar] [CrossRef]
- Hocher, A.; Borrel, G.; Fadhlaoui, K.; Brugère, J.F.; Gribaldo, S.; Warnecke, T. Growth temperature and chromatinization in archaea. Nat. Microbiol. 2022, 7, 1932–1942. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Dhar, A.; Trostel, A.; Kouzine, F.; Seshasayee, A.S.; Adhya, S. Genome scale patterns of supercoiling in a bacterial chromosome. Nat. Commun. 2016, 7, 11055. [Google Scholar] [CrossRef] [PubMed]
- Strzałka, A.; Kois-Ostrowska, A.; Kędra, M.; Łebkowski, T.; Bieniarz, G.; Szafran, M.J.; Jakimowicz, D. Enhanced binding of an HU homologue under increased DNA supercoiling preserves chromosome organisation and sustains Streptomyces hyphal growth. Nucleic Acids Res. 2022, 50, 12202–12216. [Google Scholar] [CrossRef] [PubMed]
- de Vasconcelos Junior, A.A.; Tirado-Vélez, J.M.; Martín-Galiano, A.J.; Megias, D.; Ferrándiz, M.-J.; Hernández, P.; Amblar, M.; de la Campa, A.G. StaR Is a Positive Regulator of Topoisomerase I Activity Involved in Supercoiling Maintenance in Streptococcus pneumoniae. Int. J. Mol. Sci. 2023, 24, 5973. [Google Scholar] [CrossRef]
- Lee, C.; Marians, K.J. Characterization of the nucleoid-associated protein YejK. J. Biol. Chem. 2013, 288, 31503–31516. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Mallick, B.; Nagaraja, V. Direct regulation of topoisomerase activity by a nucleoid-associated protein. Nucleic Acids Res. 2014, 42, 11156–11165. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.S.; Haakonsen, D.L.; Zeng, W.; Schumacher, M.A.; Laub, M.T. A Bacterial Chromosome Structuring Protein Binds Overtwisted DNA to Stimulate Type II Topoisomerases and Enable DNA Replication. Cell 2018, 175, 583–597.e523. [Google Scholar] [CrossRef]
- Couturier, M.; Gadelle, D.; Forterre, P.; Nadal, M.; Garnier, F. The reverse gyrase TopR1 is responsible for the homeostatic control of DNA supercoiling in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol. Microbiol. 2020, 113, 356–368. [Google Scholar] [CrossRef]
- Zhang, C.; Phillips, A.P.R.; Wipfler, R.L.; Olsen, G.J.; Whitaker, R.J. The essential genome of the crenarchaeal model Sulfolobus islandicus. Nat. Commun. 2018, 9, 4908. [Google Scholar] [CrossRef]
- Bendia, A.G.; Lemos, L.N.; Mendes, L.W.; Signori, C.N.; Bohannan, B.J.M.; Pellizari, V.H. Metabolic potential and survival strategies of microbial communities across extreme temperature gradients on Deception Island volcano, Antarctica. Environ. Microbiol. 2021, 23, 4054–4073. [Google Scholar] [CrossRef]
- Lulchev, P.; Klostermeier, D. Reverse gyrase—Recent advances and current mechanistic understanding of positive DNA supercoiling. Nucleic Acids Res. 2014, 42, 8200–8213. [Google Scholar] [CrossRef]
- Han, W.; Feng, X.; She, Q. Reverse Gyrase Functions in Genome Integrity Maintenance by Protecting DNA Breaks In Vivo. Int. J. Mol. Sci. 2017, 18, 1340. [Google Scholar] [CrossRef]
