An Investigation of RNA Methylations with Biophysical Approaches in a Cervical Cancer Cell Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture, Flow Cytometry and Western Blotting
2.2. Cell Transfection
2.3. Total RNA Isolation and Global m6A Detection
2.4. SELECT (Single-Base Elongation- and Ligation-Based qPCR Amplification Method)
2.5. FT-IR Spectroscopy Measurements
2.6. CD Spectroscopy Measurements
3. Results
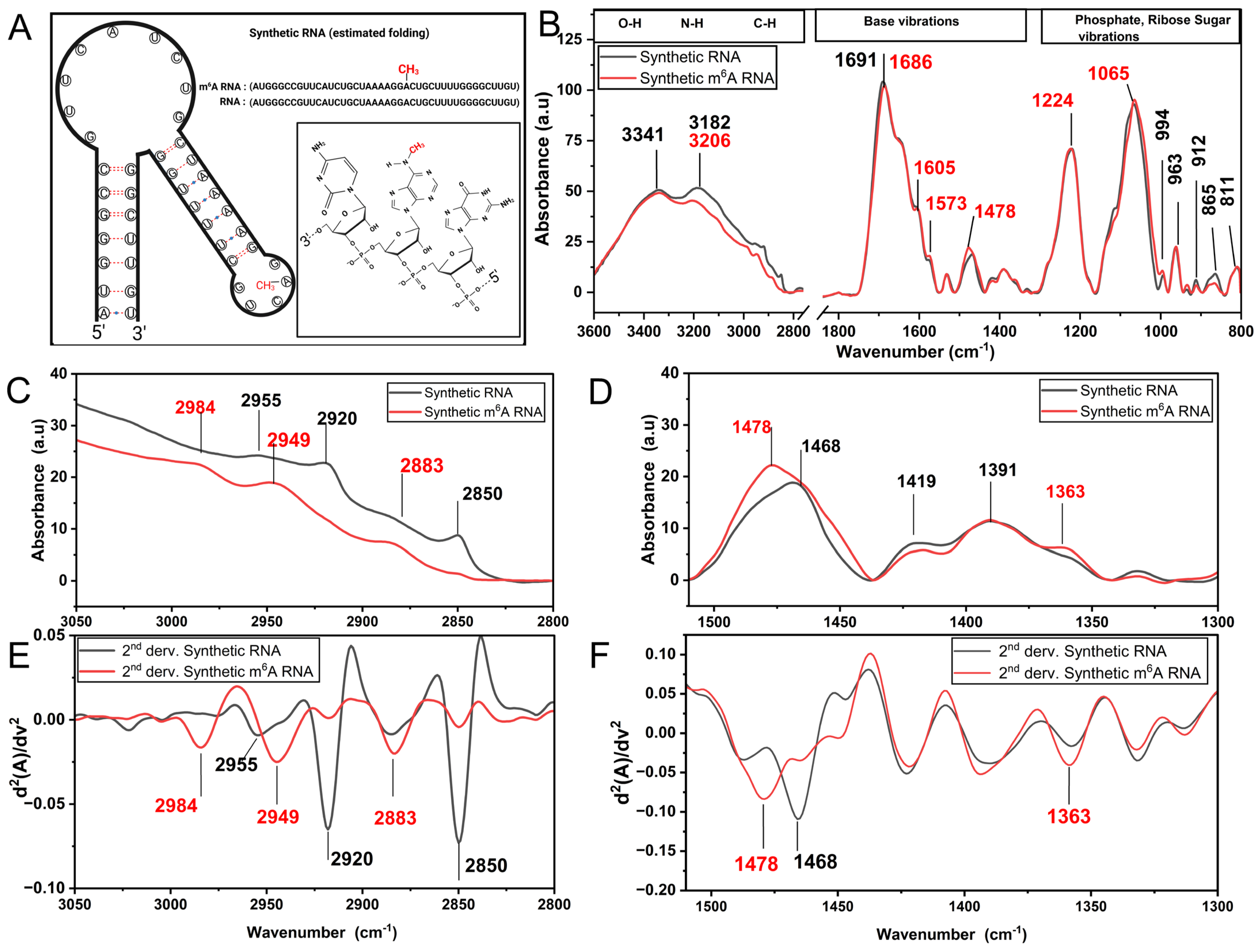
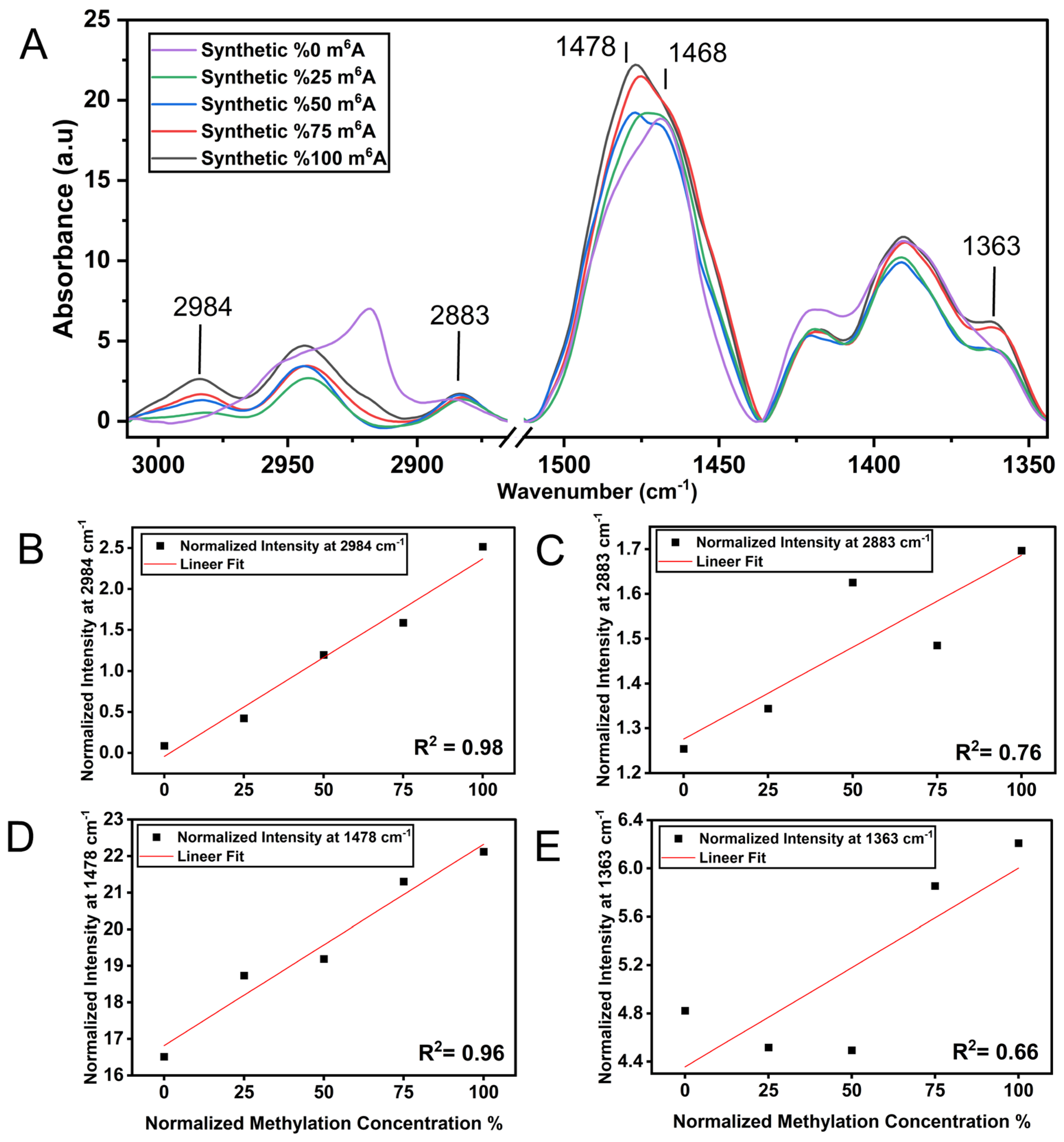
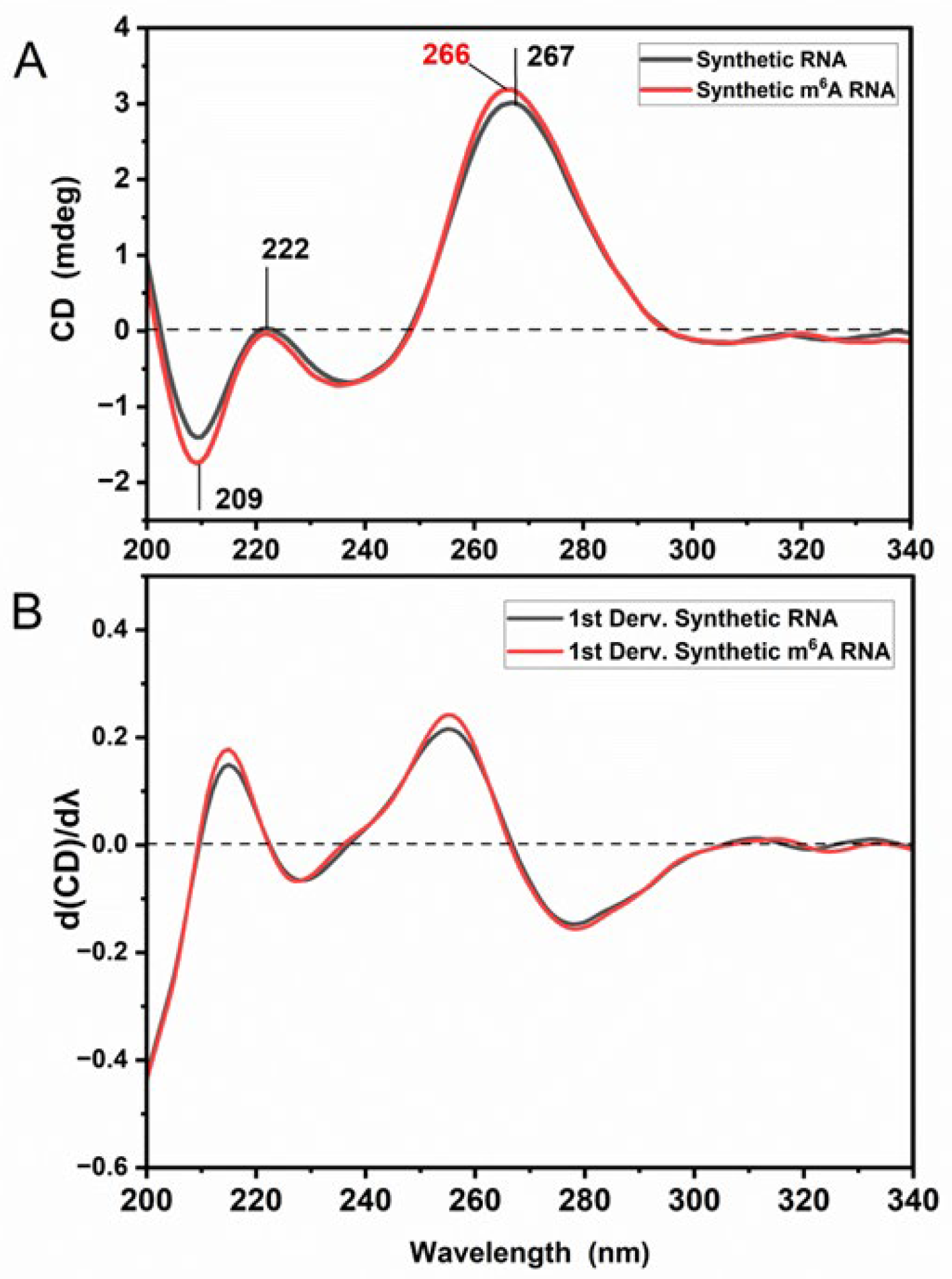
3.1. m6A Methylation Induces a Unique Spectroscopy Profile in Synthetic RNA Oligonucleotides
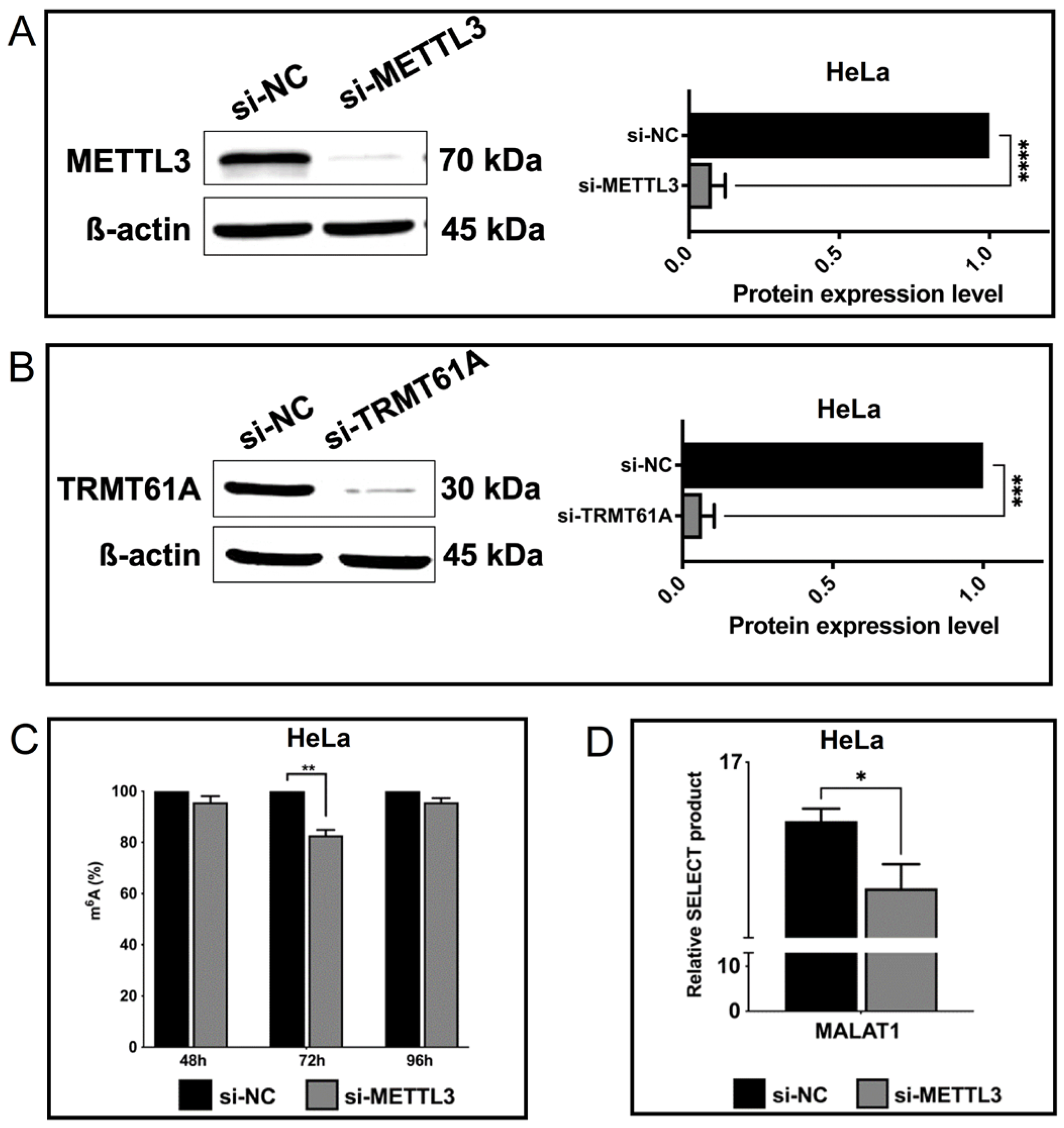
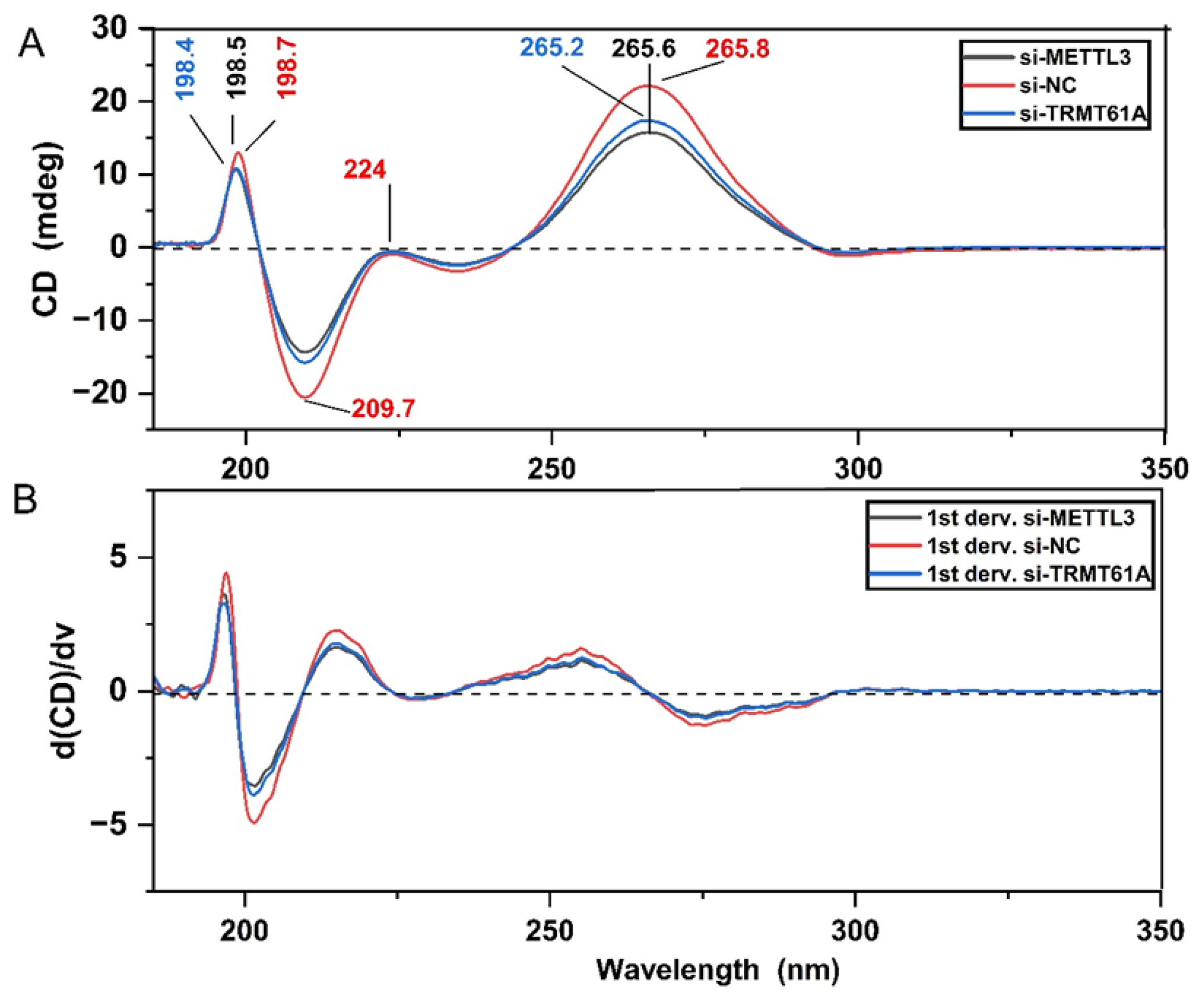
3.2. Spectroscopic Analysis of Perturbations in Cellular m6A Marks
3.3. TNF-α-Mediated Changes in the Biophysical Properties of RNAs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gilbert, W.V.; Bell, T.A.; Schaening, C. Messenger RNA Modifications: Form, Distribution, and Function. Science 2016, 352, 1408–1412. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Wei, J.; He, C. Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cell 2019, 74, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Alasar, A.A.; Tüncel, Ö.; Gelmez, A.B.; Sağlam, B.; Vatansever, İ.E.; Akgül, B. Genomewide M6A Mapping Uncovers Dynamic Changes in the M6A Epitranscriptome of Cisplatin-Treated Apoptotic HeLa Cells. Cells 2022, 11, 3905. [Google Scholar] [CrossRef] [PubMed]
- Akçaöz-Alasar, A.; Tüncel, Ö.; Sağlam, B.; Gazaloğlu, Y.; Atbinek, M.; Cagiral, U.; Iscan, E.; Ozhan, G.; Akgül, B. Epitranscriptomics M6A Analyses Reveal Distinct M6A Marks under Tumor Necrosis Factor α (TNF-α)-Induced Apoptotic Conditions in HeLa Cells. J. Cell. Physiol. 2024, 239, e31176. