Differentiation States of Phenotypic Transition of Melanoma Cells Are Revealed by 3D Cell Cultures
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Real Time PCR
2.3. Immunofluorescence
2.4. Western Blot
2.5. SDS-Zymography
2.6. Statistical Analysis
3. Results
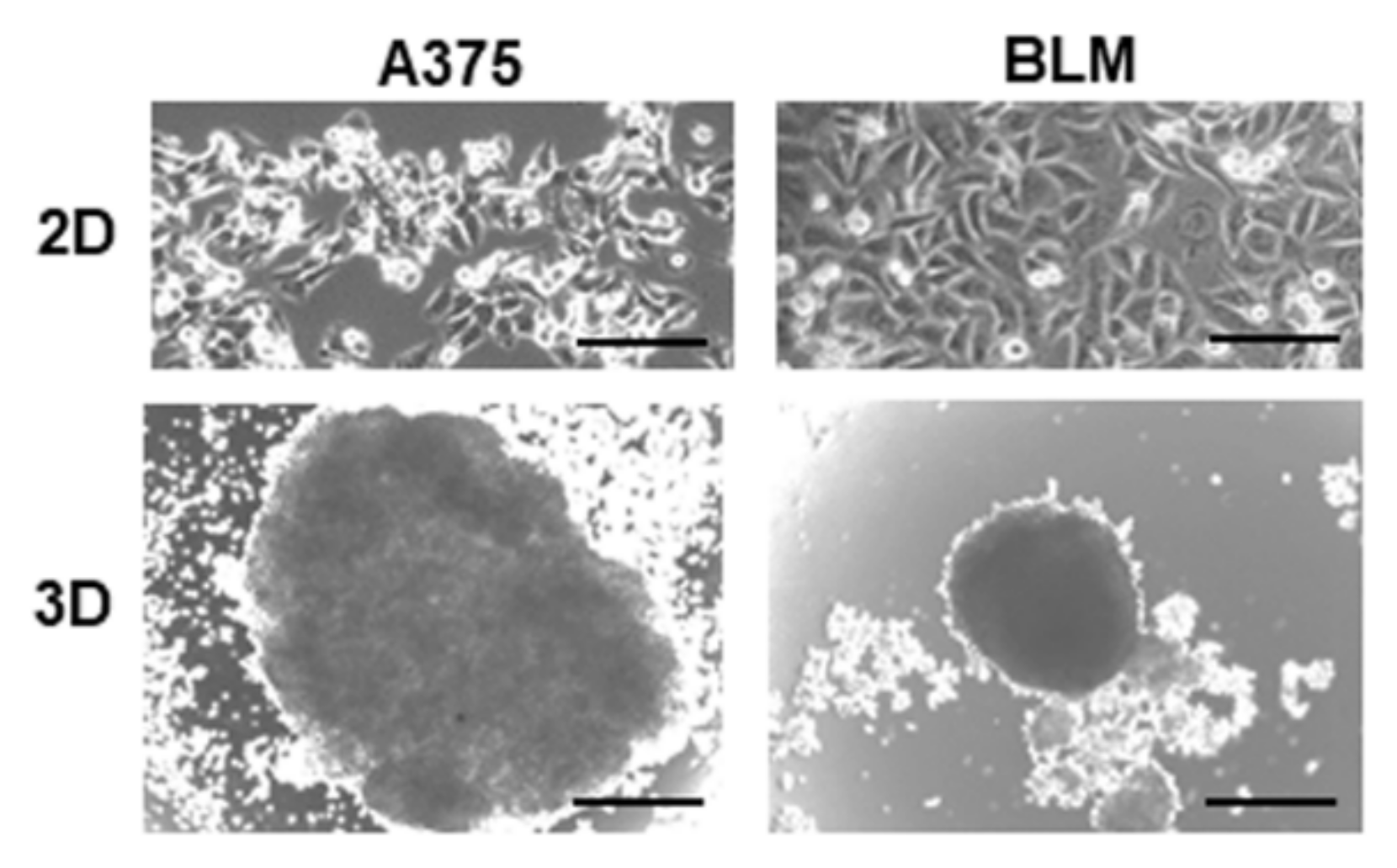
3.1. Characterization of Melanoma Cell Morphology and EMT-Related Phenotype in 2D and 3D Cultures
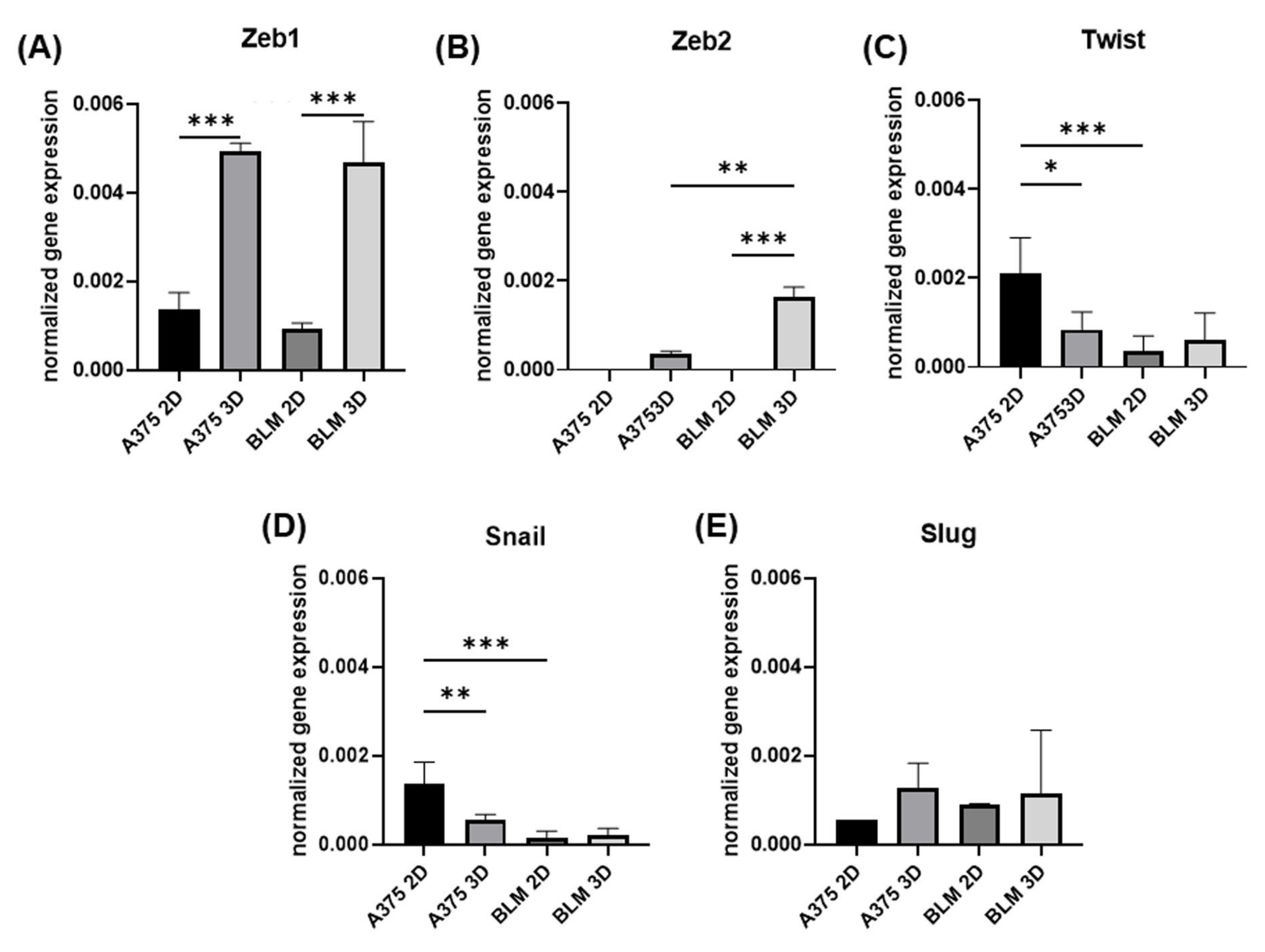
3.2. Zeb1, Zeb2, Twist, Snail and e Slug Expression
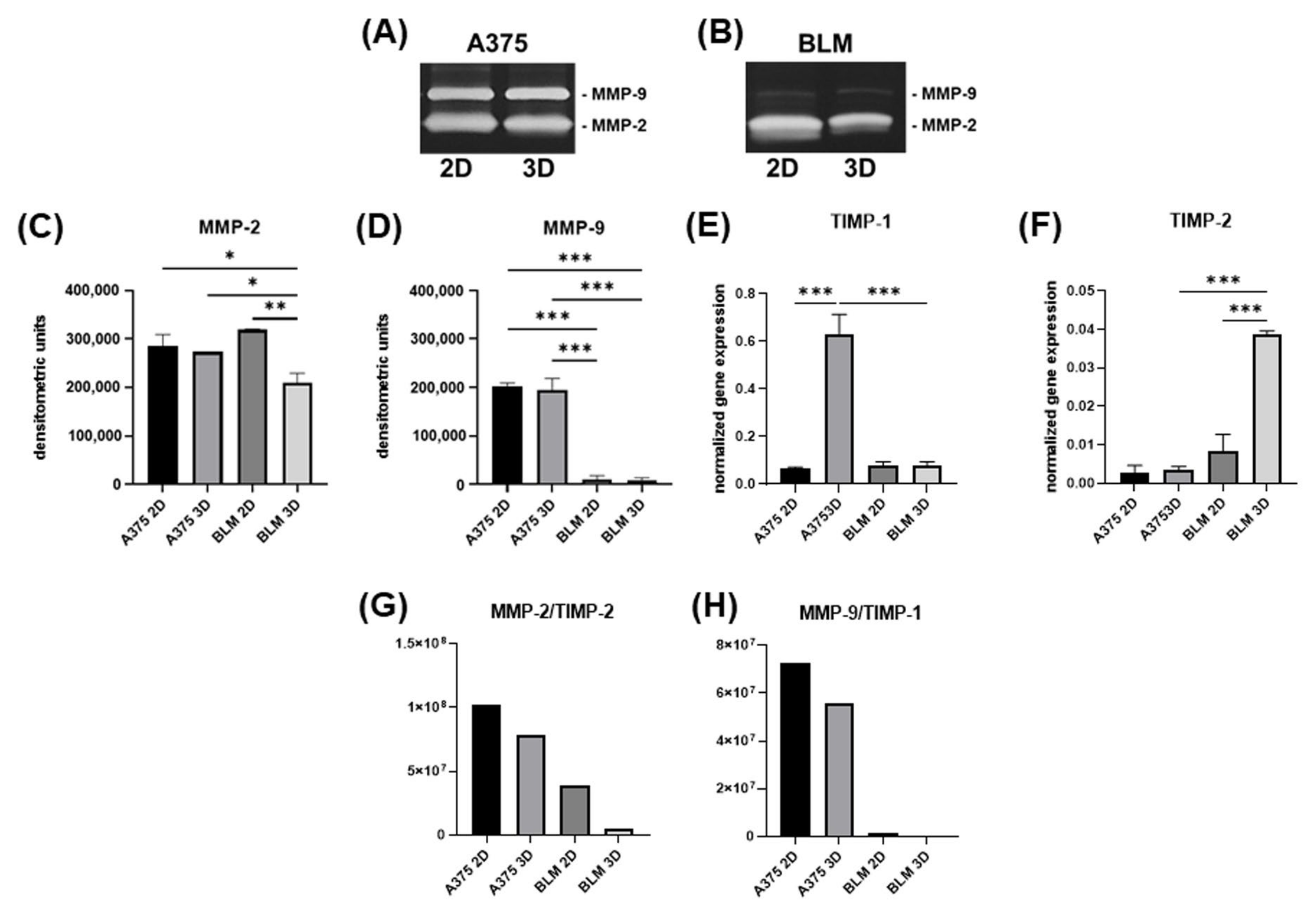
3.3. Invasive Potential
3.4. TGF-β1 and LOX Gene Expression
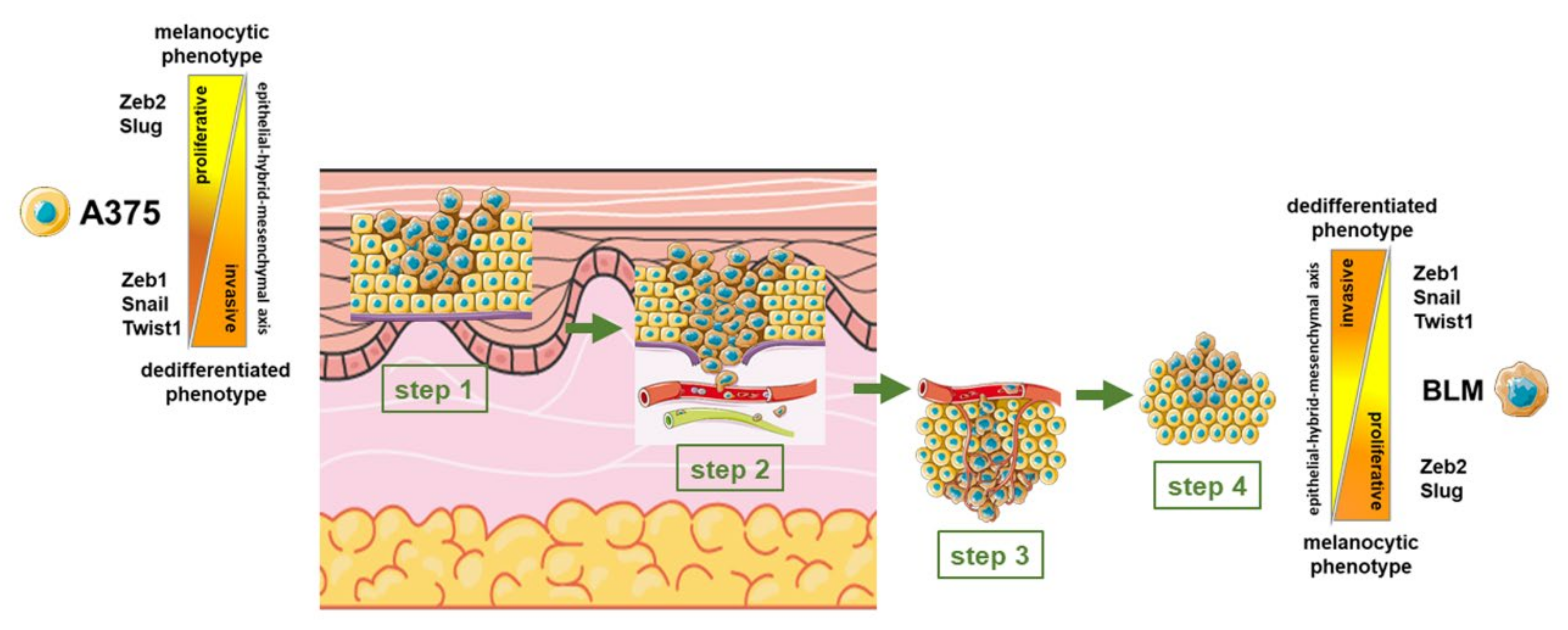
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Carr, S.; Smith, C.; Wernberg, J. Epidemiology and Risk Factors of Melanoma. Surg. Clin. N. Am. 2020, 100, 1–12. [Google Scholar] [CrossRef]
- Tímár, J.; Ladányi, A. Molecular Pathology of Skin Melanoma: Epidemiology, Differential Diagnostics, Prognosis and Therapy Prediction. Int. J. Mol. Sci. 2022, 23, 5384. [Google Scholar] [CrossRef] [PubMed]
- Timis, T.; Bergthorsson, J.T.; Greiff, V.; Cenariu, M.; Cenariu, D. Pathology and Molecular Biology of Melanoma. Curr. Issues Mol. Biol. 2023, 45, 5575–5597. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.J.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Hong, W.; Wei, X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J. Hematol. Oncol. 2022, 15, 129. [Google Scholar] [CrossRef] [PubMed]
- Zeisberg, M.; Neilson, E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Investig. 2009, 119, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Pedri, D.; Karras, P.; Landeloos, E.; Marine, J.-C.; Rambow, F. Epithelial-to-mesenchymal-like transition events in melanoma. FEBS J. 2022, 289, 1352–1368. [Google Scholar] [CrossRef]
- Alonso, S.R.; Tracey, L.; Ortiz, P.; Pérez-Gómez, B.; Palacios, J.; Pollán, M.; Linares, J.; Serrano, S.; Sáez-Castillo, A.I.; Sánchez, L.; et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007, 67, 3450–3460. [Google Scholar] [CrossRef] [PubMed]
- Pearlman, R.L.; Montes de Oca, M.K.; Pal, H.C.; Afaq, F. Potential therapeutic targets of epithelial-mesenchymal transition in melanoma. Cancer Lett. 2017, 391, 125–140. [Google Scholar] [CrossRef]
- Hodorogea, A.; Calinescu, A.; Antohe, M.; Balaban, M.; Nedelcu, R.I.; Turcu, G.; Ion, D.A.; Badarau, I.A.; Popescu, C.M.; Popescu, R.; et al. Epithelial-Mesenchymal Transition in Skin Cancers: A Review. Anal. Cell. Pathol. 2019, 2019, 3851576. [Google Scholar] [CrossRef] [PubMed]
- Vandyck, H.H.; Hillen, L.M.; Bosisio, F.M.; van den Oord, J.; zur Hausen, A.; Winnepenninckx, V. Rethinking the biology of metastatic melanoma: A holistic approach. Cancer Metastasis Rev. 2021, 40, 603–624. [Google Scholar] [CrossRef]