- Li, W.; Wang, J.C. Mammalian DNA topoisomerase IIIα is essential in early embryogenesis. Proc. Natl. Acad. Sci. USA 1998, 95, 1010–1013. [Google Scholar] [CrossRef]
- Soniat, M.M.; Nguyen, G.; Kuo, H.C.; Finkelstein, I.J. The MRN complex and topoisomerase IIIa-RMI1/2 synchronize DNA resection motor proteins. J. Biol. Chem. 2023, 299, 102802. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.A.; Sarlós, K.; Logan, C.V.; Thakur, R.S.; Parry, D.A.; Bizard, A.H.; Leitch, A.; Cleal, L.; Ali, N.S.; Al-Owain, M.A.; et al. Mutations in TOP3A Cause a Bloom Syndrome-like Disorder. Am. J. Hum. Genet. 2018, 103, 221–231. [Google Scholar] [CrossRef]
- de Nonneville, A.; Salas, S.; Bertucci, F.; Sobinoff, A.P.; Adélaïde, J.; Guille, A.; Finetti, P.; Noble, J.R.; Churikov, D.; Chaffanet, M.; et al. TOP3A amplification and ATRX inactivation are mutually exclusive events in pediatric osteosarcomas using ALT. EMBO Mol. Med. 2022, 14, e15859. [Google Scholar] [CrossRef]
- Huang, Y.; Luo, J.; Zhang, Y.; Zhang, T.; Fei, X.; Chen, L.; Zhu, Y.; Li, S.; Zhou, C.; Xu, K.; et al. Identification of MKNK1 and TOP3A as ovarian endometriosis risk-associated genes using integrative genomic analyses and functional experiments. Comput. Struct. Biotechnol. J. 2023, 21, 1510–1522. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lyu, Y.L.; Wang, J.C. Dual localization of human DNA topoisomerase IIIalpha to mitochondria and nucleus. Proc. Natl. Acad. Sci. USA 2002, 99, 12114–12119. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Feng, L.; Hsieh, T.S. Drosophila topo IIIalpha is required for the maintenance of mitochondrial genome and male germ-line stem cells. Proc. Natl. Acad. Sci. USA 2010, 107, 6228–6233. [Google Scholar] [CrossRef]
- Tsai, H.Z.; Lin, R.K.; Hsieh, T.S. Drosophila mitochondrial topoisomerase III alpha affects the aging process via maintenance of mitochondrial function and genome integrity. J. Biomed. Sci. 2016, 23, 38. [Google Scholar] [CrossRef]
- Nicholls, T.J.; Nadalutti, C.A.; Motori, E.; Sommerville, E.W.; Gorman, G.S.; Basu, S.; Hoberg, E.; Turnbull, D.M.; Chinnery, P.F.; Larsson, N.G.; et al. Topoisomerase 3alpha Is Required for Decatenation and Segregation of Human mtDNA. Mol. Cell 2018, 69, 9–23.e26. [Google Scholar] [CrossRef]
- Menger, K.E.; Chapman, J.; Díaz-Maldonado, H.; Khazeem, M.M.; Deen, D.; Erdinc, D.; Casement, J.W.; Di Leo, V.; Pyle, A.; Rodríguez-Luis, A.; et al. Two type I topoisomerases maintain DNA topology in human mitochondria. Nucleic Acids Res. 2022, 50, 11154–11174. [Google Scholar] [CrossRef]
- Hangas, A.; Kekäläinen, N.J.; Potter, A.; Michell, C.; Aho, K.J.; Rutanen, C.; Spelbrink, J.N.; Pohjoismäki, J.L.; Goffart, S. Top3α is the replicative topoisomerase in mitochondrial DNA replication. Nucleic Acids Res. 2022, 50, 8733–8748. [Google Scholar] [CrossRef]