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Xu, G.; Zhang, X.; Ye, Y.; Cai, W.; Shao, Q. RNA M6A Modification Orchestrates the Rhythm of Immune Cell Development from Hematopoietic Stem Cells to T and B Cells. Front. Immunol. 2022, 13, 839291. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Xu, Y.; Gao, S.; Qin, L.; Austria, Q.; Siedlak, S.L.; Pajdzik, K.; Dai, Q.; He, C.; Wang, W.; et al. METTL3-Dependent RNA M6A Dysregulation Contributes to Neurodegeneration in Alzheimer’s Disease through Aberrant Cell Cycle Events. Mol. Neurodegener. 2021, 16, 70. [Google Scholar] [CrossRef]
- Barbieri, I.; Kouzarides, T. Role of RNA Modifications in Cancer. Nat. Rev. Cancer 2020, 20, 303–322. [Google Scholar] [CrossRef]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6 -Methyladenosine Modification Destabilizes Developmental Regulators in Embryonic Stem Cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef]
- Akgül, B.; Akçaöz-Alasar, A.; Sağlam, B. RNA M6A Methylation at the Juxtaposition of Apoptosis and RNA Therapeutics. Trends Cell Biol. 2024, 34, 801–804. [Google Scholar] [CrossRef] [PubMed]
- Sağlam, B.; Akgül, B. An Overview of Current Detection Methods for RNA Methylation. Int. J. Mol. Sci. 2024, 25, 3098. [Google Scholar] [CrossRef] [PubMed]
- Helm, M.; Motorin, Y. Detecting RNA Modifications in the Epitranscriptome: Predict and Validate. Nat. Rev. Genet. 2017, 18, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Ensinck, I.; Sideri, T.; Modic, M.; Capitanchik, C.; Vivori, C.; Toolan-Kerr, P.; Van Werven, F.J. M6A-ELISA, a Simple Method for Quantifying N6-Methyladenosine from MRNA Populations. RNA 2023, 29, 705–712. [Google Scholar] [CrossRef]
- Güler, G.; Acikgoz, E.; Mukhtarova, G.; Oktem, G. Biomolecular Fingerprints of the Effect of Zoledronic Acid on Prostate Cancer Stem Cells: Comparison of 2D and 3D Cell Culture Models. Arch. Biochem. Biophys. 2024, 753, 109920. [Google Scholar] [CrossRef]