- Nieto, M.A.; Cano, A. The epithelial–mesenchymal transition under control: Global programs to regulate epithelial plasticity. Semin. Cancer Biol. 2012, 22, 361–368. [Google Scholar] [CrossRef]
- Puisieux, A.; Brabletz, T.; Caramel, J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 2014, 16, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Chuang, K.-T.; Chiou, S.-S.; Hsu, S.-H. Recent Advances in Transcription Factors Biomarkers and Targeted Therapies Focusing on Epithelial-Mesenchymal Transition. Cancers 2023, 15, 3338. [Google Scholar] [CrossRef]
- Ozdamar, B.; Bose, R.; Barrios-Rodiles, M.; Wang, H.-R.; Zhang, Y.; Wrana, J.L. Regulation of the polarity protein Par6 by TGFbeta receptors controls epithelial cell plasticity. Science 2005, 307, 1603–1609. [Google Scholar] [CrossRef] [PubMed]
- Papageorgis, P. TGFβ Signaling in Tumor Initiation, Epithelial-to-Mesenchymal Transition, and Metastasis. J. Oncol. 2015, 2015, 587193. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Chapman, A.; Del Fernandez Ama, L.; Ferguson, J.; Kamarashev, J.; Wellbrock, C.; Hurlstone, A. Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep. 2014, 8, 688–695. [Google Scholar] [CrossRef] [PubMed]
- Hoek, K.S.; Eichhoff, O.M.; Schlegel, N.C.; Döbbeling, U.; Kobert, N.; Schaerer, L.; Hemmi, S.; Dummer, R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008, 68, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Rambow, F.; Marine, J.-C.; Goding, C.R. Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes Dev. 2019, 33, 1295–1318. [Google Scholar] [CrossRef]
- Tang, Y.; Durand, S.; Dalle, S.; Caramel, J. EMT-Inducing Transcription Factors, Drivers of Melanoma Phenotype Switching, and Resistance to Treatment. Cancers 2020, 12, 2154. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Kalraiya, R.D. Invasive potential of melanoma cells correlates with the expression of MT1-MMP and regulated by modulating its association with motility receptors via N-glycosylation on the receptors. Biomed. Res. Int. 2014, 2014, 804680. [Google Scholar] [CrossRef] [PubMed]
- El Kharbili, M.; Cario, M.; Béchetoille, N.; Pain, C.; Boucheix, C.; Degoul, F.; Masse, I.; Berthier-Vergnes, O. Tspan8 Drives Melanoma Dermal Invasion by Promoting ProMMP-9 Activation and Basement Membrane Proteolysis in a Keratinocyte-Dependent Manner. Cancers 2020, 12, 1297. [Google Scholar] [CrossRef]
- Napoli, S.; Scuderi, C.; Gattuso, G.; Di Bella, V.; Candido, S.; Basile, M.S.; Libra, M.; Falzone, L. Functional Roles of Matrix Metalloproteinases and Their Inhibitors in Melanoma. Cells 2020, 9, 1151. [Google Scholar] [CrossRef]
- Kielosto, M.; Eriksson, J.; Nummela, P.; Yin, M.; Hölttä, E. Divergent roles of lysyl oxidase family members in ornithine decarboxylase- and RAS-transformed mouse fibroblasts and human melanoma cells. Oncotarget 2018, 9, 37733–37752. [Google Scholar] [CrossRef]
