Systematic Comparison of CRISPR and shRNA Screens to Identify Essential Genes Using a Graph-Based Unsupervised Learning Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Description
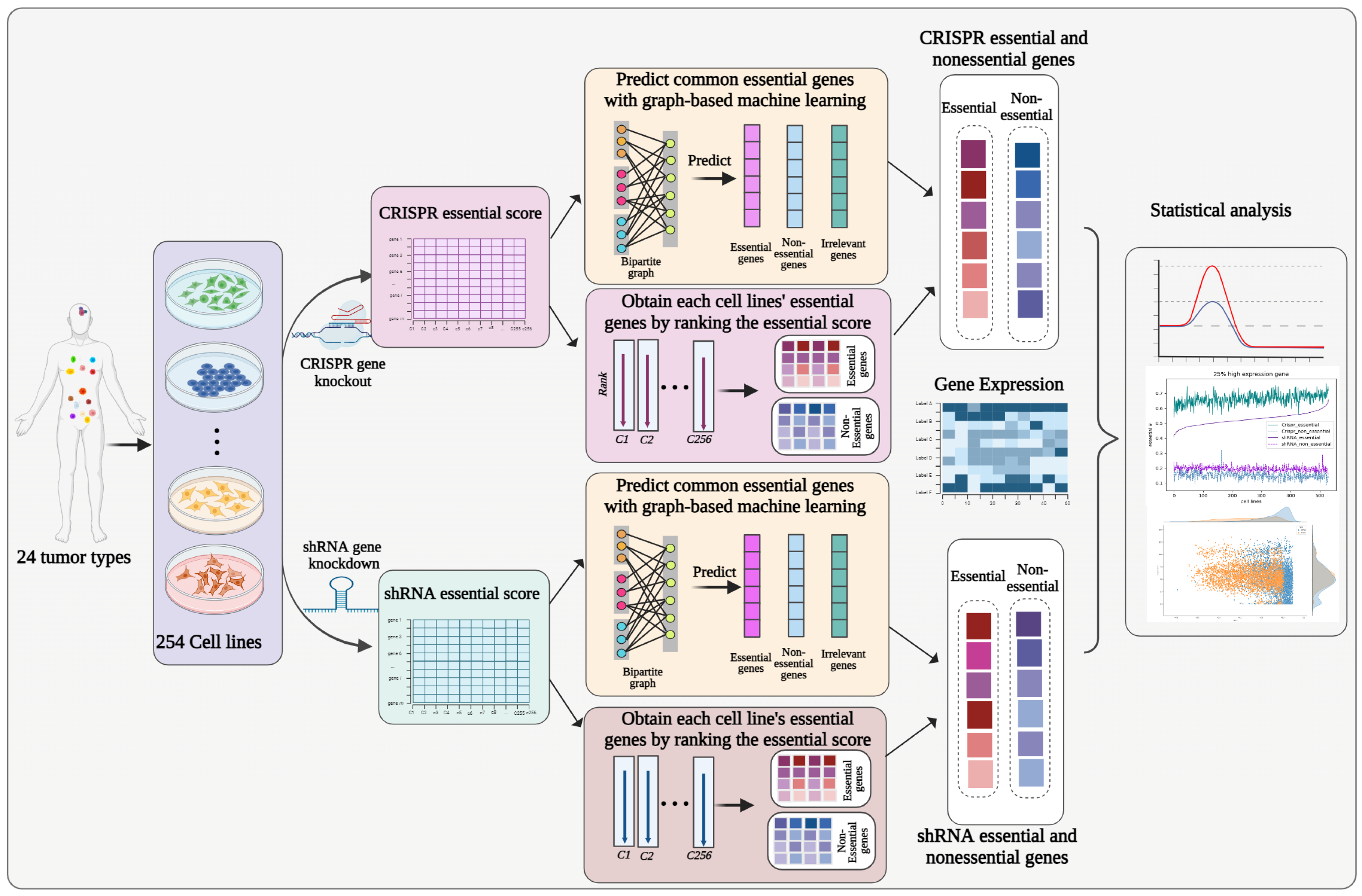
2.2. Overall Description of Our Workflow
2.3. Graph-Based Unsupervised Learning Model
2.4. Iterative Updating to Obtain Common Essential Genes
2.5. The Final Essential Genes for Each Cell Line Were Obtained
2.6. Statistical Analysis of shRNA- and CRISPR-Based Essential Genes with Different Expression Levels
3. Results
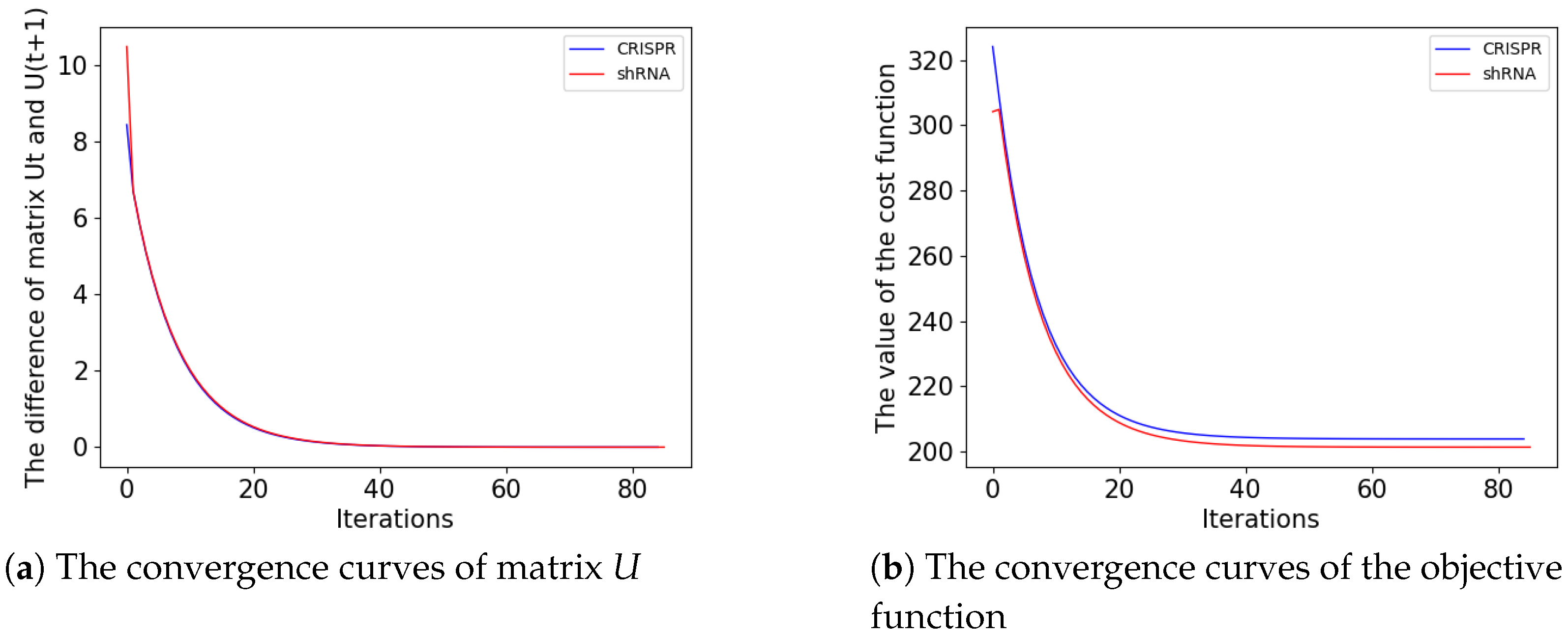
3.1. Graph-Based Machine Learning Models Converge on Both shRNAs and CRIPSRs with the Same Trends
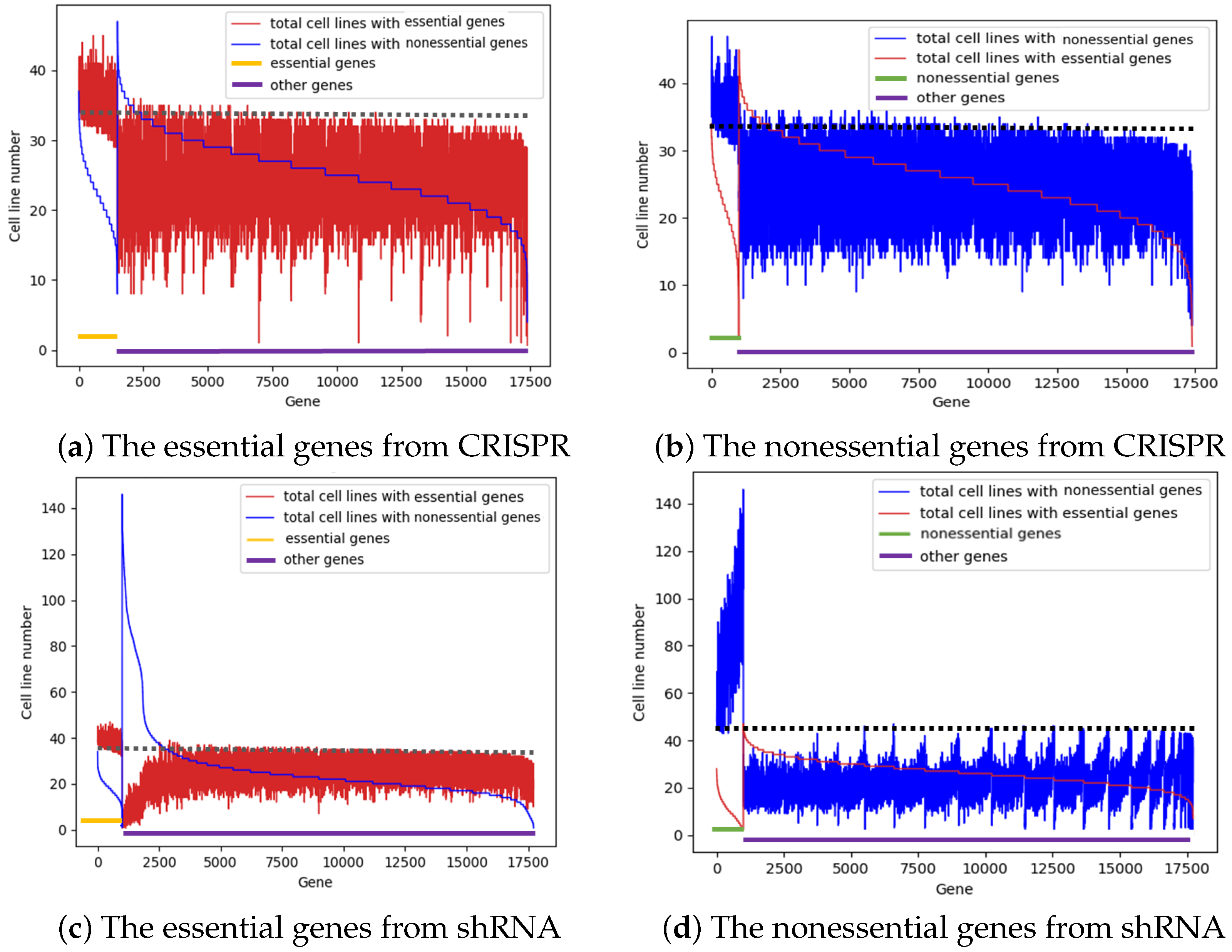
3.2. Not a Single Gene Is Essential in All Cancer Cell Lines
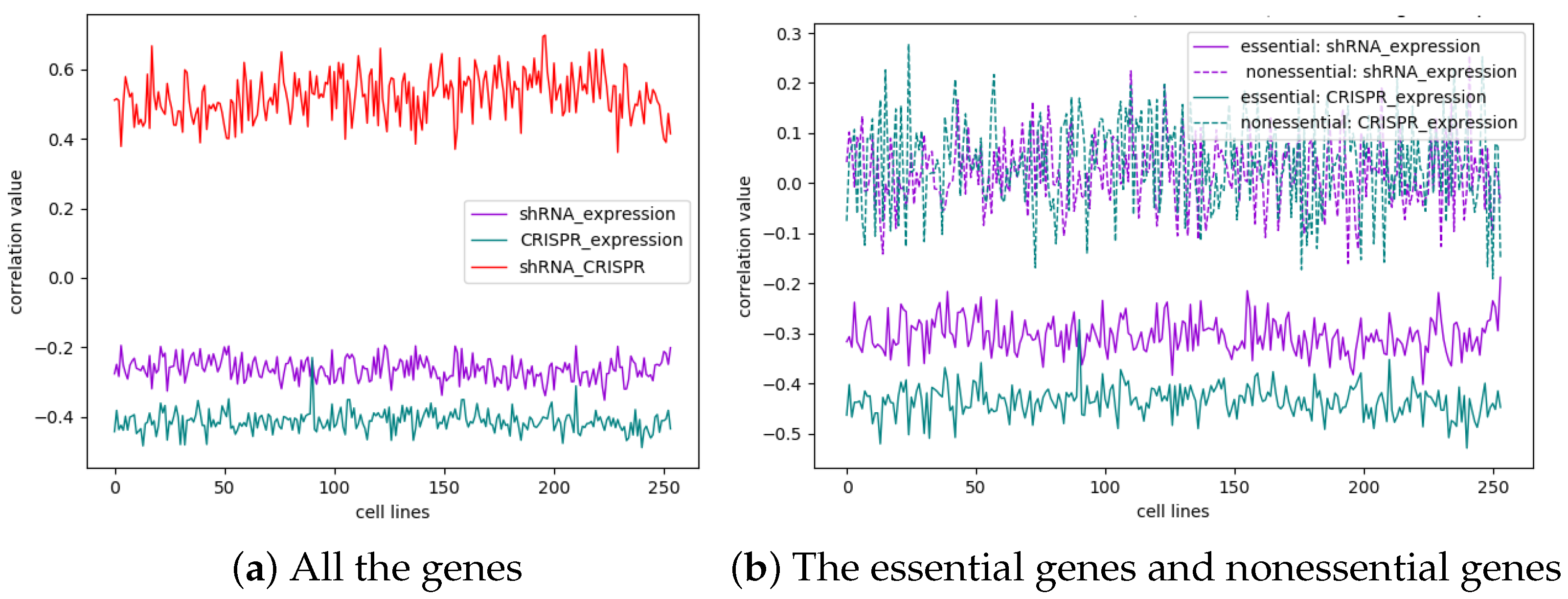
3.3. Gene Expression and Gene Essentiality Are Positively Correlated According to Both shRNA and CRISPR Screening Methodologies
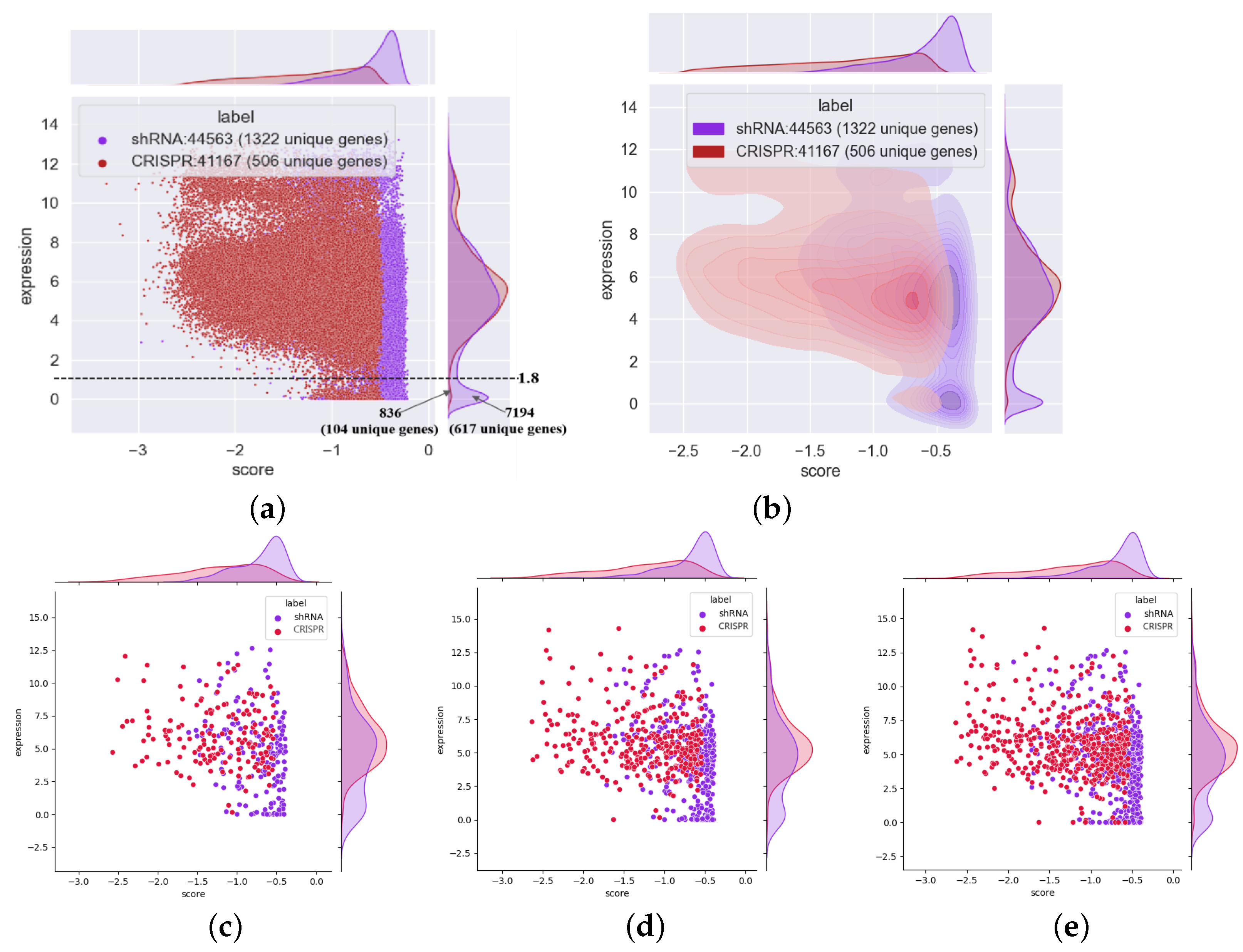