- Güler, G.; Gärtner, R.M.; Ziegler, C.; Mäntele, W. Lipid-Protein Interactions in the Regulated Betaine Symporter BetP Probed by Infrared Spectroscopy. J. Biol. Chem. 2016, 291, 4295–4307. [Google Scholar] [CrossRef] [PubMed]
- Vorob’ev, M.M.; Açıkgöz, B.D.; Güler, G.; Golovanov, A.V.; Sinitsyna, O.V. Proteolysis of Micellar β-Casein by Trypsin: Secondary Structure Characterization and Kinetic Modeling at Different Enzyme Concentrations. Int. J. Mol. Sci. 2023, 24, 3874. [Google Scholar] [CrossRef] [PubMed]
- Zucchiatti, P.; Mitri, E.; Kenig, S.; Bille, F.; Kourousias, G.; Bedolla, D.E.; Vaccari, L. Contribution of Ribonucleic Acid (RNA) to the Fourier Transform Infrared (FTIR) Spectrum of Eukaryotic Cells. Anal. Chem. 2016, 88, 12090–12098. [Google Scholar] [CrossRef] [PubMed]
- Bourassa, P.; Tajmir-Riahi, H.A. Folic Acid Binds DNA and RNA at Different Locations. Int. J. Biol. Macromol. 2015, 74, 337–342. [Google Scholar] [CrossRef]
- Simsek Ozek, N.; Tuna, S.; Erson-Bensan, A.E.; Severcan, F. Characterization of MicroRNA-125b Expression in MCF7 Breast Cancer Cells by ATR-FTIR Spectroscopy. Analyst 2010, 135, 3094–3102. [Google Scholar] [CrossRef] [PubMed]
- Sarić, A.; Rajić, J.; Tolić, A.; Dučić, T.; Vidaković, M. Synchrotron-Based FTIR Microspectroscopy Reveals DNA Methylation Profile in DNA-HALO Structure. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2023, 302, 123090. [Google Scholar] [CrossRef]
- Kelly, J.G.; Najand, G.M.; Martin, F.L. Characterisation of DNA Methylation Status Using Spectroscopy (Mid-IR versus Raman) with Multivariate Analysis. J. Biophotonics 2011, 4, 345–354. [Google Scholar] [CrossRef]
