Abstract
Regulated necrosis, termed necroptosis, represents a potential therapeutic target for refractory cancer. Ceramide nanoliposomes (CNLs), considered potential chemotherapeutic agents, induce necroptosis by targeting the activating protein mixed lineage kinase domain-like protein (MLKL). In the present study, we examined the potential of pronecroptotic therapy using CNLs for refractory triple-negative breast cancer (TNBC), for which there is a lack of definite and effective therapeutic targets among the various immunohistological subtypes of breast cancer. MLKL mRNA expression in tumor tissues was significantly higher in TNBC patients than in those with non-TNBC subtypes. Similarly, among the 50 breast cancer cell lines examined, MLKL expression was higher in TNBC-classified cell lines. TNBC cell lines were more susceptible to the therapeutic effects of CNLs than the non-TNBC subtypes of breast cancer cell lines. In TNBC-classified MDA-MB-231 cells, the knockdown of MLKL suppressed cell death induced by CNLs or the active substance short-chain C6-ceramide. Accordingly, TNBC cells were prone to CNL-evoked necroptotic cell death. These results will contribute to the development of CNL-based pronecroptotic therapy for TNBC.
1. Introduction
Breast cancer is the most commonly diagnosed type of cancer in women and the second highest cause of cancer-related death. In GLOBOCAN 2020, the incidence and mortality of breast cancer were reported to be 2,261,419 new cases and 684,996 deaths, respectively [1]. Breast cancer is a genetically and clinically heterogeneous disease that is clinically classified into four subtypes by the expression of receptors, such as the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). Subclassification is fundamental for personalized pharmacotherapy [2,3]. Among the subtypes, triple-negative breast cancer (TNBC), which is defined by the lack of ER and PR expression and the absence of HER2 amplification, accounts for 10–20% of annually diagnosed breast cancer cases [4]. The clinical features of TNBC patients include high invasiveness, a high metastatic potential, proneness to relapse, and poor prognoses [4]. The primary therapeutic regimen for TNBC is limited to radiotherapy and conventional chemotherapy because of the lack of therapeutic molecular targets, such as ER, PR, and HER2 [2,4]. Therefore, novel therapeutic targets need to be developed.
Necroptosis is a form of regulated cell death with the hallmarks of necrosis and is a therapeutic target for cancer treatment [5,6,7]. The induction of necroptosis has attracted attention as an alternative approach for cancer treatment to overcome chemoresistance and promote anti-tumor immunity [8,9,10]. The signaling pathway of necroptosis includes regulatory proteins, such as receptor-interacting protein kinase (RIPK) 1 and RIPK3, and activating pore-forming mixed lineage kinase domain protein (MLKL). RIPK3 phosphorylates MLKL, and phosphorylated MLKL, in turn, forms an oligomer that translocates to the plasma membrane to form pores, which eventually increase plasma membrane permeability [7,11,12]. Epigenetically, RIPK3 expression is often silenced in cancer cells [13]. MLKL expression is transcriptionally up-regulated by interferon-γ-regulatory factor 1 (IRF1) and the signal transducer and activator of transcription 1 (STAT1) in MDA-MB-231 breast cancer and HeLa cervical cancer cell lines [14]. The up-regulation of MLKL is presumed to sensitize cancer cells to necroptotic cell death, and pronecroptotic therapy is potentially effective for cancer cells that highly express MLKL.
Ceramide is a central lipid in sphingolipid metabolism and a potent tumor suppressor that augments apoptotic and non-apoptotic cell death [15,16,17,18]. The potential of ceramide-based therapeutics for the treatment of cancer has been attracting increasing interest [19,20,21,22]. We developed a non-toxic and biologically stable nanoliposome formulation of short-chain C6-ceramide termed ceramide nanoliposomes (CNLs) as potential chemotherapeutic agents [22]. CNLs exerted therapeutic effects in preclinical cancer models [23,24,25] and are currently being investigated clinically [26].
CNLs activate MLKL-governed necroptosis independent of RIPK3 in ovarian cancer cells [25]. Importantly, the anti-tumor activities of CNLs correlated with MLKL expression in ovarian cancer cell lines, implying the involvement of MLKL expression in the therapeutic effectiveness of CNLs. In the present study, we investigated the potential of pronecroptotic therapy for breast cancer using CNLs.
2. Materials and Methods
2.1. Materials
Horseradish peroxidase-conjugated antibodies against mouse or rabbit IgG and human serum were purchased from Jackson ImmunoResearch (West Grove, PA, USA). Mouse monoclonal antibodies specific for β-actin (clone 15G5A11/E2, MA1-140), SuperSignal West Dura Extended Duration Substrate, Triton X-100, BCA protein assay reagents, OPTI MEM, goat anti-rabbit IgG conjugated with Alexa555, RNAiMAX, control siRNAs (4390843 or 4390846), and human MLKL siRNAs (MLKL siRNA-1, s47088; MLKL siRNA-2, s47089) were obtained from Thermo Fisher Scientific (Rockford, IL, USA). Roswell Park Memorial Institute 1640 medium, high glucose Dulbecco’s Modified Eagle’s Medium, phosphate-buffered saline (PBS), EDTA, glycine, NaCl, Tris, sodium dodecyl sulfate (SDS), and trypsin were purchased from Nacalai Tesque (Kyoto, Japan). Fetal bovine serum was obtained from Biowest (Nuaillé, France). A rabbit anti-MLKL antibody (ab184718) was purchased from Abcam (Cambridge, MA, USA). Gradient polyacrylamide gels (4–20%) and nitrocellulose membranes were obtained from Bio-Rad (Hercules, CA, USA). 4’,6-Diamidino-2-phenylindole was obtained from Dojindo (Kumamoto, Japan). C6-Ceramide (N-hexanoyl-D-erythro-sphingosine) was purchased from Avanti Polar Lipids (Alabaster, AL, USA). Glass bottom culture dishes (35 mm) were from MATTEK (Ashland, MA, USA). MDA-MB-231, MDA-MB-453, MDA-MB-468, MCF-7, ZR-751, T47D, HCC1937, and BT-20 cells were obtained from ATCC (Manassas, VA, USA). CellTiter-Glo2.0 was purchased from Promega (Madison, WI, USA).
2.2. Mixed Lineage Kinase Domain-like Protein Expression Profiles of Breast Cancer Tissues
To examine MLKL expression in the tumor tissues of patients with breast cancer, we searched The Cancer Genome Atlas (TCGA) database obtained from cBioportal (https://www.cbioportal.org/) and characterized immunohistological subtypes.
2.3. Gene Expression Profiles of Breast Cancer Cell Lines
Breast cancer cell lines were subclassified into two groups: TNBC and non-TNBC. Twenty-four TNBC cell lines (BT20, BT-549, CAL120, CAL148, CAL51, CAL851, DU4475, HCC1143, HCC1187, HCC1395, HCC1599, HCC1806, HCC1937, HCC2157, HCC38, HCC70, HDQ-P1, HMC18, HS578T, MDA-MB-157, MDA-MB-231, MDA-MB-436, MDA-MB-453, and MDA-MB-468) were clinicopathologically characterized and subclassified [27,28,29]. The remaining non-TNBC cell lines (AU565, BT-474, BT-483, CAMA1, EFM19, EFM192A, HCC1419, HCC1428, HCC1500, HCC1569, HCC1954, HCC202, HCC2218, KPL1, MCF-7, MDA-MB-134VI, MDA-MB-175VII, MDA-MB-361, MDA-MB-415, SKBR3, T47D, JIMT1, UACC812, UACC893, ZR751, and ZR7530) were subclassified in accordance with the DepMap (https://depmap.org/portal/)-based gene expression profiles of ER, PR, and HER2.