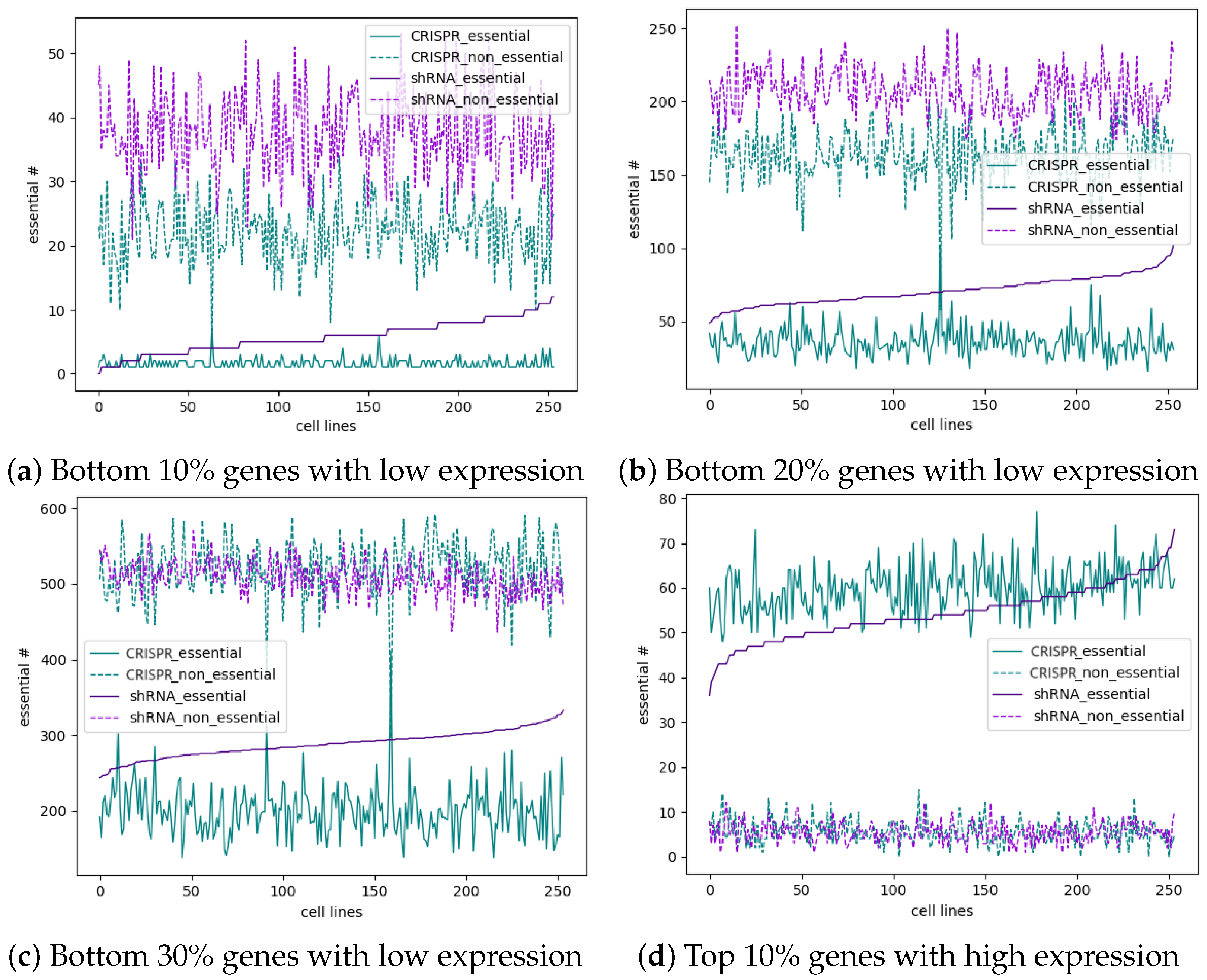
3.4. shRNA Outperforms CRISPR in the Identification of Essential Genes with Low Expression Levels
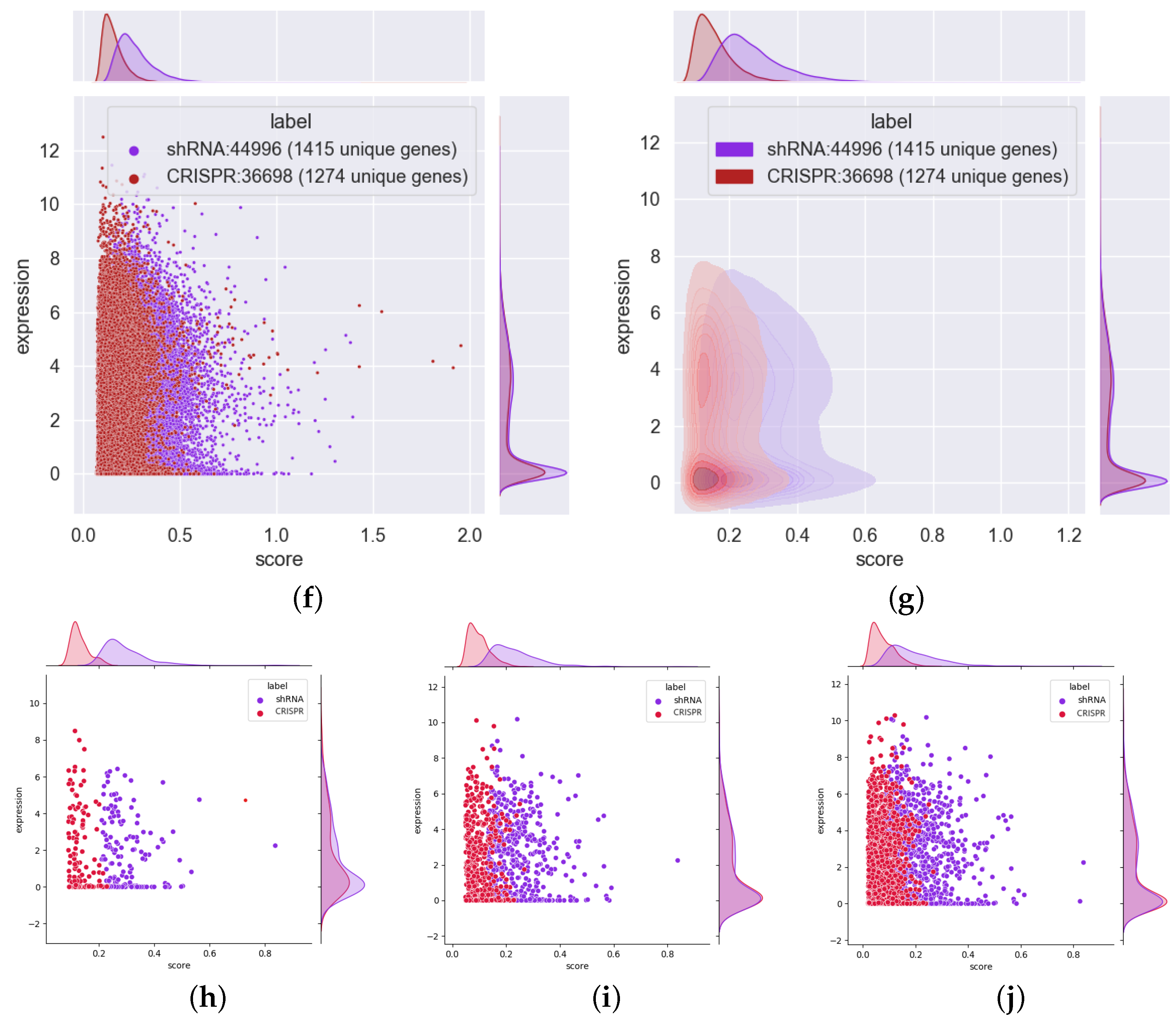
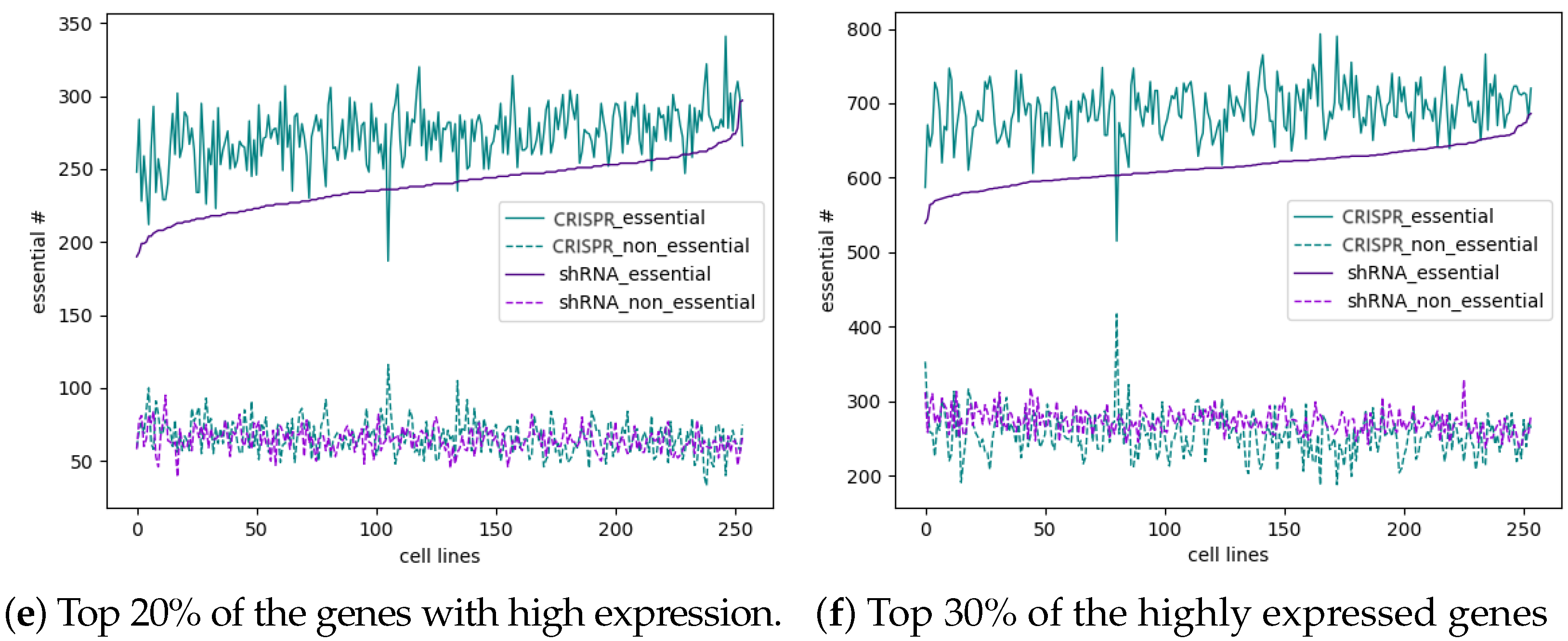
3.5. Most Essential Genes Are Highly Expressed, and CRISPR Has a Slightly Better Ability to Identify Highly Expressed Essential Genes
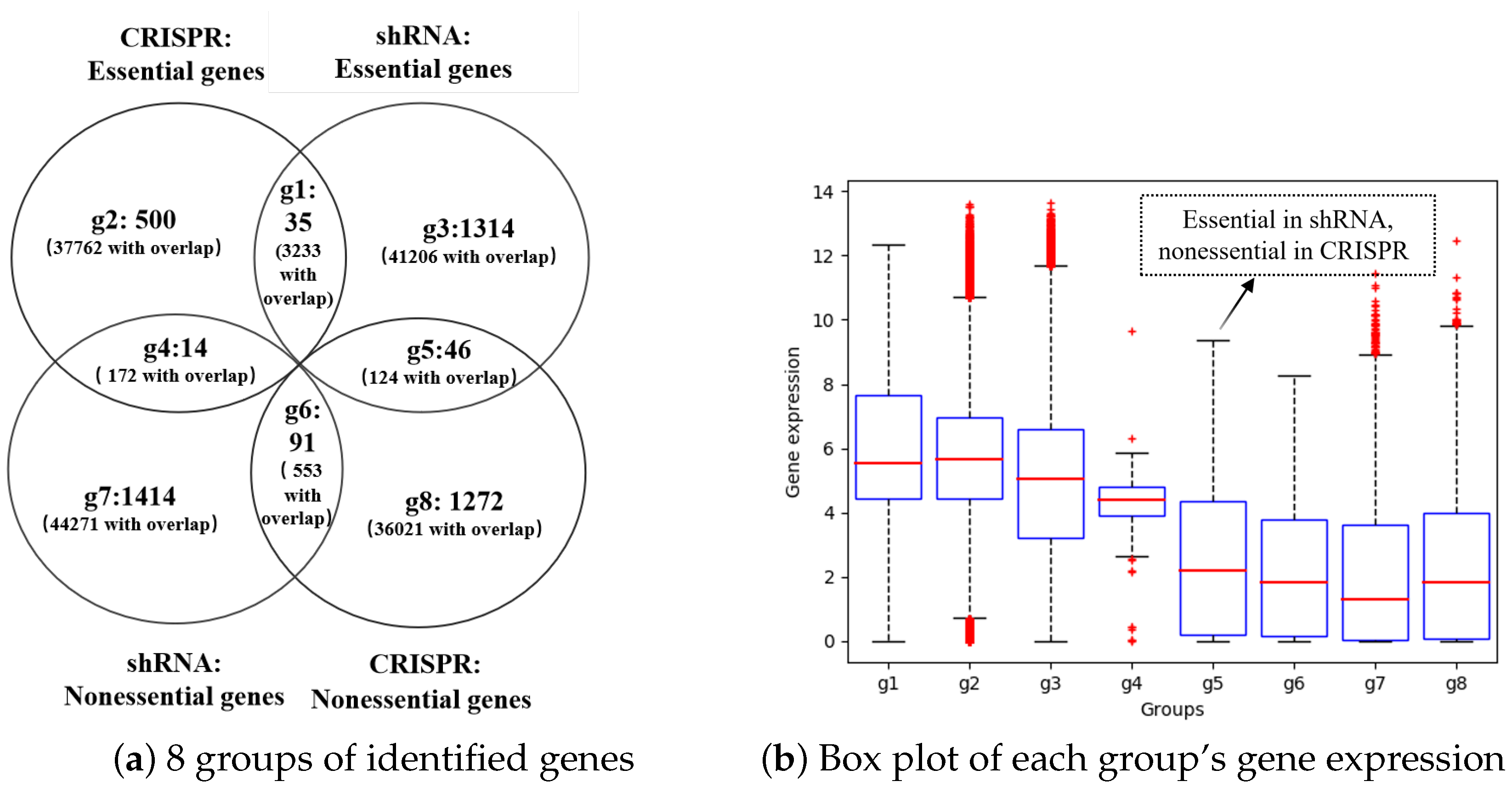
3.6. The Essential Genes Identified via shRNA and CRISPR Have Little Overlap, and Most of the Genes Expressed at Low Levels can Be Identified Only by shRNA
4. Conclusions and Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, T.; Yu, H.; Hughes, N.W.; Liu, B.; Kendirli, A.; Klein, K.; Chen, W.W.; Lander, E.S.; Sabatini, D.M. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic Ras. Cell 2017, 168, 890–903. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Z.; Jiang, S.; Li, R.; Li, W.; Zhao, C.; Hong, H.; Huang, X.; Li, H.; Bo, X. New insights on human essential genes based on integrated analysis and the construction of the HEGIAP web-based platform. Brief. Bioinform. 2023, 21, 1397–1410. [Google Scholar] [CrossRef] [PubMed]