- Duan, M.; Li, Y.; Zhang, F.; Huang, Q. Assessing B-Z DNA Transitions in Solutions via Infrared Spectroscopy. Biomolecules 2023, 13, 964. [Google Scholar] [CrossRef] [PubMed]
- Banyay, M.; Gräslund, A. Structural Effects of Cytosine Methylation on DNA Sugar Pucker Studied by FTIR. J. Mol. Biol. 2002, 324, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Sousa, V.K.; Pedro, J.A.F.; Kumagai, P.S.; Lopes, J.L.S. Effect of Setting Data Collection Parameters on the Reliability of a Circular Dichroism Spectrum. Eur. Biophys. J. 2021, 50, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Le Brun, E.; Arluison, V.; Wien, F. Application of Synchrotron Radiation Circular Dichroism for RNA Structural Analysis. Methods Mol. Biol. 2020, 2113, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Nichols, P.J.; Bevers, S.; Henen, M.; Kieft, J.S.; Vicens, Q.; Vögeli, B. Recognition of Non-CpG Repeats in Alu and Ribosomal RNAs by the Z-RNA Binding Domain of ADAR1 Induces A-Z Junctions. Nat. Commun. 2021, 12, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Güler, G.; Džafić, E.; Vorob’Ev, M.M.; Vogel, V.; Mäntele, W. Real Time Observation of Proteolysis with Fourier Transform Infrared (FT-IR) and UV-Circular Dichroism Spectroscopy: Watching a Protease Eat a Protein. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2011, 79, 104–111. [Google Scholar] [CrossRef]
- Meiser, N.; Mench, N.; Hengesbach, M. RNA Secondary Structure Dependence in METTL3–METTL14 MRNA Methylation Is Modulated by the N-Terminal Domain of METTL. Biol. Chem. 2020, 402, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Szabat, M.; Gudanis, D.; Kotkowiak, W.; Gdaniec, Z.; Kierzek, R.; Pasternak, A. Thermodynamic Features of Structural Motifs Formed by β-L-RNA. PLoS ONE 2016, 11, e0149478. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Toh, J.D.W.; Wong, K.H.Q.; Gao, Y.G.; Hong, W.; Woon, E.C.Y. N 6 -Methyladenosine: A Conformational Marker That Regulates the Substrate Specificity of Human Demethylases FTO and ALKBH5. Sci. Rep. 2016, 6, 25677. [Google Scholar] [CrossRef] [PubMed]