2.4. Cell Culture
MDA-MB-231, MDA-MB-453, and MDA-MB-468 cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum. MCF-7, ZR-751, T47D, HCC1937, and BT-20 cells were cultured in Roswell Park Memorial Institute 1640 medium supplemented with 10% fetal bovine serum. Cells were maintained at <80% confluence under standard incubator conditions (37 °C in a humidified atmosphere with 5% CO2).
2.5. Ceramide Nanoliposomes Preparation
CNLs were prepared as described in Sakae et al. [30]. Briefly, dioleoylphosphatidylethanolamine, distearoylphosphatidylcholine, distearoylphosphatidylethanolamine-polyethylene glycol 2000, polyethylene glycol 750-N-octanoylsphingosine, and C6-ceramide were mixed in chloroform, dried to a thin film under nitrogen, and then hydrated by the addition of sterilized 0.9% NaCl solution at 60 °C with sonicating and vortexing. Lipid solutions were then extruded at 60 °C by passing through 100 nm polycarbonate filters. Size (80 nM) and charge (−7 mV) were validated using Malvern Zetasizer Nano (Malvern Panalytical, Malvern, UK).
2.6. Cell Viability Assay
Cells (4 × 103 cells/well) seeded on a 96-well culture plate were treated with ceramide-free ghost nanoliposomes or CNLs for 24 h. Cell viability was assessed using the CellTiter-Glo luminescent cell viability assay in accordance with the manufacturer’s protocol. Luminescence was measured with a GloMax Navigator Microplate Luminometer (Promega, Madison, WI, USA).
2.7. siRNA Transfection
Cells were seeded in 6-well plates (4 × 105 cells/well) and transfected with 5 nM siRNAs for control or MLKL using RNAiMAX transfection reagent in accordance with the manufacturer’s instructions. For each well, 5 pmol of siRNAs, 150 μL of OPTI MEM, and 1 μL of RNAiMAX were used. After transfection for 24 h, cells were treated with CNL or ghost nanoliposomes.
2.8. Trypan Blue Exclusion Assay
Harvested cells were resuspended in 100 µL of PBS. A 1:1 dilution of a cell suspension (15 μL) and 0.4% trypan blue solution (15 μL) was prepared and loaded into a hemocytometer. The numbers of trypan blue-excluding live cells and dead cells were counted.
2.9. Immunofluorescence Microscopy
MDA-MB-231 cells (1 or 2 × 105 cells) were seeded on 35 mm glass-bottomed dishes. Cells were fixed with 4% formaldehyde at room temperature for 10 min. Fixed cells were further permeabilized with 0.1% Triton X-100 for 10 min. After washing with PBS, cells were blocked at room temperature for 1 h with 20% human serum. Cells were incubated with an MLKL antibody (1:500 dilution) in PBS containing 20% human serum overnight at 4 °C. After washing with PBS, cells were incubated with an Alexa555-conjugated anti-rabbit IgG antibody (1:500 dilution) at 4 °C for 1 h. Nuclear staining was performed with 4’,6-diamidino-2-phenylindole (1:1000 dilution). All images were captured using a confocal microscope (Olympus FV1000D system, Olympus, Tokyo, Japan).
2.10. Western Blotting
Cells were harvested with ice-cold PBS containing 10 mM EDTA. The protein contents of samples were measured by a BCA protein assay. Proteins were denatured at 98 °C for 10 min. Proteins (5 μg) were subjected to SDS-polyacrylamide gel electrophoresis (4–20% gradient polyacrylamide gels) and then electrophoretically transferred onto nitrocellulose membranes. Membranes were blocked with PBS/0.1% Tween 20 containing 5% nonfat dried milk at room temperature for 30 min. After washing with PBS/0.1% Tween 20, membranes were incubated with antibodies against MLKL (1:1000) or β-actin (1:10,000) in PBS/0.1% Tween 20 containing 5% bovine serum albumin at 4 °C overnight. Membranes were washed with PBS/0.1% Tween 20 and incubated with secondary antibodies conjugated with horseradish peroxidase in PBS/0.1% Tween 20 containing 5% nonfat dried milk at 4 °C for 1 h. After washing, proteins were visualized using enhanced chemiluminescence reagents. The quantification of chemiluminescent signals was performed with a ChemiDoc digital imaging system (Bio-Rad, Hercules, CA, USA).
2.11. Statistical Analysis
Data were analyzed using multiple t-tests and a robust or simple linear regression by GraphPad Prism software version 8.4.3. p values < 0.05 were considered to be significant.
3. Results
3.1. High Mixed Lineage Kinase Domain-like Protein Expression in Triple-Negative Breast Cancer Patients
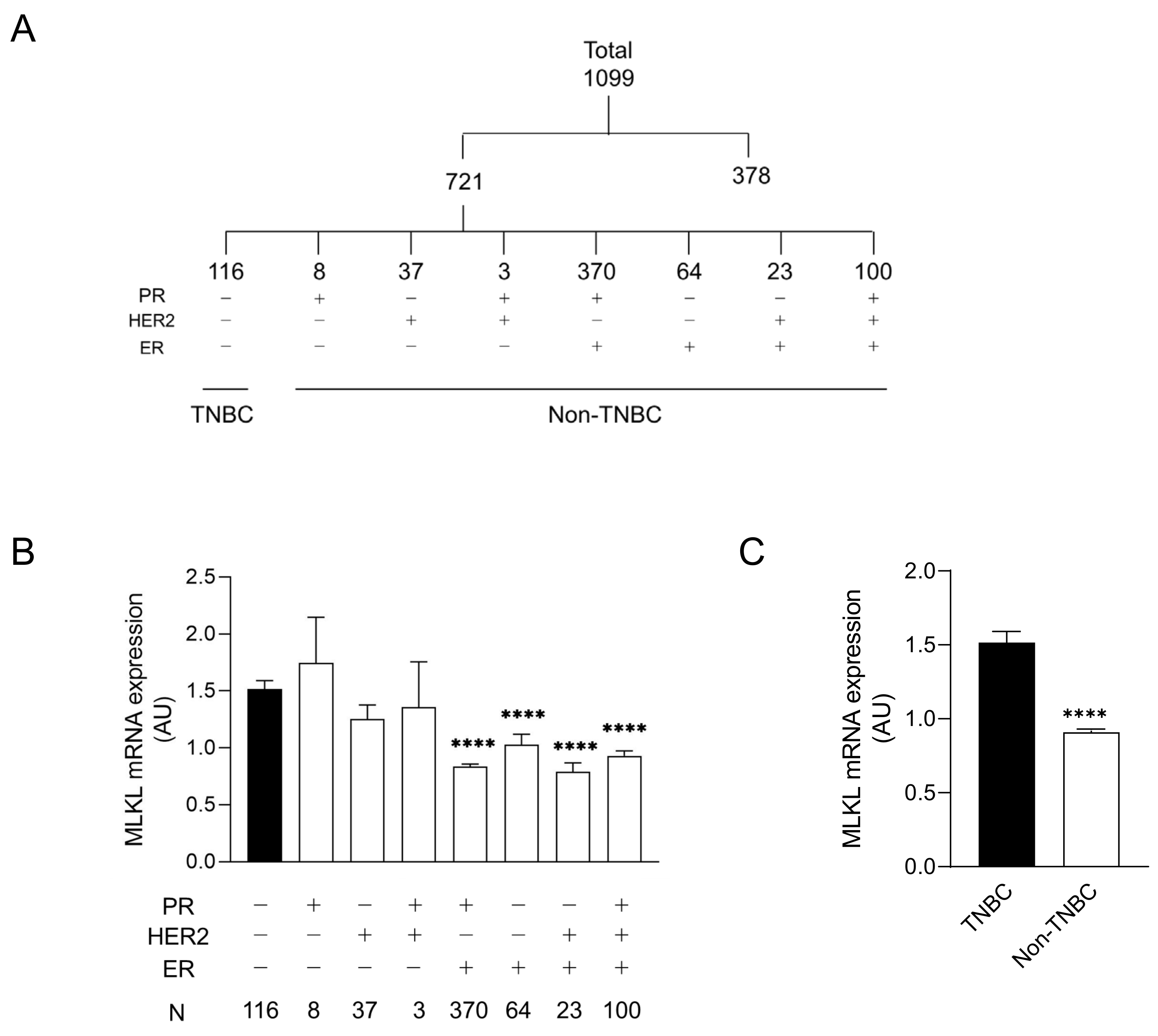
CNLs are regarded as pronecroptotic chemotherapeutic reagents, and MLKL is predictive of the therapeutic effectiveness of CNLs. To predict the susceptibility of the immunohistological subtypes of breast cancer to CNL-induced necroptosis, MLKL expression in tumor tissues was analyzed using the TCGA database. Among 1099 cases, 721 were defined as “positive” and/or “negative” for ER, PR, and HER2 based on immunohistological profiles in the TCGA database. The profiles of the remaining 378 cases were not immunohistologically subclassified because of (1) full or partial defects in the profile in the TCGA database or (2) they were defined as “equivocal” and/or “indeterminate”. The 721 cases were further divided into eight subtypes by distinct receptor expression (Figure 1A). Figure 1B shows MLKL mRNA expression in distinct subtypes. A statistical analysis of seven subtypes relative to TNBC was performed. MLKL mRNA expression was significantly higher in the TNBC group than in the PR (+)/HER2 (−)/ER (+), PR (−)/HER2 (−)/ER (+), PR (−)/HER2 (+)/ER (+), and PR (+)/HER2 (+)/ER (+) groups. MLKL mRNA expression was also significantly higher in the TNBC group than in the non-TNBC groups (Figure 1C).