- Vázquez-Naharro, A.; Bustos-Tauler, J.; Floristán, A.; Yuste, L.; Oltra, S.S.; Vinyals, A.; Moreno-Bueno, G.; Fabra, À.; Portillo, F.; Cano, A.; et al. Loxl3 Promotes Melanoma Progression and Dissemination Influencing Cell Plasticity and Survival. Cancers 2022, 14, 1200. [Google Scholar] [CrossRef]
- Wang, W.; Wang, X.; Yao, F.; Huang, C. Lysyl Oxidase Family Proteins: Prospective Therapeutic Targets in Cancer. Int. J. Mol. Sci. 2022, 23, 2270. [Google Scholar] [CrossRef]
- Fontana, F.; Marzagalli, M.; Sommariva, M.; Gagliano, N.; Limonta, P. In Vitro 3D Cultures to Model the Tumor Microenvironment. Cancers 2021, 13, 2970. [Google Scholar] [CrossRef]
- El Harane, S.; Zidi, B.; El Harane, N.; Krause, K.-H.; Matthes, T.; Preynat-Seauve, O. Cancer Spheroids and Organoids as Novel Tools for Research and Therapy: State of the Art and Challenges to Guide Precision Medicine. Cells 2023, 12, 1001. [Google Scholar] [CrossRef]
- Manduca, N.; Maccafeo, E.; de Maria, R.; Sistigu, A.; Musella, M. 3D cancer models: One step closer to in vitro human studies. Front. Immunol. 2023, 14, 1175503. [Google Scholar] [CrossRef] [PubMed]
- Ohguro, H.; Watanabe, M.; Sato, T.; Hikage, F.; Furuhashi, M.; Okura, M.; Hida, T.; Uhara, H. 3D Spheroid Configurations Are Possible Indictors for Evaluating the Pathophysiology of Melanoma Cell Lines. Cells 2023, 12, 759. [Google Scholar] [CrossRef] [PubMed]
- Gagliano, N.; Sforza, C.; Sommariva, M.; Menon, A.; Conte, V.; Sartori, P.; Procacci, P. 3D-spheroids: What can they tell us about pancreatic ductal adenocarcinoma cell phenotype? Exp. Cell Res. 2017, 357, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Gagliano, N.; Celesti, G.; Tacchini, L.; Pluchino, S.; Sforza, C.; Rasile, M.; Valerio, V.; Laghi, L.; Conte, V.; Procacci, P. Epithelial-to-mesenchymal transition in pancreatic ductal adenocarcinoma: Characterization in a 3D-cell culture model. World J. Gastroenterol. 2016, 22, 4466–4483. [Google Scholar] [CrossRef]
- Fontana, F.; Raimondi, M.; Marzagalli, M.; Sommariva, M.; Limonta, P.; Gagliano, N. Epithelial-To-Mesenchymal Transition Markers and CD44 Isoforms Are Differently Expressed in 2D and 3D Cell Cultures of Prostate Cancer Cells. Cells 2019, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Raimondi, M.; Marzagalli, M.; Sommariva, M.; Gagliano, N.; Limonta, P. Three-Dimensional Cell Cultures as an In Vitro Tool for Prostate Cancer Modeling and Drug Discovery. Int. J. Mol. Sci. 2020, 21, 6806. [Google Scholar] [CrossRef]
- Kutle, I.; Polten, R.; Hachenberg, J.; Klapdor, R.; Morgan, M.; Schambach, A. Tumor Organoid and Spheroid Models for Cervical Cancer. Cancers 2023, 15, 2518. [Google Scholar] [CrossRef]