- Arluison, V.; Wien, F. RNA Spectroscopy Methods and Protocols Methods in Molecular Biology; Humana: Louisville, KY, USA, 2020. [Google Scholar]
- Ranjbar, B.; Gill, P. Circular Dichroism Techniques: Biomolecular and Nanostructural Analyses- A Review. Chem. Biol. Drug Des. 2009, 74, 101–120. [Google Scholar] [CrossRef] [PubMed]
- Cheong, A.; Low, J.J.A.; Lim, A.; Yen, P.M.; Woon, E.C.Y. A Fluorescent Methylation-Switchable Probe for Highly Sensitive Analysis of FTO N6-Methyladenosine Demethylase Activity in Cells. Chem. Sci. 2018, 9, 7174–7185. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Yin, P. Structural Insights into N6-Methyladenosine (m6A) Modification in the Transcriptome. Genom. Proteom. Bioinform. 2018, 16, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Wang, Y.; Tang, Q.; Wei, L.; Zhang, X.; Jia, G. An Elongation- and Ligation-Based QPCR Amplification Method for the Radiolabeling-Free Detection of Locus-Specific N 6 -Methyladenosine Modification. Angew. Chem. 2018, 130, 16227–16232. [Google Scholar] [CrossRef]
- Senguen, F.T.; Doran, T.M.; Anderson, E.A.; Nilsson, B.L. Clarifying the Influence of Core Amino Acid Hydrophobicity, Secondary Structure Propensity, and Molecular Volume on Amyloid-β 16-22 Self-Assembly. Mol. Biosyst. 2011, 7, 497–510. [Google Scholar] [CrossRef] [PubMed]
- Andrade, J.; Pereira, C.G.; Ranquine, T.; Azarias, C.A.; Bell, M.J.V.; De Carvalho Dos Anjos, V. Long-Term Ripening Evaluation of Ewes’ Cheeses by Fourier-Transformed Infrared Spectroscopy under Real Industrial Conditions. J. Spectrosc. 2018, 2018, 1381864. [Google Scholar] [CrossRef]
