Uncovering the Interaction between TRAF1 and MAVS in the RIG-I Pathway to Enhance the Upregulation of IRF1/ISG15 during Classical Swine Fever Virus Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, and Construction of Recombinant Plasmids
2.2. Antibodies and Reagents
2.3. Small Interfering RNA (siRNA) Knockdown
2.4. Transfection/Infection Assays
2.5. Immunoprecipitation Assay
2.6. Indirect Immunofluorescence Assay (IFA)
2.7. WB Analysis
2.8. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
2.9. Statistical Analysis
3. Results
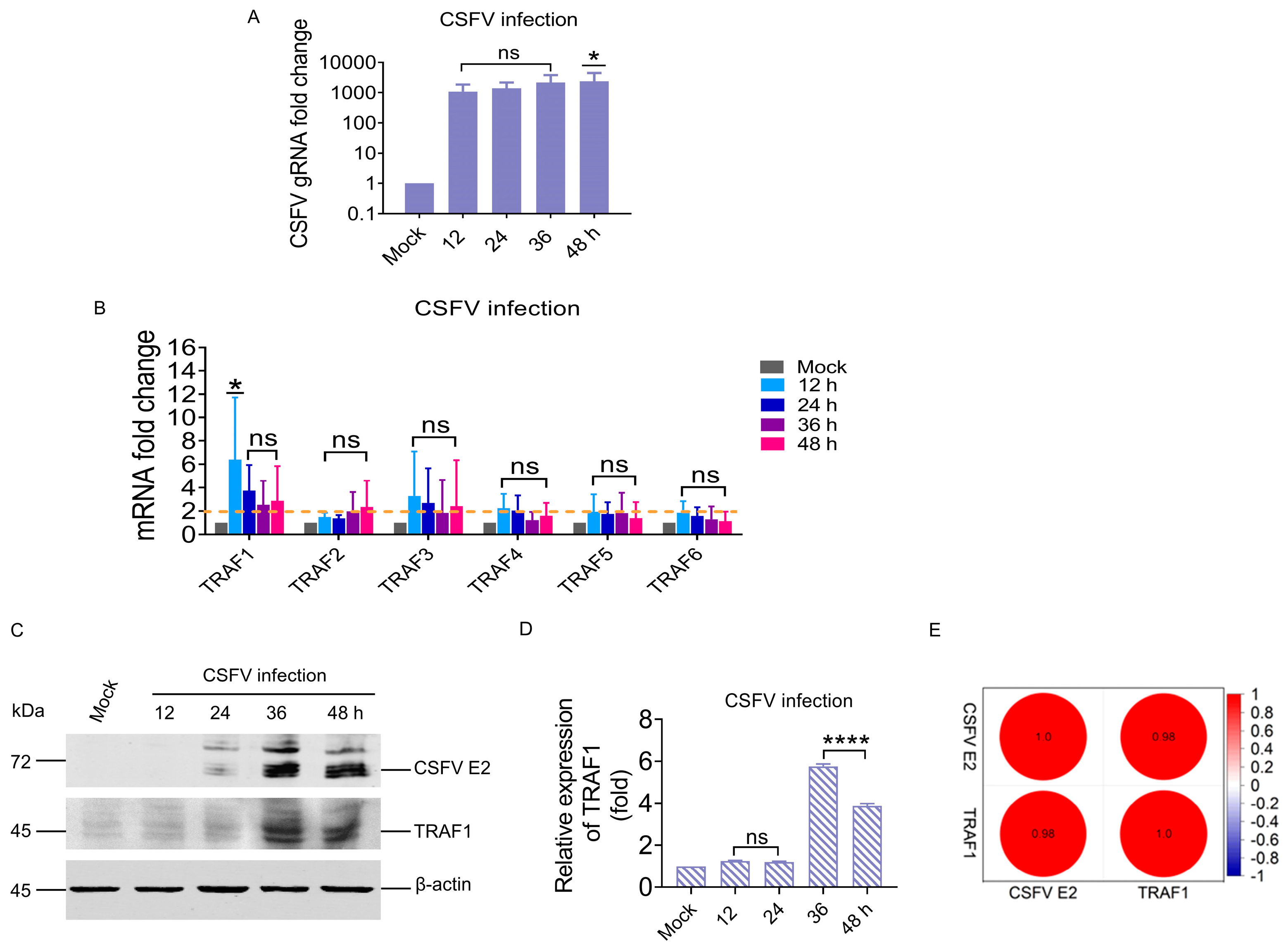
3.1. CSFV Infection Significantly Upregulated TRAF1 in the TRAF Family
3.2. TRAF1 Expression Positively Correlated with the RIG-I/MAVS Pathway during CSFV Infection
3.3. Interaction of TRAF1 with MAVS
3.4. TRAF1 Downregulation by siRNA-Enhanced CSFV Replication
3.5. TRAF1 Overexpression Inhibits CSFV Replication by the Positive Regulation of IRF1 and ISG15
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luo, Y.; Ji, S.; Liu, Y.; Lei, J.L.; Xia, S.L.; Wang, Y.; Du, M.L.; Shao, L.; Meng, X.Y.; Zhou, M.; et al. Isolation and Characterization of a Moderately Virulent Classical Swine Fever Virus Emerging in China. Transbound. Emerg. Dis. 2017, 64, 1848–1857. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Wu, K.; Zhao, M.; Yuan, J.; Ma, S.; Zhu, E.; Chen, Y.; Ding, H.; Yi, L.; Chen, J. LDHB inhibition induces mitophagy and facilitates the progression of CSFV infection. Autophagy 2021, 17, 2305–2324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, P.; Liu, Y.; Cheng, G. TRAF-mediated regulation of immune and inflammatory responses. Sci. China Life Sci. 2010, 53, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Park, H.H. Structure of TRAF Family: Current Understanding of Receptor Recognition. Front. Immunol. 2018, 9, 1999. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Liu, Y.; Jin, C.; Wang, X.; Song, W.; Zhang, Q. Genome-wide identification, characterization and expression profiling of TRAF family genes in Sebastes schlegelii. Fish Shellfish Immunol. 2022, 127, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Scheurich, P. Tumor necrosis factor receptor-associated factor (TRAF) 2 and its role in TNF signaling. Int. J. Biochem. Cell Biol. 2001, 33, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Ji, X.; Lv, Y.; Cui, D.; Xie, J. The Roles of TRAF3 in Immune Responses. Dis. Markers 2023, 1, 7787803. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Tian, X.; Zhang, J.; Wang, Z.; Chen, G. Roles of TRAF6 in Central Nervous System. Curr. Neuropharmacol. 2018, 16, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Chen, L.; Tian, S.; Lin, Y.; Tang, Q.; Zhou, X.; Li, D.; Yeung CK, L.; Che, T.; Jin, L.; et al. Comprehensive variation discovery and recovery of missing sequence in the pig genome using multiple de novo assemblies. Genome Res. 2017, 27, 865–874. [Google Scholar] [CrossRef]