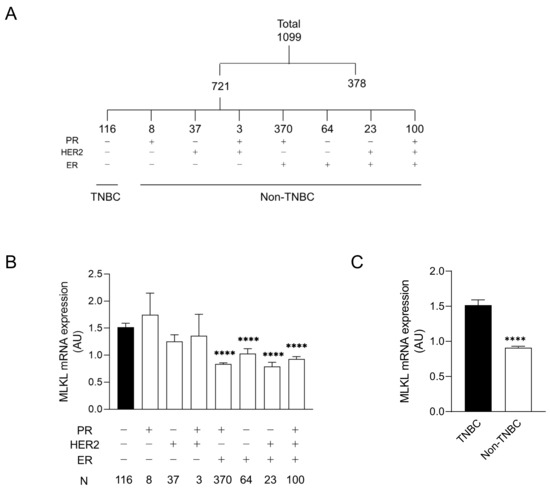
Figure 1.
mRNA expression of mixed lineage kinase domain-like protein (MLKL) in tumor tissues of patients with breast cancer. (A) Among 1099 patients registered in The Cancer Genome Atlas (TCGA), 721 were divided into eight molecular subtypes by the expression of receptors, including estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). (B) Eight subgroups were further divided into two groups: triple-negative breast cancer (TNBC) and the remaining seven molecular subtypes (non-TNBC). Data represent means ± standard error. ****, p < 0.0001, compared to TNBC. (C) MLKL mRNA expression in tumor tissues was compared between TNBC and non-TNBC. Data represent means ± standard error. ****, p < 0.0001.
To obtain an understanding of the relevance of MLKL mRNA expression to the prognosis of patients with breast cancer, we performed an online analysis with Kaplan–Meier Plotter (http://www.kmplot.com) [31]. Patients were divided into two groups: high and low MLKL expression. Progression-free survival periods were plotted to generate Kaplan–Meier curves. As shown in Supplementary Figure S1, progression-free survival was longer in the MLKL-high patient group than in the MLKL-low group. MLKL expression in tumor tissues can possibly affect therapeutic effectiveness.
3.2. High Mixed Lineage Kinase Domain-like Protein Expression in Triple-Negative Breast Cancer Cell Lines
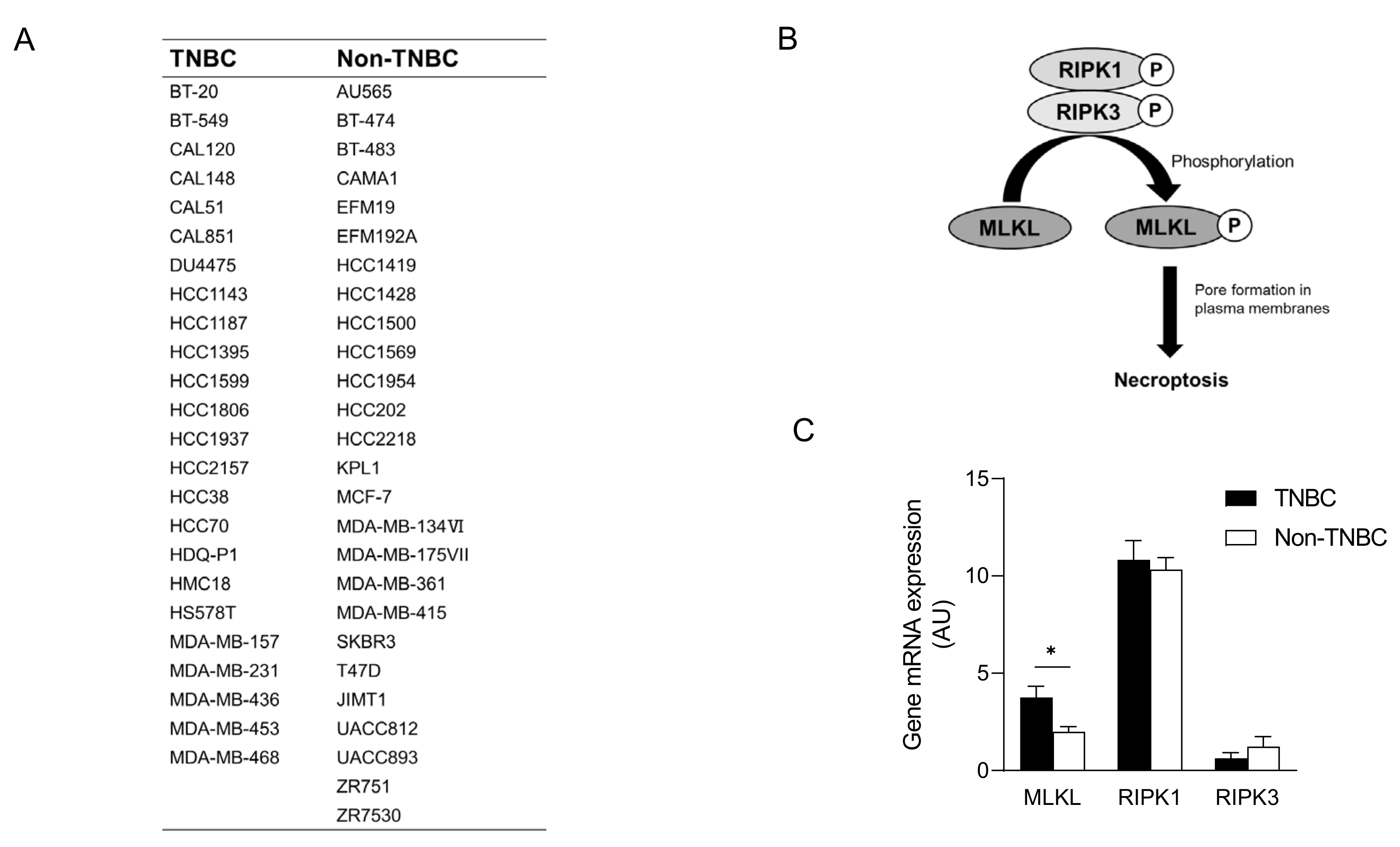
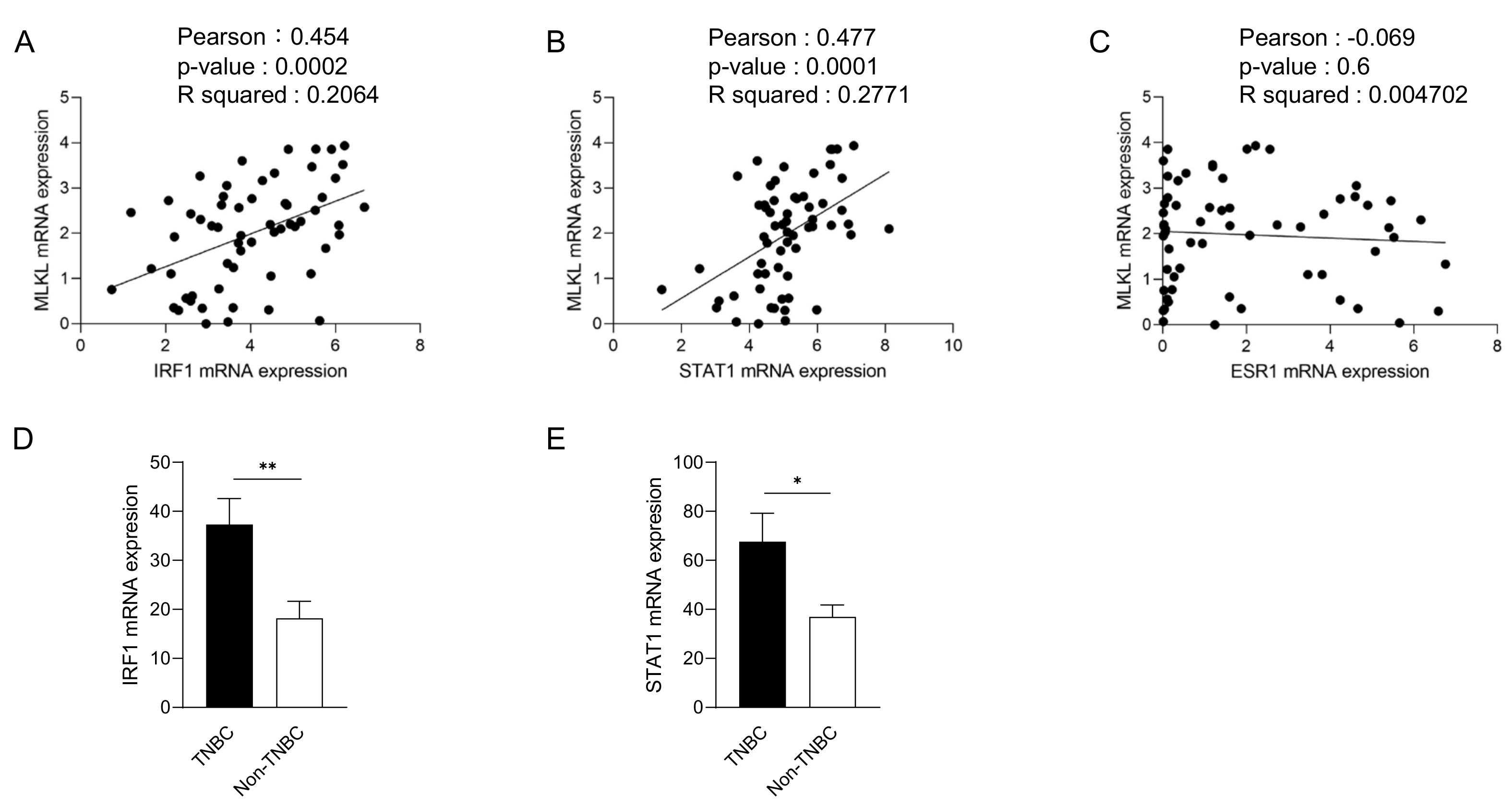
To recapitulate the high expression of MLKL in TNBC, we investigated MLKL mRNA expression in breast cancer cell lines. These cell lines were subclassified into TNBC (n = 24) [27,28,29] and non-TNBC (n = 26), and the gene expression profiles of the cell lines were obtained from DepMap (Figure 2A). In addition to MLKL, the necroptosis-associated genes RIPK1 and RIPK3 (Figure 2B) were