- Fedorenko, I.V.; Gibney, G.T.; Smalley, K.S.M. NRAS mutant melanoma: Biological behavior and future strategies for therapeutic management. Oncogene 2013, 32, 3009–3018. [Google Scholar] [CrossRef]
- Jackson, H.W.; Defamie, V.; Waterhouse, P.; Khokha, R. TIMPs: Versatile extracellular regulators in cancer. Nat. Rev. Cancer 2017, 17, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Centeno, P.P.; Pavet, V.; Marais, R. The journey from melanocytes to melanoma. Nat. Rev. Cancer 2023, 23, 372–390. [Google Scholar] [CrossRef] [PubMed]
- Belote, R.L.; Le, D.; Maynard, A.; Lang, U.E.; Sinclair, A.; Lohman, B.K.; Planells-Palop, V.; Baskin, L.; Tward, A.D.; Darmanis, S.; et al. Human melanocyte development and melanoma dedifferentiation at single-cell resolution. Nat. Cell Biol. 2021, 23, 1035–1047. [Google Scholar] [CrossRef] [PubMed]
- Shain, A.H.; Yeh, I.; Kovalyshyn, I.; Sriharan, A.; Talevich, E.; Gagnon, A.; Dummer, R.; North, J.; Pincus, L.; Ruben, B.; et al. The Genetic Evolution of Melanoma from Precursor Lesions. N. Engl. J. Med. 2015, 373, 1926–1936. [Google Scholar] [CrossRef] [PubMed]
- Sanna, A.; Harbst, K.; Johansson, I.; Christensen, G.; Lauss, M.; Mitra, S.; Rosengren, F.; Häkkinen, J.; Vallon-Christersson, J.; Olsson, H.; et al. Tumor genetic heterogeneity analysis of chronic sun-damaged melanoma. Pigment Cell Melanoma Res. 2020, 33, 480–489. [Google Scholar] [CrossRef] [PubMed]
- Karras, P.; Bordeu, I.; Pozniak, J.; Nowosad, A.; Pazzi, C.; van Raemdonck, N.; Landeloos, E.; van Herck, Y.; Pedri, D.; Bervoets, G.; et al. A cellular hierarchy in melanoma uncouples growth and metastasis. Nature 2022, 610, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Ennen, M.; Keime, C.; Gambi, G.; Kieny, A.; Coassolo, S.; Thibault-Carpentier, C.; Margerin-Schaller, F.; Davidson, G.; Vagne, C.; Lipsker, D.; et al. MITF-High and MITF-Low Cells and a Novel Subpopulation Expressing Genes of Both Cell States Contribute to Intra- and Intertumoral Heterogeneity of Primary Melanoma. Clin. Cancer Res. 2017, 23, 7097–7107. [Google Scholar] [CrossRef]
- Krengel, S.; Grotelüschen, F.; Bartsch, S.; Tronnier, M. Cadherin expression pattern in melanocytic tumors more likely depends on the melanocyte environment than on tumor cell progression. J. Cutan. Pathol. 2004, 31, 1–7. [Google Scholar] [CrossRef]
- Murtas, D.; Maxia, C.; Diana, A.; Pilloni, L.; Corda, C.; Minerba, L.; Tomei, S.; Piras, F.; Ferreli, C.; Perra, M.T. Role of epithelial-mesenchymal transition involved molecules in the progression of cutaneous melanoma. Histochem. Cell Biol. 2017, 148, 639–649. [Google Scholar] [CrossRef]