- Kim, C.M.; Park, H.H. Comparison of Target Recognition by TRAF1 and TRAF2. Int. J. Mol. Sci. 2020, 21, 2895. [Google Scholar] [CrossRef]
- Chen, H.; Jiang, Z. The essential adaptors of innate immune signaling. Protein Cell 2013, 4, 27–39. [Google Scholar] [CrossRef]
- Kell, A.M.; Gale, M., Jr. RIG-I in RNA virus recognition. Virology 2015, 479–480, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hur, S. How RIG-I like receptors activate MAVS. Curr. Opin. Virol. 2015, 12, 91–98. [Google Scholar] [CrossRef]
- Seth, R.B.; Sun, L.; Ea, C.K.; Chen, Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 2005, 122, 669–682. [Google Scholar] [CrossRef]
- Bradley, J.R.; Pober, J.S. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 2001, 20, 6482–6491. [Google Scholar] [CrossRef] [PubMed]
- Edilova, M.I.; Abdul-Sater, A.A.; Watts, T.H. TRAF1 Signaling in Human Health and Disease. Front. Immunol. 2018, 9, 2969. [Google Scholar] [CrossRef]
- Wang, C.Y.; Mayo, M.W.; Korneluk, R.G.; Goeddel, D.V.; Baldwin, A.S., Jr. NF-kappaB antiapoptosis: Induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 1998, 281, 1680–1683. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Q.; Li, X.N.; Liang, J.J.; Cai, X.B.; Tao, Q.; Li, Y.X.; Qin, Q.; Xu, S.P.; Luo, T.R. IRF1 up-regulates isg15 gene expression in dsRNA stimulation or CSFV infection by targeting nucleotides-487 to-325 in the 5′ flanking region. Mol. Immunol. 2018, 94, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.B.; Hou, H.L.; Zhang, L.Y.; Tang, R.Z.; Shan, Y.; Li, X.Q.; Zhang, K.; Zhao, Q.N.; Ying, X.; Su, L.J.; et al. Preparation of swine ISG15 polyclonal antibody and its application in classical swine fever virus. Thai J. Vet. Med. 2020, 50, 417–429. [Google Scholar] [CrossRef]
- Zhao, Q.N.; Ying, X.; Li, X.Q.; Zhang, K.; Li, X.N.; Liang, J.J.; Li, X.N.; Luo, T.N. Construction of porcine mavs gene prokaryotic expression vector and preparation of polyclonal antibody. Chin. J. Vet. Med. 2017, 53, 39–42. [Google Scholar]
- Chai, M.; Xu, G.L.; Sun, W. Optimization of method of titration rabies virus with fluorescence focus units assay. Zhongguo Yi Miao He Mian Yi 2010, 16, 258–260. (In Chinese) [Google Scholar] [PubMed]
- Inoue, J.I.; Ishida, T.; Tsukamoto, N.; Kobayashi, N.; Naito, A.; Azuma, S.; Yamamoto, T. Tumor necrosis factor receptor-associated factor (TRAF) family: Adapter proteins that mediate cytokine signaling. Exp. Cell Res. 2000, 254, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Loo, Y.M.; Gale, M., Jr. Immune signaling by RIG-I-like receptors. Immunity 2011, 34, 680–692. [Google Scholar] [CrossRef] [PubMed]