also examined. MLKL gene expression was significantly higher in TNBC than in the non-TNBC; however, no significant differences were observed in RIPK1 and RIPK3 expression (Figure 2C). To investigate the possible mechanisms by which MLKL is up-regulated in TNBC cells, further database searches were performed. The STAT1-IRF1 pathway transcriptionally promotes MLKL expression in multiple types of cancer cell lines in response to interferon-γ [14]. MLKL mRNA expression correlated with IRF1 and STAT1 mRNA expression, but not with ESR1 expression, in breast cancer cell lines (Figure 3A–C). Moreover, the mRNA expression of IRF1 and STAT1 was significantly higher in TNBC than in the non-TNBC (Figure 3D,E). Therefore, the STAT1-IRF1 pathway was assumed to contribute to the high expression of MLKL in TNBC cells.
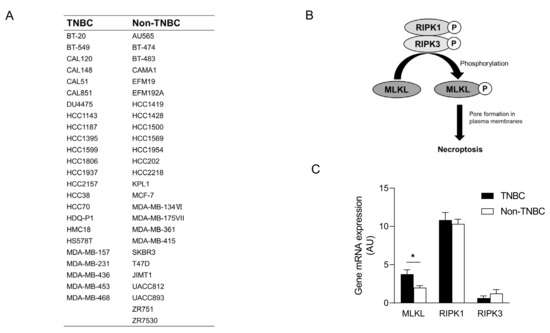
Figure 2.
mRNA expression of mixed lineage kinase domain-like protein (MLKL) in breast cancer cell lines. (A) List of breast cancer cell lines and their subclassification into triple-negative breast cancer (TNBC) (n = 24) and non-TNBC (n = 26). (B) Molecular mechanisms underlying necroptosis. (C) MLKL mRNA expression was compared between TNBC and non-TNBC. Data represent means ± standard error. *, p < 0.05 by the Student’s t-test.
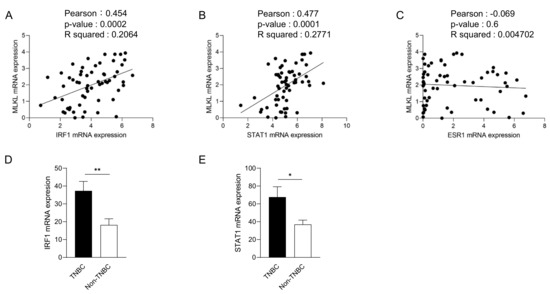
Figure 3.
Correlations between mixed lineage kinase domain-like protein (MLKL) mRNA expression and estrogen receptor 1 (ESR1), interferon regulatory factor 1 (IRF1) or signal transducer and activator of transcription 1 (STAT1) mRNA expression in breast cancer cell lines. The mRNA expression of ESR1, IRF1, or STAT1 is plotted with MLKL mRNA expression. R2 values were obtained using GraphPad Prism (A–C). (D,E) IRF1 (D) and STAT1 (E) mRNA expression was compared between TNBC and non-TNBC. Data represent means ± standard error. *, p < 0.05; **, p < 0.01.