- Primiano, G.; Torraco, A.; Verrigni, D.; Sabino, A.; Bertini, E.; Carrozzo, R.; Silvestri, G.; Servidei, S. Novel TOP3A Variant Associated with Mitochondrial Disease: Expanding the Clinical Spectrum of Topoisomerase III Alpha-Related Diseases. Neurol. Genet. 2022, 8, e200007. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Jia, N.; Guo, C.; Wen, J.; Wu, L.; Ogi, T.; Zhang, H. Predominant cellular mitochondrial dysfunction in the TOP3A gene-caused Bloom syndrome-like disorder. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166106. [Google Scholar] [CrossRef]
- Llauradó, A.; Rovira-Moreno, E.; Codina-Solà, M.; Martínez-Saez, E.; Salvadó, M.; Sanchez-Tejerina, D.; Sotoca, J.; López-Diego, V.; Restrepo-Vera, J.L.; Garcia-Arumi, E.; et al. Chronic progressive external ophthalmoplegia plus syndrome due to homozygous missense variant in TOP3A gene. Clin. Genet. 2023, 103, 492–494. [Google Scholar] [CrossRef] [PubMed]
- Erdinc, D.; Rodríguez-Luis, A.; Fassad, M.R.; Mackenzie, S.; Watson, C.M.; Valenzuela, S.; Xie, X.; Menger, K.E.; Sergeant, K.; Craig, K.; et al. Pathological variants in TOP3A cause distinct disorders of mitochondrial and nuclear genome stability. EMBO Mol. Med. 2023, 15, e16775. [Google Scholar] [CrossRef]
- Stoll, G.; Pietilainen, O.P.H.; Linder, B.; Suvisaari, J.; Brosi, C.; Hennah, W.; Leppa, V.; Torniainen, M.; Ripatti, S.; Ala-Mello, S.; et al. Deletion of TOP3beta, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 2013, 16, 1228–1237. [Google Scholar] [CrossRef]
- Richter, J.D.; Zhao, X. The molecular biology of FMRP: New insights into fragile X syndrome. Nat. Rev. Neurosci. 2021, 22, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Rahman, F.U.; Kim, Y.R.; Kim, E.K.; Kim, H.R.; Cho, S.M.; Lee, C.S.; Kim, S.J.; Araki, K.; Yamamura, K.I.; Lee, M.N.; et al. Topoisomerase IIIβ Deficiency Induces Neuro-Behavioral Changes and Brain Connectivity Alterations in Mice. Int. J. Mol. Sci. 2021, 22, 12806. [Google Scholar] [CrossRef] [PubMed]
- Joo, Y.; Xue, Y.; Wang, Y.; McDevitt, R.A.; Sah, N.; Bossi, S.; Su, S.; Lee, S.K.; Peng, W.; Xie, A.; et al. Topoisomerase 3beta knockout mice show transcriptional and behavioural impairments associated with neurogenesis and synaptic plasticity. Nat. Commun. 2020, 11, 3143. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Joo, Y.; Bossi, S.; McDevitt, R.; Xie, A.; Wang, Y.; Xue, Y.; Su, S.; Lee, S.K.; Sah, N.; et al. Tdrd3-null mice show post-transcriptional and behavioral impairments associated with neurogenesis and synaptic plasticity. Res. Sq. 2023, 233, 102568. [Google Scholar] [CrossRef]
- Lee, S.K.; Xue, Y.; Shen, W.; Zhang, Y.; Joo, Y.; Ahmad, M.; Chinen, M.; Ding, Y.; Ku, W.L.; De, S.; et al. Topoisomerase 3beta interacts with RNAi machinery to promote heterochromatin formation and transcriptional silencing in Drosophila. Nat. Commun. 2018, 9, 4946. [Google Scholar] [CrossRef]
- Huang, L.; Wang, Z.; Narayanan, N.; Yang, Y. Arginine methylation of the C-terminus RGG motif promotes TOP3B topoisomerase activity and stress granule localization. Nucleic Acids Res. 2018, 46, 3061–3074. [Google Scholar] [CrossRef]