- Loh, C.-Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Hsu, M.; Andl, T.; Li, G.; Meinkoth, J.L.; Herlyn, M. Cadherin repertoire determines partner-specific gap junctional communication during melanoma progression. J. Cell Sci. 2000, 113 Pt 9, 1535–1542. [Google Scholar] [CrossRef] [PubMed]
- Lade-Keller, J.; Riber-Hansen, R.; Guldberg, P.; Schmidt, H.; Hamilton-Dutoit, S.J.; Steiniche, T. E- to N-cadherin switch in melanoma is associated with decreased expression of phosphatase and tensin homolog and cancer progression. Br. J. Dermatol. 2013, 169, 618–628. [Google Scholar] [CrossRef]
- Li, G.; Schaider, H.; Satyamoorthy, K.; Hanakawa, Y.; Hashimoto, K.; Herlyn, M. Downregulation of E-cadherin and Desmoglein 1 by autocrine hepatocyte growth factor during melanoma development. Oncogene 2001, 20, 8125–8135. [Google Scholar] [CrossRef]
- Qi, J.; Chen, N.; Wang, J.; Siu, C.-H. Transendothelial migration of melanoma cells involves N-cadherin-mediated adhesion and activation of the beta-catenin signaling pathway. Mol. Biol. Cell 2005, 16, 4386–4397. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Mège, R.M. N-Cadherin and Fibroblast Growth Factor Receptors crosstalk in the control of developmental and cancer cell migrations. Eur. J. Cell Biol. 2016, 95, 415–426. [Google Scholar] [CrossRef]
- Silye, R.; Karayiannakis, A.J.; Syrigos, K.N.; Poole, S.; van Noorden, S.; Batchelor, W.; Regele, H.; Sega, W.; Boesmueller, H.; Krausz, T.; et al. E-cadherin/catenin complex in benign and malignant melanocytic lesions. J. Pathol. 1998, 186, 350–355. [Google Scholar] [CrossRef]
- Sanders, D.S.; Blessing, K.; Hassan, G.A.; Bruton, R.; Marsden, J.R.; Jankowski, J. Alterations in cadherin and catenin expression during the biological progression of melanocytic tumours. Mol. Pathol. 1999, 52, 151–157. [Google Scholar] [CrossRef]
- Danen, E.H.; de Vries, T.J.; Morandini, R.; Ghanem, G.G.; Ruiter, D.J.; van Muijen, G.N. E-cadherin expression in human melanoma. Melanoma Res. 1996, 6, 127–131. [Google Scholar] [CrossRef]
- Kim, J.E.; Leung, E.; Baguley, B.C.; Finlay, G.J. Heterogeneity of expression of epithelial-mesenchymal transition markers in melanocytes and melanoma cell lines. Front. Genet. 2013, 4, 97. [Google Scholar] [CrossRef]
- Cook, D.P.; Vanderhyden, B.C. Context specificity of the EMT transcriptional response. Nat. Commun. 2020, 11, 2142. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, M.J.; Seftor, E.A.; Chu, Y.W.; Seftor, R.E.; Nagle, R.B.; McDaniel, K.M.; Leong, S.P.; Yohem, K.H.; Leibovitz, A.M.; Meyskens, F.L. Coexpression of vimentin and keratins by human melanoma tumor cells: Correlation with invasive and metastatic potential. J. Natl. Cancer Inst. 1992, 84, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.W.; Seftor, E.A.; Romer, L.H.; Hendrix, M.J. Experimental coexpression of vimentin and keratin intermediate filaments in human melanoma cells augments motility. Am. J. Pathol. 1996, 148, 63–69. [Google Scholar] [PubMed]