3.3. Therapeutic Effects of Ceramide Nanoliposomes in Breast Cancer Cell Lines
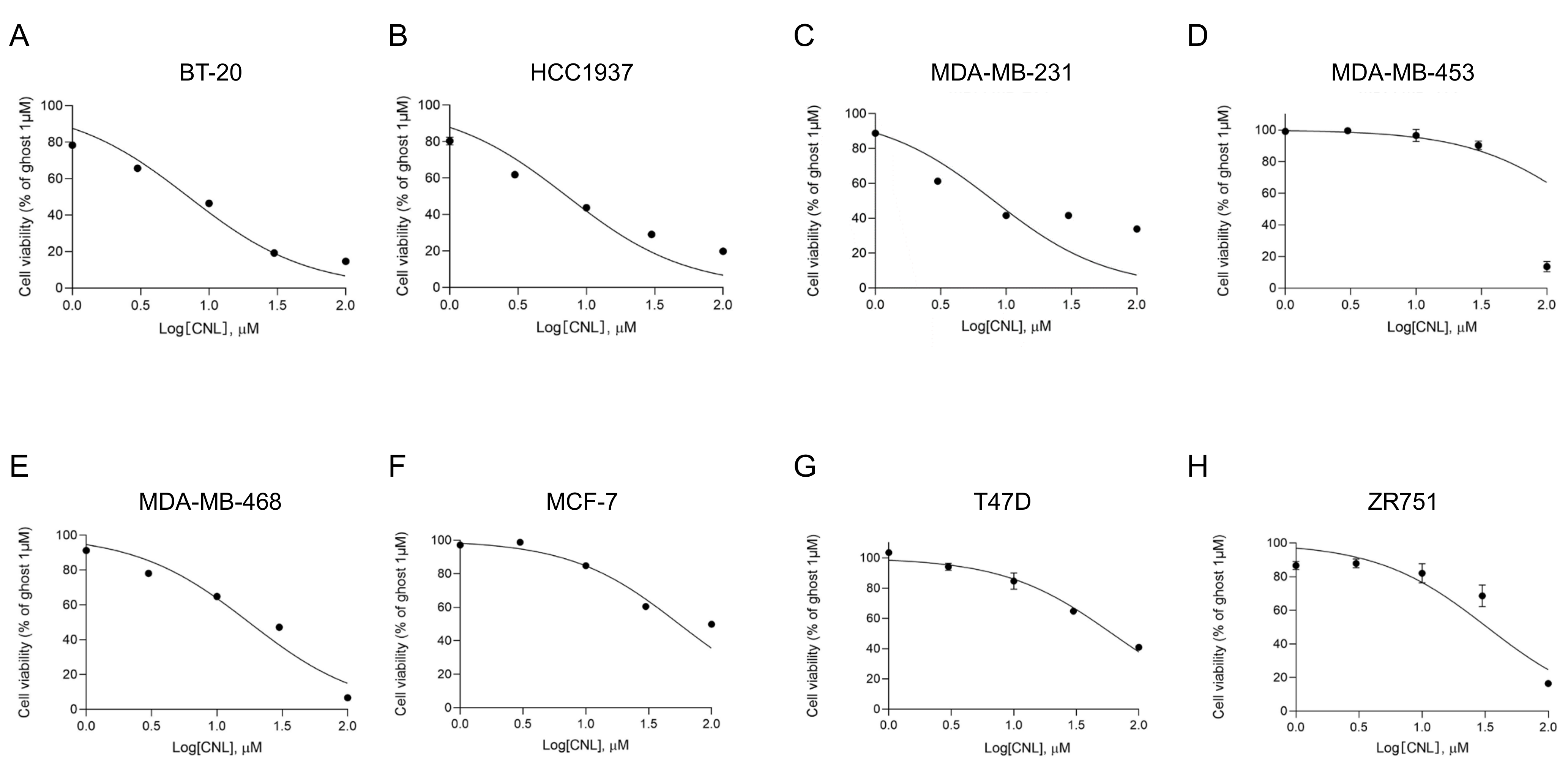
In ovarian cancers, MLKL increases the susceptibility of cells to necroptosis induced by CNLs [25]. To assess the susceptibility of MLKL-high TNBC cell lines to CNLs, we examined the effects of CNLs on the viability of breast cancer cell lines (Figure 4) and measured IC50 values (Table 1). Among the TNBC cell lines examined, the IC50 values of BT-20, HCC1937, MDA-MB-231, and MDA-MB-468 cells were < 20 μM, whereas that of MDA-MB-453 cells was > 200 μM. Notably, TNBC cell lines, except for MDA-MB-453 cells, showed lower IC50 values than the non-TNBC subtypes, including MCF-7, T47D, and ZR751 cells. The susceptibility of TNBC cell lines to CNL-induced cell death appeared to be higher than that of the non-TNBC cell lines.
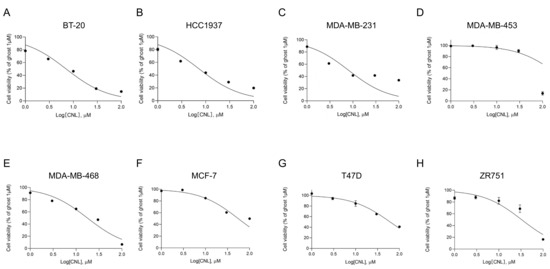
Figure 4.
Effects of ceramide nanoliposomes (CNLs) on the cell viability of breast cancer cell lines. BT-20 (A), HCC1937 (B), MDA-MB-231 (C), MDA-MB-453 (D), MDA-MB-468 (E), MCF-7 (F), T47D (G), or ZR751 (H) cells were seeded on 96-well plates (4000 cells/well). Cells were treated with CNLs up to 100 µM or 1 µM ceramide-free ghost nanoliposomes for 24 h. Cell viability was assessed using a CellTiter-Glo luminescent cell viability assay. Cell viability is expressed as the percentage of 1 μM ceramide-free ghost nanoliposomes. Data represent means ± standard error (n = 3).

Table 1.
IC50 values of ceramide nanoliposomes (CNLs) in breast cancer cell lines. Individual IC50 values of eight breast cancer cell lines for CNLs were determined by GraphPad prism.