- Yuan, W.; Al-Hadid, Q.; Wang, Z.; Shen, L.; Cho, H.; Wu, X.; Yang, Y. TDRD3 promotes DHX9 chromatin recruitment and R-loop resolution. Nucleic Acids Res. 2021, 49, 8573–8591. [Google Scholar] [CrossRef]
- Saha, S.; Yang, X.; Huang, S.N.; Agama, K.; Baechler, S.A.; Sun, Y.; Zhang, H.; Saha, L.K.; Su, S.; Jenkins, L.M.; et al. Resolution of R-loops by topoisomerase III-β (TOP3B) in coordination with the DEAD-box helicase DDX5. Cell Rep. 2022, 40, 111067. [Google Scholar] [CrossRef]
- Zhang, T.; Wallis, M.; Petrovic, V.; Challis, J.; Kalitsis, P.; Hudson, D.F. Loss of TOP3B leads to increased R-loop formation and genome instability. Open Biol. 2019, 9, 190222. [Google Scholar] [CrossRef]
- Saha, S.; Sun, Y.; Huang, S.N.; Baechler, S.A.; Pongor, L.S.; Agama, K.; Jo, U.; Zhang, H.; Tse-Dinh, Y.C.; Pommier, Y. DNA and RNA Cleavage Complexes and Repair Pathway for TOP3B RNA- and DNA-Protein Crosslinks. Cell Rep 2020, 33, 108569. [Google Scholar] [CrossRef]
- Prasanth, K.R.; Hirano, M.; Fagg, W.S.; McAnarney, E.T.; Shan, C.; Xie, X.; Hage, A.; Pietzsch, C.A.; Bukreyev, A.; Rajsbaum, R.; et al. Topoisomerase III-ss is required for efficient replication of positive-sense RNA viruses. Antivir. Res. 2020, 182, 104874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Su, S.; Altouma, V.; Zhu, X.; Xue, Y.; Shen, W.; Wilgenburg, B.; Wang, W. Topoisomerase 3b is dispensable for replication of a positive-sense RNA virus--murine coronavirus. Antivir. Res. 2022, 208, 105451. [Google Scholar] [CrossRef] [PubMed]
- Rani, P.; Kalladi, S.; Bansia, H.; Rao, S.; Jha, R.K.; Jain, P.; Bhaduri, T.; Nagaraja, V. A Type IA DNA/RNA topoisomerase with RNA hydrolysis activity participates in ribosomal RNA processing. J. Mol. Biol. 2020, 432, 5614–5631. [Google Scholar] [CrossRef] [PubMed]
- Mills, M.; Tse-Dinh, Y.C.; Neuman, K.C. Direct observation of topoisomerase IA gate dynamics. Nat. Struct. Mol. Biol. 2018, 25, 1111–1118. [Google Scholar] [CrossRef]
- Spakman, D.; Bakx, J.A.M.; Biebricher, A.S.; Peterman, E.J.G.; Wuite, G.J.L.; King, G.A. Unravelling the mechanisms of Type 1A topoisomerases using single-molecule approaches. Nucleic Acids Res. 2021, 49, 5470–5492. [Google Scholar] [CrossRef]


| Species | Protein | Gene(s) | Length (# AA) | UniProtKB Accession Number |
|---|---|---|---|---|
| Archaea | ||||
| Sulfolobus solfataricus | Reverse gyrase 1 | rgy1 | 1,242 | Q97ZZ8 |
| Sulfolobus solfataricus | Reverse gyrase 2 | rgy2 | 1,166 | Q97ZF5 |
| Sulfolobus solfataricus | Topo III | topA | 668 | Q97ZJ8 |
| Nanoarchaeum equitans | Reverse gyrase | NEQ434, NEQ318 | 575, 701 | Q74M79,Q74MA4 |
| Nanoarchaeum equitans | Topo III | NEQ045, NEQ324 | 408, 172 | Q74N66,Q74M99 |
| Methanosarcina mazei | Topo III | MSMAW_1096 | 832 | A0A0E3PWI6 |
| Methanosarcina mazei | Topo III | MSMAW_1564 | 752 | A0A0E3PXP2 |
| Thermoplasma volcanium | Topo III | topA | 770 | Q97CT2 |
| Nitrososphaera viennensis | Topo III | topA | 697 | A0A060HRZ8 |