- Li, M.; Zhang, B.; Sun, B.; Wang, X.; Ban, X.; Sun, T.; Liu, Z.; Zhao, X. A novel function for vimentin: The potential biomarker for predicting melanoma hematogenous metastasis. J. Exp. Clin. Cancer Res. 2010, 29, 109. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Poser, I.; Domínguez, D.; de Herreros, A.G.; Varnai, A.; Buettner, R.; Bosserhoff, A.K. Loss of E-cadherin expression in melanoma cells involves up-regulation of the transcriptional repressor Snail. J. Biol. Chem. 2001, 276, 24661–24666. [Google Scholar] [CrossRef]
- Batlle, E.; Sancho, E.; Francí, C.; Domínguez, D.; Monfar, M.; Baulida, J.; García De Herreros, A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2000, 2, 84–89. [Google Scholar] [CrossRef]
- Cano, A.; Pérez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; Del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Shirley, S.H.; Greene, V.R.; Duncan, L.M.; Torres Cabala, C.A.; Grimm, E.A.; Kusewitt, D.F. Slug expression during melanoma progression. Am. J. Pathol. 2012, 180, 2479–2489. [Google Scholar] [CrossRef]
- Wels, C.; Joshi, S.; Koefinger, P.; Bergler, H.; Schaider, H. Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J. Investig. Dermatol. 2011, 131, 1877–1885. [Google Scholar] [CrossRef]
- Iwakami, Y.; Yokoyama, S.; Watanabe, K.; Hayakawa, Y. STAM-binding protein regulates melanoma metastasis through SLUG stabilization. Biochem. Biophys. Res. Commun. 2018, 507, 484–488. [Google Scholar] [CrossRef]
- Vandamme, N.; Berx, G. Melanoma cells revive an embryonic transcriptional network to dictate phenotypic heterogeneity. Front. Oncol. 2014, 4, 352. [Google Scholar] [CrossRef] [PubMed]
- Caramel, J.; Papadogeorgakis, E.; Hill, L.; Browne, G.J.; Richard, G.; Wierinckx, A.; Saldanha, G.; Osborne, J.; Hutchinson, P.; Tse, G.; et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 2013, 24, 466–480. [Google Scholar] [CrossRef]
- Denecker, G.; Vandamme, N.; Akay, O.; Koludrovic, D.; Taminau, J.; Lemeire, K.; Gheldof, A.; de Craene, B.; van Gele, M.; Brochez, L.; et al. Identification of a ZEB2-MITF-ZEB1 transcriptional network that controls melanogenesis and melanoma progression. Cell Death Differ. 2014, 21, 1250–1261. [Google Scholar] [CrossRef]
- Vandamme, N.; Denecker, G.; Bruneel, K.; Blancke, G.; Akay, Ö.; Taminau, J.; de Coninck, J.; de Smedt, E.; Skrypek, N.; van Loocke, W.; et al. The EMT Transcription Factor ZEB2 Promotes Proliferation of Primary and Metastatic Melanoma While Suppressing an Invasive, Mesenchymal-Like Phenotype. Cancer Res. 2020, 80, 2983–2995. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wu, J.; Zhang, Y.; Liu, X.; Xu, N.; Zuo, F.; Xu, J. SOX4 promotes melanoma cell migration and invasion though the activation of the NF-κB signaling pathway. Int. J. Mol. Med. 2017, 40, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghì, A.; Spandidos, D.A.; Mccubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef]