- Banyay, M.; Sarkar, M.; Graslund, A. A Library of IR Bands of Nucleic Acids in Solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Micura, R.; Pils, W.; Höbartner, C.; Grubmayr, K.; Ebert, M.-O.; Jaun, B. Methylation of the Nucleobases in RNA Oligonucleotides Mediates Duplex–Hairpin Conversion. Nucleic Acids Res. 2001, 29, 3997–4005. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.J.T.; Pan, T.; Kalsotra, A. RNA Modifications and Structures Cooperate to Guide RNA-Protein Interactions. Nat. Rev. Mol. Cell Biol. 2017, 18, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Kierzek, E.; Zhang, X.; Watson, R.M.; Kennedy, S.D.; Szabat, M.; Kierzek, R.; Mathews, D.H. Secondary Structure Prediction for RNA Sequences Including N6-Methyladenosine. Nat. Commun. 2022, 13, 1271. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the Human and Mouse M6A RNA Methylomes Revealed by M6A-Seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Dovbeshko, G.I.; Gridina, N.Y.; Kruglova, E.B.; Pashchuk, O.P. FTIR Spectroscopy Studies of Nucleic Acid Damage. Talanta 2000, 53, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Geinguenaud, F.; Militello, V.; Arluison, V. Application of FTIR Spectroscopy to Analyze RNA Structure. RNA Spectrosc. Methods Protoc. 2020, 2113, 119–133. [Google Scholar] [CrossRef]
- Wood, B.R. The Importance of Hydration and DNA Conformation in Interpreting Infrared Spectra of Cells and Tissues. Chem. Soc. Rev. 2016, 45, 1980–1998. [Google Scholar] [CrossRef]
- Hwang, W.; Arluison, V.; Hohng, S. Dynamic Competition of DsrA and RpoS Fragments for the Proximal Binding Site of Hfq as a Means for Efficient Annealing. Nucleic Acids Res. 2011, 39, 5131–5139. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.A.; Shambaugh, M.E.; Polayes, D.; Matera, A.G.; Rottman, F.M. Purification and CDNA Cloning of the AdoMet-Binding Subunit of the Human MRNA (N6-Adenosine)-Methyltransferase. RNA 1997, 3, 1233–1247. [Google Scholar] [PubMed]