| Nitrosopumilus maritimus * | ? | Nmar_1311, Nmar_1632 | 124, 152 | A9A2G8, A9A650 |
| Thermoplasmatales archaeon | Topo I | BRD56_01955 | 1067 | A0A2P6VYX4 |
| Thermoplasmatales archaeon | Topo I | BRD56_11190 | 732 | A0A2P6VTI6 |
| Bacteria | ||||
| Escherichia coli | Topo I | topA | 865 | P06612 |
| Escherichia coli | Topo III | topB | 653 | P14294 |
| Thermotoga maritima | Reverse gyrase | rgy | 1104 | O51934 |
| Thermotoga maritima | Topo I | topA | 633 | P46799 |
| Mycobacterium tuberculosis | Topo I | topA | 934 | P9WG49 |
| Deinococcus radiodurans | Topo I | topA | 1021 | Q9RUL0 |
| Agrobacterium tumefaciens | Topo I | topA | 892 | A9CJ93 |
| Caulobacter crescentus | Topo I | topA | 899 | Q9A5J6 |
| Chlamydia trachomatis | Topo I | topA | 857 | A0A0H3MCD9 |
| Helicobacter pylori | Topo I | topA_4 | 746 | A0A402E4A0 |
| Helicobacter pylori | Topo I | topA_3 | 686 | A0A402E2T6 |
| Helicobacter pylori | Topo I | topA_5 | 644 | A0A402E598 |
| Neisseria gonorrhoeae | Topo I | topA | 768 | A0A7H9WPJ8 |
| Neisseria gonorrhoeae | Topo III | topB_1 | 679 | Q5EP76 |
| Neisseria gonorrhoeae | Topo III | topB_2 | 753 | A0A7H9WG49 |
| Eukarya | ||||
| Saccharomyces cerevisiae | Topo III | TOP3 | 656 | P13099 |
| Ustilago maydis | Topo III | UMAG_11929 | 986 | A0A0D1C790 |
| Choanephora cucurbitarum | Topo IIIα | TOP3A | 749 | A0A1C7N0U0 |
| Choanephora cucurbitarum | Topo IIIβ | TOP3B | 548, 290 | A0A1C7NLX2, A0A1C7NMP8 |
| Trypanosoma brucei brucei | Topo IIIα | Tb11.01.1280 | 918 | Q383X7 |
| Trypanosoma brucei brucei | Topo IIIβ | Tb11.01.0910 | 841 | Q384B1 |
| Trypanosoma brucei brucei | Topo IAmt | Tb10.70.5940 | 806 | Q38C52 |
| Giardia intestinalis | Topo IIIα | DHA2_154636 | 896 | V6TB71 |
| Giardia intestinalis | Topo IIIβ | DHA2_15190 | 973 | V6TB61 |
| Arabidopsis thaliana | Topo IIIα | TOP3A | 926 | Q9LVP1 |
| Arabidopsis thaliana | Topo IIIβ | TOP3B | 865 | F4ISQ7 |
| Arabidopsis thaliana | Topo IAmt | AT4G31210 | 1284 | F4JRX3 |
| Homo sapiens | Topo IIIα | TOP3A | 1001 | Q13472 |
| Homo sapiens | Topo IIIβ | TOP3B | 862 | O95985 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, K.; Tse-Dinh, Y.-C. Variation of Structure and Cellular Functions of Type IA Topoisomerases across the Tree of Life. Cells 2024, 13, 553. https://doi.org/10.3390/cells13060553
Tan K, Tse-Dinh Y-C. Variation of Structure and Cellular Functions of Type IA Topoisomerases across the Tree of Life. Cells. 2024; 13(6):553. https://doi.org/10.3390/cells13060553
Chicago/Turabian StyleTan, Kemin, and Yuk-Ching Tse-Dinh. 2024. "Variation of Structure and Cellular Functions of Type IA Topoisomerases across the Tree of Life" Cells 13, no. 6: 553. https://doi.org/10.3390/cells13060553
APA StyleTan, K., & Tse-Dinh, Y.-C. (2024). Variation of Structure and Cellular Functions of Type IA Topoisomerases across the Tree of Life. Cells, 13(6), 553. https://doi.org/10.3390/cells13060553