3.4. Pronecroptotic Effects of Ceramide Nanoliposomes in Triple-Negative Breast Cancer Cell Lines
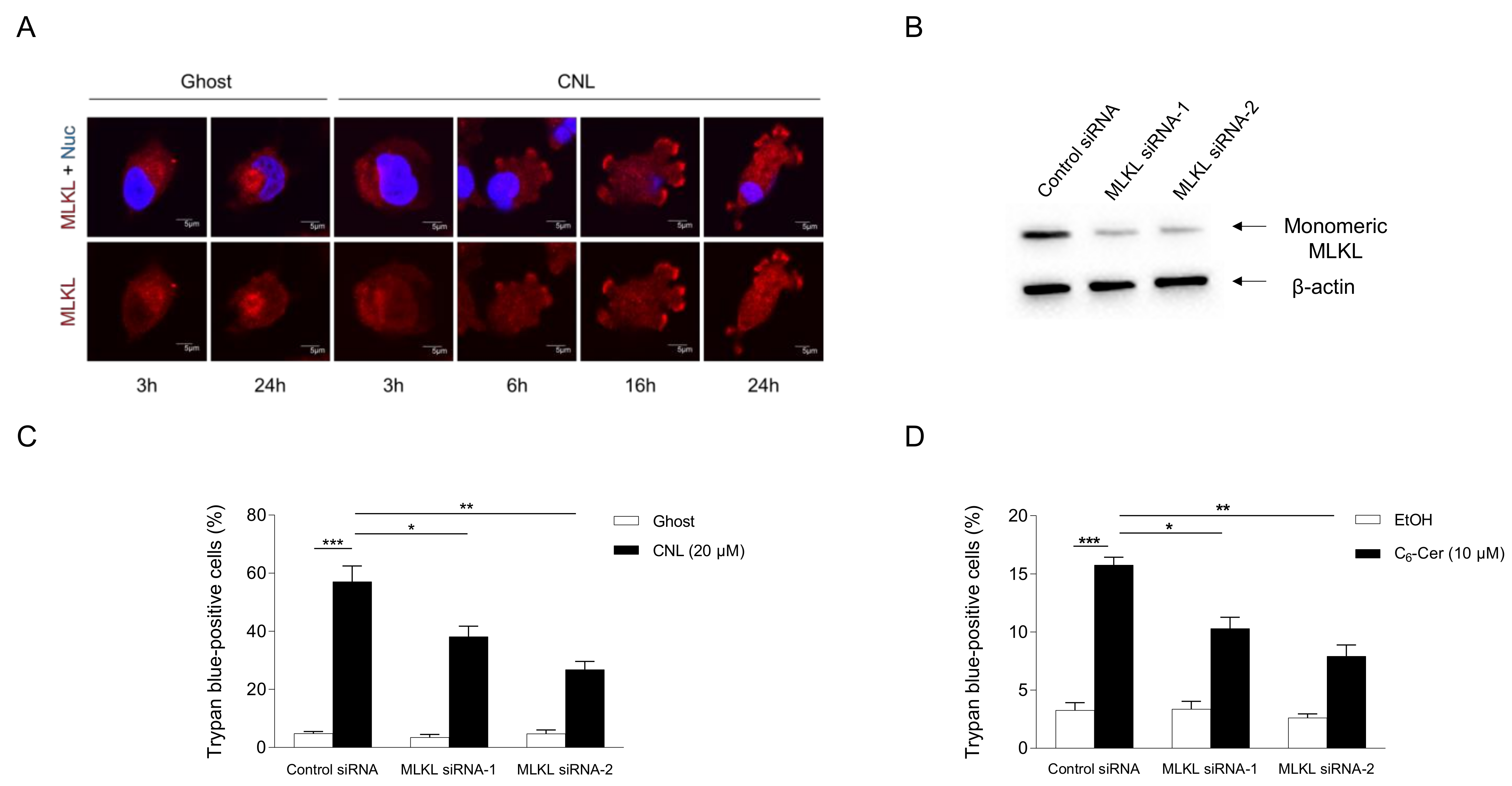
The involvement of MLKL in CNL-induced cell death was assessed in TNBC cell lines. To evaluate the activation of MLKL, its subcellular localization was examined by immunofluorescence microscopy (Figure 5A). MLKL localized to the cytosol under steady-state conditions. In response to CNLs, MLKL relocalized to the plasma membrane in a time-dependent manner. Two siRNA sequences against MLKL were verified to efficiently knockdown MLKL proteins in MDA-MB-231 cells (Figure 5B). In control siRNA-treated cells, CNLs were confirmed to induce cell death. Importantly, the knockdown of MLKL significantly suppressed CNL-induced cell death (Figure 5C), suggesting its involvement in CNL-induced cell death. The necroptotic effects of CNLs were also observed in TNBC-subclassified MDA-MB-468 cells (Supplementary Figure S2). We then investigated the involvement of C6-ceramide in necroptotic cell death because CNLs are a nanoliposomal formulation of C6-ceramide. Similar to CNL, 10 μM C6-ceramide killed MDA-MB-231 cells, and this was significantly suppressed by the knockdown of MLKL proteins (Figure 5D). These results indicate that C6-ceramide in CNLs triggered necroptosis in TNBC cells.
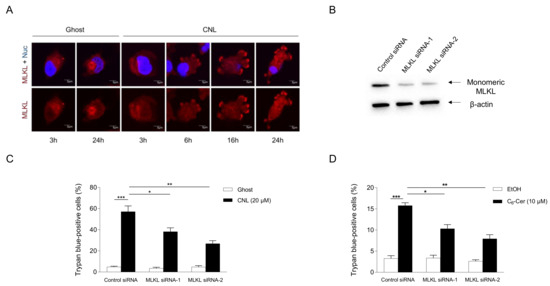
Figure 5.
Ceramide involvement in necroptosis. MDA-MB-231 cells (1 × 105 cells/35 mm glass-bottomed dish) were treated with 20 μM ceramide nanoliposomes (CNLs) or ceramide-free ghost nanoliposomes (Ghost) for up to 24 h. (A) Fixed cells were stained with a mixed lineage kinase domain-like protein (MLKL) antibody (red) and 4’,6-diamidino-2-phenylindole (blue). Imaging was performed by confocal microscopy, and representative images are shown. White arrows show the accumulation of MLKL on the plasma membrane. (B–D) MDA-MB-231 cells (2 × 105 cells/well) were transfected with 5 nM control or two distinct MLKL siRNAs (MLKL siRNA-1 or MLKL siRNA-2). After transfection for 24 h, cells were treated with 20 µM Ghost or CNLs for 24 h. Cells were harvested and cellular proteins were extracted. Proteins were subjected to Western blotting using antibodies specific for MLKL or β-actin (B). The cell death rate was assessed by the trypan blue assay, and data represent the mean ± standard error of three values (C). (D) MDA-MB-231 cells were transfected with 5 nM control or MLKL siRNAs for 24 h. After transfection, cells were treated with vehicle ethanol or 10 μM C6-ceramide for 24 h. Cell death was assessed by trypan blue exclusion assay and data represent the mean ± standard error of three or six values. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
4. Discussion
The induction and/or manipulation of necroptosis is a promising therapeutic approach. We demonstrated for the first time that the tumor tissues and cell lines of TNBC were characterized by the high expression of MLKL and the use of CNLs as necroptosis-inducing reagents was therapeutically effective in TNBC cells highly expressing MLKL.
Low expression of MLKL is associated with poor prognosis in cervical squamous cell cancer [32], ovarian cancer [33], gastric cancer [34], and pancreatic adenocarcinoma [35]. Those reports are consistent with our findings on breast cancer (Supplementary Figure S1). MLKL might serve as a candidate tumor suppressor and a potential prognostic biomarker in certain types of cancers. Although the clinical significance of the up-regulated expression of MLKL mRNAs in TNBC currently remains unclear, the understanding could play important roles in predicting the effectiveness of MLKL-activating pronecroptotic therapy for TNBC.
CNLs were therapeutically ineffective against MDA-MB-453 cells, subclassified as a TNBC cell line. MLKL mRNA expression in MDA-MB-453 cells was the lowest among the TNBC cell lines examined in the present study (Supplementary Table S1), which may have resulted in poor susceptibility to CNLs. Moreover, unknown mechanisms also need to be considered for a more detailed understanding of poor susceptibility.