- Taheri, G.; Habibi, M. Identification of essential genes associated with SARS-CoV-2 infection as potential drug target candidates with machine learning algorithms. Sci. Rep. 2023, 13, 15141. [Google Scholar] [CrossRef]
- Zhong, J.C.; Qu, Z.H.; Zhong, Y.; Tang, C.; Pan, Y. Continuous and Discrete Similarity Coefficient for Identifying Essential Proteins Using Gene Expression Data. Big Data Min. Anal. 2023, 6, 185–200. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Zhang, F. High-throughput functional genomics using CRISPR–Cas9. Nat. Rev. Genet. 2015, 16, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Tamaddondoust, R.N.; Wong, A.; Chandrashekhar, M.; Azzam, E.I.; Alain, T.; Wang, Y. Identification of novel regulators of radiosensitivity using high-throughput genetic screening. Int. J. Mol. Sci. 2022, 23, 8774. [Google Scholar] [CrossRef]
- Bernards, R. Finding effective cancer therapies through loss of function genetic screens. Curr. Opin. Genet. Dev. 2014, 24, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Amberkar, S.; Kiani, N.A.; Bartenschlager, R.; Alvisi, G.; Kaderali, L. High-throughput RNA interference screens integrative analysis: Towards a comprehensive understanding of the virus-host interplay. World J. Virol. 2013, 2, 18. [Google Scholar] [CrossRef]
- Hart, T.; Chandrashekhar, M.; Aregger, M.; Steinhart, Z.; Brown, K.R.; MacLeod, G.; Mis, M.; Zimmermann, M.; Fradet-Turcotte, A.; Sun, S.; et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 2023, 163, 1515–1526. [Google Scholar] [CrossRef]
- Morgens, D.W.; Deans, R.M.; Li, A.; Bassik, M.C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 2016, 34, 634–636. [Google Scholar] [CrossRef]
- Kaelin, W.G., Jr. Use and abuse of RNAi to study mammalian gene function. Science 2012, 337, 421–422. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Birmingham, A.; Wiemann, S.; Beijersbergen, R.L.; Hornung, V.; Smith, A.v.B. Advances in CRISPR-Cas9 genome engineering: Lessons learned from RNA interference. Nucleic Acids Res. 2015, 43, 3407–3419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nguyen, T.M.; Zhang, X.O.; Wang, L.; Phan, T.; Clohessy, J.G.; Pandolfi, P.P. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2023, 22, 1–22. [Google Scholar] [CrossRef]
- Grimm, D.; Streetz, K.L.; Jopling, C.L.; Storm, T.A.; Pandey, K.; Davis, C.R.; Marion, P.; Salazar, F.; Kay, M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 2006, 441, 537–541. [Google Scholar] [CrossRef]
- Frock, R.L.; Hu, J.; Meyers, R.M.; Ho, Y.J.; Kii, E.; Alt, F.W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015, 33, 179–186. [Google Scholar] [CrossRef]
- Høiberg, H.C.; Sparvath, S.M.; Andersen, V.L.; Kjems, J.; Andersen, E.S. An RNA origami octahedron with intrinsic siRNAs for potent gene knockdown. Biotechnol. J. 2019, 14, 1700634. [Google Scholar] [CrossRef]
- Evers, B.; Jastrzebski, K.; Heijmans, J.P.; Grernrum, W.; Beijersbergen, R.L.; Bernards, R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 2016, 34, 631–633. [Google Scholar] [CrossRef]
- Kampmann, M.; Horlbeck, M.A.; Chen, Y.; Tsai, J.C.; Bassik, M.C.; Gilbert, L.A.; Villalta, J.E.; Kwon, S.C.; Chang, H.; Kim, V.N.; et al. Next-generation libraries for robust RNA interference-based genome-wide screens. Proc. Natl. Acad. Sci. USA 2015, 112, E3384–E3391. [Google Scholar] [CrossRef]
- Behan, F.M.; Iorio, F.; Picco, G.; Gonçalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M.; et al. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef]
- Hart, T.; Brown, K.R.; Sircoulomb, F.; Rottapel, R.; Moffat, J. Measuring error rates in genomic perturbation screens: Gold standards for human functional genomics. Mol. Syst. Biol. 2014, 10, 733. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Fang, L.; Liu, F.; Wang, X.; Chen, J.; Chou, K.C. Identification of real microRNA precursors with a pseudo structure status composition approach. PLoS ONE 2015, 10, e0121501. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Behan, F.M.; Gonçalves, E.; Bhosle, S.G.; Chen, E.; Shepherd, R.; Beaver, C.; Ansari, R.; Pooley, R.; Wilkinson, P.; et al. Unsupervised correction of gene-independent cell responses to CRISPR-Cas9 targeting. BMC Genom. 2018, 19, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a cancer dependency map. Cell 2017, 170, 564–576. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., III; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the cancer cell line encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Lin, W.; Wang, J.; Zhang, W.J.; Wu, F.X. An unsupervised machine learning method for assessing quality of tandem mass spectra. In Proceedings of the Proteome Science; Springer: Berlin/Heidelberg, Germany, 2012; Volume 10, pp. 1–8. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Denomy, C.; Freywald, A.; Pan, Y.; Vizeacoumar, F.J.; Vizeacoumar, F.S.; Wu, F.-X. Systematic Comparison of CRISPR and shRNA Screens to Identify Essential Genes Using a Graph-Based Unsupervised Learning Model. Cells 2024, 13, 1653. https://doi.org/10.3390/cells13191653
Ding Y, Denomy C, Freywald A, Pan Y, Vizeacoumar FJ, Vizeacoumar FS, Wu F-X. Systematic Comparison of CRISPR and shRNA Screens to Identify Essential Genes Using a Graph-Based Unsupervised Learning Model. Cells. 2024; 13(19):1653. https://doi.org/10.3390/cells13191653
Chicago/Turabian StyleDing, Yulian, Connor Denomy, Andrew Freywald, Yi Pan, Franco J. Vizeacoumar, Frederick S. Vizeacoumar, and Fang-Xiang Wu. 2024. "Systematic Comparison of CRISPR and shRNA Screens to Identify Essential Genes Using a Graph-Based Unsupervised Learning Model" Cells 13, no. 19: 1653. https://doi.org/10.3390/cells13191653
APA StyleDing, Y., Denomy, C., Freywald, A., Pan, Y., Vizeacoumar, F. J., Vizeacoumar, F. S., & Wu, F.-X. (2024). Systematic Comparison of CRISPR and shRNA Screens to Identify Essential Genes Using a Graph-Based Unsupervised Learning Model. Cells, 13(19), 1653. https://doi.org/10.3390/cells13191653