- Anderson, J.; Phan, L.; Hinnebusch, A.G. The Gcd10pGcd14p Complex Is the Essential Two-Subunit TRNA(1-Methyladenosine) Methyltransferase of Saccharomyces Cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 5173–5178. [Google Scholar] [CrossRef]
- Lindqvist, M.; Sarkar, M.; Winqvist, A.; Rozners, E.; Strömberg, R.; Gräslund, A. Optical Spectroscopic Study of the Effects of a Single Deoxyribose Substitution in a Ribose Backbone: Implications in RNA-RNA Interaction. Biochemistry 2000, 39, 1693–1701. [Google Scholar] [CrossRef]
- Güler, G.; Vorob’Ev, M.M.; Vogel, V.; Mäntele, W. Proteolytically-Induced Changes of Secondary Structural Protein Conformation of Bovine Serum Albumin Monitored by Fourier Transform Infrared (FT-IR) and UV-Circular Dichroism Spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2016, 161, 8–18. [Google Scholar] [CrossRef]
- Nandiyanto, A.B.D.; Ragadhita, R.; Fiandini, M. Interpretation of Fourier Transform Infrared Spectra (FTIR): A Practical Approach in the Polymer/Plastic Thermal Decomposition. Indones. J. Sci. Technol. 2023, 8, 113–126. [Google Scholar] [CrossRef]
- Banyay, M.; Sandbrink, J.; Strömberg, R.; Gräslund, A. Characterization of an RNA Bulge Structure by Fourier Transform Infrared Spectroscopy. Biochem. Biophys. Res. Commun. 2004, 324, 634–639. [Google Scholar] [CrossRef]
- Liu, Z.-G.; Hsu, H.; Goeddel, D.V. Dissection of TNF Receptor 1 Effector Functions: JNK Activation Is Not Linked to Apoptosis While NF-B Activation Prevents Cell Death. Cell 1996, 87, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Riazance, J.H.; Baase, W.A.; Johnson, W.C.; Hall, K.; Cruz, P.; Tinoco, I. Evidence for Z-form RNA by vacuum UV circular dichroism. Nucleic Acids Res. 1985, 13, 4983–4989. [Google Scholar] [CrossRef] [PubMed]