Since Obeid et al. found that C6-ceramide is an apoptotic lipid [15], extensive attention has been focused on the development of ceramide-based therapeutics for cancer [18,21,22]. Several types of liposomal formulations of C6-ceramide have been developed: a stealth type, such as CNLs [22], and a receptor-targeted type, such as transferrin-conjugated ceramide liposomes [20]. The chemotherapeutic effects of CNLs are well-conserved across cancer types. The mechanisms by which CNLs induce cell death in cancer cells involve apoptosis [24], necrosis [36], and necroptosis [25]. In the present study, we showed that CNLs and C6-ceramide were pronecroptotic in TNBC cell lines; however, the underlying molecular mechanisms remain largely unknown. The metabolic fate of short-chain C6-ceramide in cells [37] may need to be considered. C6-ceramide is subjected to deacylation by ceramidase to form sphingosine. Free sphingosine, in turn, undergoes ceramide synthase (CERS)-dependent acylation [38,39]. Six isoforms of CERS encoded in human genes exhibit distinct substrate preferences for certain chain-length fatty acyl CoAs, thereby generating well-defined ceramide molecular species with specific acyl-chain lengths [39]. Individual ceramide molecular species may be associated with the execution of a distinct type of cell death. Some ceramide molecular species, including C16-ceramide, have been implicated in the execution of apoptosis [40,41]; however, the types of ceramide molecular species involved in necroptotic cell death remain unknown. Further studies that clarify the metabolic fate of C6-ceramide in CNLs will be instrumental for revealing the biological roles of ceramide molecular species in necroptosis and developing molecular bases for CNL precision medicine.
Our preclinical analyses of CNLs were based on a cell model and we did not validate the therapeutic effectiveness of CNLs in a TNBC animal model. A previous study demonstrated that treatment with liposomal C6-ceramide reduced the size of tumors in mouse xenograft models of the TNBC cell line MDA-MB-231 [42]. The in vivo effectiveness of CNLs has also been validated in various types of cancers, including gastric cancer [24], hepatocellular carcinoma [43], ovarian cancer [25], and leukemia [44]. In future studies, we will examine the pronecroptotic effects of CNLs in a TNBC xenograft model.
In conclusion, TNBC is characterized by the high expression of MLKL and pronecroptotic therapy utilizing CNLs is a potential therapeutic option for TNBC. The present results provide insights into the development of novel precision medicine for TNBC.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/cells13050405/s1, Figure S1: Correlation of MLKL mRNA expression with the progression-free survival probability in breast cancer patients; Figure S2: Ceramide nanoliposomes (CNLs)-evoked cell death; Table S1: Expression of Mixed Lineage Kinase Domain-like Protein (MLKL) mRNA in Breast Cancer Cell Lines.
Author Contributions
Conceptualization, Y.O. (Yuki Ohya), Y.O. (Yuri Ogiso) and K.K.; methodology, Y.O. (Yuki Ohya), Y.O. (Yuri Ogiso) and K.K.; validation, Y.O. (Yuki Ohya), Y.O. (Yuri Ogiso) and K.K.; formal analysis, Y.O. (Yuki Ohya) and Y.O. (Yuri Ogiso); investigation, Y.O. (Yuki Ohya), Y.O. (Yuri Ogiso), M.M., H.S. and K.K.; resources, T.E.F., M.K., W.S. and K.K.; data curation, Y.O. (Yuki Ohya) and Y.O. (Yuri Ogiso); writing—original draft preparation, Y.O. (Yuki Ohya), Y.O. (Yuri Ogiso), Y.O. (Yuri Ogiso) and K.K.; writing—review and editing, Y.O. (Yuki Ohya), Y.O. (Yuri Ogiso), M.M., H.S., K.N., Y.M., T.E.F., M.K., W.S., T.N. and K.K.; visualization, Y.O. (Yuki Ohya), Y.O. (Yuri Ogiso), K.N. and K.K.; supervision, K.K.; project administration, Y.O. (Yuki Ohya), Y.O. (Yuri Ogiso) and K.K.; funding acquisition, K.K. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported in part by a JSPS KAKENHI Grant (22K06623 to K.K.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Acknowledgments
We thank Jun-pei Imazu for technical assistance and the members of the Immunopharmacology Laboratory of the Faculty of Pharmaceutical Sciences at Setsunan University for critical discussion.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Nedeljkovic, M.; Damjanovic, A. Mechanisms of Chemotherapy Resistance in Triple-Negative Breast Cancer-How We Can Rise to the Challenge. Cells 2019, 8, 957. [Google Scholar] [CrossRef] [PubMed]
- Manjunath, M.; Choudhary, B. Triple-negative breast cancer: A run-through of features, classification and current therapies. Oncol. Lett. 2021, 22, 512. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.Y.; Park, S.; Kwon, Y. Recent therapeutic trends and promising targets in triple negative breast cancer. Pharmacol. Ther. 2019, 199, 30–57. [Google Scholar] [CrossRef] [PubMed]
- Linkermann, A.; Green, D.R. Necroptosis. N. Engl. J. Med. 2014, 370, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, X.; Gueydan, C.; Han, J. Plasma membrane changes during programmed cell deaths. Cell Res. 2018, 28, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Berghe, T.V.; Vandenabeele, P.; Kroemer, G. The molecular machinery of regulated cell death. Cell Res. 2019, 29, 347–364. [Google Scholar] [CrossRef]
- Su, Z.; Yang, Z.; Xie, L.; DeWitt, J.P.; Chen, Y. Cancer therapy in the necroptosis era. Cell Death Differ. 2016, 23, 748–756. [Google Scholar] [CrossRef]
- Van Hoecke, L.; Van Lint, S.; Roose, K.; Van Parys, A.; Vandenabeele, P.; Grooten, J.; Tavernier, J.; De Koker, S.; Saelens, X. Treatment with mRNA coding for the necroptosis mediator MLKL induces antitumor immunity directed against neo-epitopes. Nat. Commun. 2018, 9, 3417. [Google Scholar] [CrossRef]
- Gong, Y.; Fan, Z.; Luo, G.; Yang, C.; Huang, Q.; Fan, K.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. The role of necroptosis in cancer biology and therapy. Mol. Cancer 2019, 18, 100. [Google Scholar] [CrossRef]
- Xia, B.; Fang, S.; Chen, X.; Hu, H.; Chen, P.; Wang, H.; Gao, Z. MLKL forms cation channels. Cell Res. 2016, 26, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Flores-Romero, H.; Ros, U.; Garcia-Saez, A.J. Pore formation in regulated cell death. EMBO J. 2020, 39, e105753. [Google Scholar] [CrossRef] [PubMed]
- Koo, G.B.; Morgan, M.J.; Lee, D.G.; Kim, W.J.; Yoon, J.H.; Koo, J.S.; Kim, S.I.; Kim, S.J.; Son, M.K.; Hong, S.S.; et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res. 2015, 25, 707–725. [Google Scholar] [CrossRef] [PubMed]
- Knuth, A.K.; Rosler, S.; Schenk, B.; Kowald, L.; van Wijk, S.J.L.; Fulda, S. Interferons Transcriptionally Up-Regulate MLKL Expression in Cancer Cells. Neoplasia 2019, 21, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Obeid, L.M.; Linardic, C.M.; Karolak, L.A.; Hannun, Y.A. Programmed cell death induced by ceramide. Science 1993, 259, 1769–1771. [Google Scholar] [CrossRef] [PubMed]