- Guarneri, C.; Bevelacqua, V.; Polesel, J.; Falzone, L.; Cannavò, P.S.; Spandidos, D.A.; Malaponte, G.; Libra, M. NF-κB inhibition is associated with OPN/MMP-9 downregulation in cutaneous melanoma. Oncol. Rep. 2017, 37, 737–746. [Google Scholar] [CrossRef]
- Liang, X.; Sun, R.; Zhao, X.; Zhang, Y.; Gu, Q.; Dong, X.; Zhang, D.; Sun, J.; Sun, B. Rictor regulates the vasculogenic mimicry of melanoma via the AKT-MMP-2/9 pathway. J. Cell. Mol. Med. 2017, 21, 3579–3591. [Google Scholar] [CrossRef]
- Väisänen, A.; Kallioinen, M.; Taskinen, P.J.; Turpeenniemi-Hujanen, T. Prognostic value of MMP-2 immunoreactive protein (72 kD type IV collagenase) in primary skin melanoma. J. Pathol. 1998, 186, 51–58. [Google Scholar] [CrossRef]
- Hofmann, U.B.; Westphal, J.R.; van Muijen, G.N.; Ruiter, D.J. Matrix metalloproteinases in human melanoma. J. Investig. Dermatol. 2000, 115, 337–344. [Google Scholar] [CrossRef]
- van den Oord, J.J.; Paemen, L.; Opdenakker, G.; de Wolf-Peeters, C. Expression of gelatinase B and the extracellular matrix metalloproteinase inducer EMMPRIN in benign and malignant pigment cell lesions of the skin. Am. J. Pathol. 1997, 151, 665–670. [Google Scholar] [PubMed]
- Bergers, G.; Brekken, R.; McMahon, G.; Vu, T.H.; Itoh, T.; Tamaki, K.; Tanzawa, K.; Thorpe, P.; Itohara, S.; Werb, Z.; et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat. Cell Biol. 2000, 2, 737–744. [Google Scholar] [CrossRef]
- Gomez, D.E.; Alonso, D.F.; Yoshiji, H.; Thorgeirsson, U.P. Tissue inhibitors of metalloproteinases: Structure, regulation and biological functions. Eur. J. Cell Biol. 1997, 74, 111–122. [Google Scholar] [PubMed]
- Sun, J.; Stetler-Stevenson, W.G. Overexpression of tissue inhibitors of metalloproteinase 2 up-regulates NF-kappaB activity in melanoma cells. J. Mol. Signal. 2009, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Strongin, A.Y.; Collier, I.; Bannikov, G.; Marmer, B.L.; Grant, G.A.; Goldberg, G.I. Mechanism of cell surface activation of 72-kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J. Biol. Chem. 1995, 270, 5331–5338. [Google Scholar] [CrossRef]
- Butler, G.S.; Butler, M.J.; Atkinson, S.J.; Will, H.; Tamura, T.; van Schade Westrum, S.; Crabbe, T.; Clements, J.; d’Ortho, M.P.; Murphy, G. The TIMP2 membrane type 1 metalloproteinase “receptor” regulates the concentration and efficient activation of progelatinase A. A kinetic study. J. Biol. Chem. 1998, 273, 871–880. [Google Scholar] [CrossRef]