- Vanloon, J.; Bennett, H.A.; Martin, A.; Wien, F.; Harroun, T.; Yan, H. Synchrotron Radiation Circular Dichroism Spectroscopy of Oligonucleotides at Millimolar Concentrations. Bioorg. Med. Chem. Lett. 2023, 92, 129376. [Google Scholar] [CrossRef] [PubMed]


| Name | Sequence (5′ → 3′) |
|---|---|
| MALAT1 m6A2515 UP | tagccagtaccgtagtgcgtgAATTACTTCCGTTACGAAAG |
| MALAT1 m6A2515 DOWN | 5phos/CCTTCACATTTTTCAAACTAAGCTACTcagaggctgagtcgctgcat |
| MALAT1 A2511 UP | tagccagtaccgtagtgcgtgAATTACTTCCGTTACGAAAGTCCT |
| MALAT1 A2511 DOWN | 5phos/CACATTTTTCAAACTAAGCTACTcagaggctgagtcgctgcat |
| SELECT qRT-PCR Forward | ATGCAGCGACTCAGCCTCTG |
| SELECT qRT-PCR Reverse | TAGCCAGTACCGTAGTGCGTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sağlam, B.; Akkuş, O.; Akçaöz-Alasar, A.; Ceylan, Ç.; Güler, G.; Akgül, B. An Investigation of RNA Methylations with Biophysical Approaches in a Cervical Cancer Cell Model. Cells 2024, 13, 1832. https://doi.org/10.3390/cells13221832
Sağlam B, Akkuş O, Akçaöz-Alasar A, Ceylan Ç, Güler G, Akgül B. An Investigation of RNA Methylations with Biophysical Approaches in a Cervical Cancer Cell Model. Cells. 2024; 13(22):1832. https://doi.org/10.3390/cells13221832
Chicago/Turabian StyleSağlam, Buket, Onur Akkuş, Azime Akçaöz-Alasar, Çağatay Ceylan, Günnur Güler, and Bünyamin Akgül. 2024. "An Investigation of RNA Methylations with Biophysical Approaches in a Cervical Cancer Cell Model" Cells 13, no. 22: 1832. https://doi.org/10.3390/cells13221832
APA StyleSağlam, B., Akkuş, O., Akçaöz-Alasar, A., Ceylan, Ç., Güler, G., & Akgül, B. (2024). An Investigation of RNA Methylations with Biophysical Approaches in a Cervical Cancer Cell Model. Cells, 13(22), 1832. https://doi.org/10.3390/cells13221832