- Kolesnick, R.N.; Kronke, M. Regulation of ceramide production and apoptosis. Annu. Rev. Physiol. 1998, 60, 643–665. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Okazaki, T. The role of sphingomyelin and sphingomyelin synthases in cell death, proliferation and migration-from cell and animal models to human disorders. Biochim. Biophys. Acta 2014, 1841, 692–703. [Google Scholar] [CrossRef] [PubMed]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2018, 18, 33–50. [Google Scholar] [CrossRef]
- van Vlerken, L.E.; Duan, Z.; Seiden, M.V.; Amiji, M.M. Modulation of intracellular ceramide using polymeric nanoparticles to overcome multidrug resistance in cancer. Cancer Res. 2007, 67, 4843–4850. [Google Scholar] [CrossRef]
- Koshkaryev, A.; Piroyan, A.; Torchilin, V.P. Increased apoptosis in cancer cells in vitro and in vivo by ceramides in transferrin-modified liposomes. Cancer Biol. Ther. 2012, 13, 50–60. [Google Scholar] [CrossRef]
- Morad, S.A.; Cabot, M.C. Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 2013, 13, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Kester, M.; Bassler, J.; Fox, T.E.; Carter, C.J.; Davidson, J.A.; Parette, M.R. Preclinical development of a C6-ceramide NanoLiposome, a novel sphingolipid therapeutic. Biol. Chem. 2015, 396, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Costa-Pinheiro, P.; Heher, A.; Raymond, M.H.; Jividen, K.; Shaw, J.J.; Paschal, B.M.; Walker, S.J.; Fox, T.E.; Kester, M. Role of SPTSSB-Regulated de Novo Sphingolipid Synthesis in Prostate Cancer Depends on Androgen Receptor Signaling. iScience 2020, 23, 101855. [Google Scholar] [CrossRef]
- Li, G.; Liu, D.; Kimchi, E.T.; Kaifi, J.T.; Qi, X.; Manjunath, Y.; Liu, X.; Deering, T.; Avella, D.M.; Fox, T.; et al. Nanoliposome C6-Ceramide Increases the Anti-tumor Immune Response and Slows Growth of Liver Tumors in Mice. Gastroenterology 2018, 154, 1024–1036.e9. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Kitatani, K.; Toyoshima, M.; Ishibashi, M.; Usui, T.; Minato, J.; Egiz, M.; Shigeta, S.; Fox, T.; Deering, T.; et al. Ceramide Nanoliposomes as a MLKL-Dependent, Necroptosis-Inducing, Chemotherapeutic Reagent in Ovarian Cancer. Mol. Cancer Ther. 2018, 17, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Ciner, A.; Gourdin, T.; Davidson, J.; Parette, M.; Walker, S.J.; Fox, T.E.; Jiang, Y. A phase I study of the ceramide nanoliposome in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2023, 93, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Espinosa Fernandez, J.R.; Eckhardt, B.L.; Lee, J.; Lim, B.; Pearson, T.; Seitz, R.S.; Hout, D.R.; Schweitzer, B.L.; Nielsen, T.J.; Lawrence, O.R.; et al. Identification of triple-negative breast cancer cell lines classified under the same molecular subtype using different molecular characterization techniques: Implications for translational research. PLoS ONE 2020, 15, e0231953. [Google Scholar] [CrossRef]
- Paroni, G.; Zanetti, A.; Barzago, M.M.; Kurosaki, M.; Guarrera, L.; Fratelli, M.; Troiani, M.; Ubezio, P.; Bolis, M.; Vallerga, A.; et al. Retinoic Acid Sensitivity of Triple-Negative Breast Cancer Cells Characterized by Constitutive Activation of the notch1 Pathway: The Role of Rarbeta. Cancers 2020, 12, 3027. [Google Scholar] [CrossRef]
- Sakae, H.; Ogiso, Y.; Matsuda, M.; Shimora, H.; Deering, T.; Fox, T.E.; Kester, M.; Nabe, T.; Kitatani, K. Ceramide Nanoliposomes as Potential Therapeutic Reagents for Asthma. Cells 2023, 12, 591. [Google Scholar] [CrossRef]
- Gyorffy, B. Discovery and ranking of the most robust prognostic biomarkers in serous ovarian cancer. Geroscience 2023, 45, 1889–1898. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Mei, L.; Zhu, Q.; Shi, G.; Wang, H. Mixed lineage kinase domain-like protein is a prognostic biomarker for cervical squamous cell cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 15035–15038. [Google Scholar] [PubMed]
- He, L.; Peng, K.; Liu, Y.; Xiong, J.; Zhu, F.F. Low expression of mixed lineage kinase domain-like protein is associated with poor prognosis in ovarian cancer patients. OncoTargets Ther. 2013, 6, 1539–1543. [Google Scholar] [CrossRef][Green Version]
- Ertao, Z.; Jianhui, C.; Kang, W.; Zhijun, Y.; Hui, W.; Chuangqi, C.; Changjiang, Q.; Sile, C.; Yulong, H.; Shirong, C. Prognostic value of mixed lineage kinase domain-like protein expression in the survival of patients with gastric caner. Tumour Biol. 2016, 37, 13679–13685. [Google Scholar] [CrossRef] [PubMed]
- Colbert, L.E.; Fisher, S.B.; Hardy, C.W.; Hall, W.A.; Saka, B.; Shelton, J.W.; Petrova, A.V.; Warren, M.D.; Pantazides, B.G.; Gandhi, K.; et al. Pronecrotic mixed lineage kinase domain-like protein expression is a prognostic biomarker in patients with early-stage resected pancreatic adenocarcinoma. Cancer 2013, 119, 3148–3155. [Google Scholar] [CrossRef] [PubMed]
- Ryland, L.K.; Doshi, U.A.; Shanmugavelandy, S.S.; Fox, T.E.; Aliaga, C.; Broeg, K.; Baab, K.T.; Young, M.; Khan, O.; Haakenson, J.K.; et al. C6-ceramide nanoliposomes target the Warburg effect in chronic lymphocytic leukemia. PLoS ONE 2013, 8, e84648. [Google Scholar] [CrossRef] [PubMed]
- Kitatani, K.; Idkowiak-Baldys, J.; Hannun, Y.A. The sphingolipid salvage pathway in ceramide metabolism and signaling. Cell. Signal. 2008, 20, 1010–1018. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, Y.; Mitsutake, S.; Tsuji, K.; Kihara, A.; Igarashi, Y. Ceramide biosynthesis in keratinocyte and its role in skin function. Biochimie 2009, 91, 784–790. [Google Scholar] [CrossRef]
- Levy, M.; Futerman, A.H. Mammalian ceramide synthases. IUBMB Life 2010, 62, 347–356. [Google Scholar] [CrossRef]
- Renert, A.F.; Leprince, P.; Dieu, M.; Renaut, J.; Raes, M.; Bours, V.; Chapelle, J.P.; Piette, J.; Merville, M.P.; Fillet, M. The proapoptotic C16-ceramide-dependent pathway requires the death-promoting factor Btf in colon adenocarcinoma cells. J. Proteome Res. 2009, 8, 4810–4822. [Google Scholar] [CrossRef]
- Stiban, J.; Perera, M. Very long chain ceramides interfere with C16-ceramide-induced channel formation: A plausible mechanism for regulating the initiation of intrinsic apoptosis. Biochim. Biophys. Acta 2015, 1848, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Stover, T.C.; Sharma, A.; Robertson, G.P.; Kester, M. Systemic delivery of liposomal short-chain ceramide limits solid tumor growth in murine models of breast adenocarcinoma. Clin. Cancer Res. 2005, 11, 3465–3474. [Google Scholar] [CrossRef] [PubMed]
- Tagaram, H.R.; Divittore, N.A.; Barth, B.M.; Kaiser, J.M.; Avella, D.; Kimchi, E.T.; Jiang, Y.; Isom, H.C.; Kester, M.; Staveley-O’Carroll, K.F. Nanoliposomal ceramide prevents in vivo growth of hepatocellular carcinoma. Gut 2011, 60, 695–701. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ryland, L.; Yang, J.; Liao, A.; Aliaga, C.; Watts, R.; Tan, S.F.; Kaiser, J.; Shanmugavelandy, S.S.; Rogers, A.; et al. Targeting of survivin by nanoliposomal ceramide induces complete remission in a rat model of NK-LGL leukemia. Blood 2010, 116, 4192–4201. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).