- Liu, Y. Epithelial to mesenchymal transition in renal fibrogenesis: Pathologic significance, molecular mechanism, and therapeutic intervention. J. Am. Soc. Nephrol. 2004, 15, 1–12. [Google Scholar] [CrossRef]
- Cantelli, G.; Orgaz, J.L.; Rodriguez-Hernandez, I.; Karagiannis, P.; Maiques, O.; Matias-Guiu, X.; Nestle, F.O.; Marti, R.M.; Karagiannis, S.N.; Sanz-Moreno, V. TGF-β-Induced Transcription Sustains Amoeboid Melanoma Migration and Dissemination. Curr. Biol. 2015, 25, 2899–2914. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, F.; Dang, J.; Cheng, F.; Zheng, F. Bidirectional regulation between tumor cell-intrinsic PD-L1 and TGF-β1 in epithelial-to-mesenchymal transition in melanoma. Transl. Cancer Res. 2022, 11, 3698–3710. [Google Scholar] [CrossRef]
- Da Silva, R.; Uno, M.; Marie, S.K.N.; Oba-Shinjo, S.M. LOX expression and functional analysis in astrocytomas and impact of IDH1 mutation. PLoS ONE 2015, 10, e0119781. [Google Scholar] [CrossRef]
- Sethi, A.; Mao, W.; Wordinger, R.J.; Clark, A.F. Transforming growth factor-beta induces extracellular matrix protein cross-linking lysyl oxidase (LOX) genes in human trabecular meshwork cells. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5240–5250. [Google Scholar] [CrossRef] [PubMed]
- Eritja, N.; Dolcet, X.; Matias-Guiu, X. Three-dimensional epithelial cultures: A tool to model cancer development and progression. Histol. Histopathol. 2013, 28, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Kunz-Schughart, L.A. Multicellular tumor spheroids: Intermediates between monolayer culture and in vivo tumor. Cell Biol. Int. 1999, 23, 157–161. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fontana, F.; Sommariva, M.; Anselmi, M.; Bianchi, F.; Limonta, P.; Gagliano, N. Differentiation States of Phenotypic Transition of Melanoma Cells Are Revealed by 3D Cell Cultures. Cells 2024, 13, 181. https://doi.org/10.3390/cells13020181
Fontana F, Sommariva M, Anselmi M, Bianchi F, Limonta P, Gagliano N. Differentiation States of Phenotypic Transition of Melanoma Cells Are Revealed by 3D Cell Cultures. Cells. 2024; 13(2):181. https://doi.org/10.3390/cells13020181
Chicago/Turabian StyleFontana, Fabrizio, Michele Sommariva, Martina Anselmi, Francesca Bianchi, Patrizia Limonta, and Nicoletta Gagliano. 2024. "Differentiation States of Phenotypic Transition of Melanoma Cells Are Revealed by 3D Cell Cultures" Cells 13, no. 2: 181. https://doi.org/10.3390/cells13020181
APA StyleFontana, F., Sommariva, M., Anselmi, M., Bianchi, F., Limonta, P., & Gagliano, N. (2024). Differentiation States of Phenotypic Transition of Melanoma Cells Are Revealed by 3D Cell Cultures. Cells, 13(2), 181. https://doi.org/10.3390/cells13020181