- Chung, J.Y.; Park, Y.C.; Ye, H.; Wu, H. All TRAFs are not created equal: Common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 2002, 115, 679–688. [Google Scholar] [CrossRef] [PubMed]
- McPherson, A.J.; Snell, L.M.; Mak, T.W.; Watts, T.H. Opposing roles for TRAF1 in the alternative versus classical NF-κB pathway in T cells. J. Biol. Chem. 2012, 287, 23010–23019. [Google Scholar] [CrossRef] [PubMed]
- Sasai, M.; Tatematsu, M.; Oshiumi, H.; Funami, K.; Matsumoto, M.; Hatakeyama, S.; Seya, T. Direct binding of TRAF2 and TRAF6 to TICAM-1/TRIF adaptor participates in activation of the Toll-like receptor 3/4 pathway. Mol. Immunol. 2010, 47, 1283–1291. [Google Scholar] [CrossRef] [PubMed]
- Oussa, N.A.; Soumounou, Y.; Sabbagh, L. TRAF1 phosphorylation on Serine 139 modulates NF-κB activity downstream of 4-1BB in T cells. Biochem. Biophys. Res. Commun. 2013, 432, 129–134. [Google Scholar] [CrossRef]
- Kato, T., Jr.; Gotoh, Y.; Hoffmann, A.; Ono, Y. Negative regulation of constitutive NF-kappaB and JNK signaling by PKN1-mediated phosphorylation of TRAF1. Genes Cells 2008, 13, 509–520. [Google Scholar] [CrossRef]
- Arron, J.R.; Pewzner-Jung, Y.; Walsh, M.C.; Kobayashi, T.; Choi, Y. Regulation of the subcellular localization of tumor necrosis factor receptor-associated factor (TRAF)2 by TRAF1 reveals mechanisms of TRAF2 signaling. J. Exp. Med. 2002, 196, 923–934. [Google Scholar] [CrossRef] [PubMed]
- Matthys, V.S.; Cimica, V.; Dalrymple, N.A.; Glennon, N.B.; Bianco, C.; Mackow, E.R. Hantavirus GnT elements mediate TRAF3 binding and inhibit RIG-I/TBK1-directed beta interferon transcription by blocking IRF3 phosphorylation. J. Virol. 2014, 88, 2246–2259. [Google Scholar] [CrossRef]
- Lee, N.R.; Ban, J.; Lee, N.J.; Yi, C.M.; Choi, J.Y.; Kim, H.; Lee, J.K.; Seong, J.; Cho, N.H.; Jung, J.U.; et al. Activation of RIG-I-Mediated Antiviral Signaling Triggers Autophagy Through the MAVS-TRAF6-Beclin-1 Signaling Axis. Front. Immunol. 2018, 9, 2096. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Goeddel, D.V. TNF-R1 signaling: A beautiful pathway. Science 2002, 296, 1634–1635. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Kontodimas, K.; Bosmann, M. The MAVS Immune Recognition Pathway in Viral Infection and Sepsis. Antioxid. Redox Signal. 2021, 35, 1376–1392. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Ding, T.; Zuo, Z.; Xu, Z.; Deng, J.; Wei, Z. Regulation of MAVS Expression and Signaling Function in the Antiviral Innate Immune Response. Front. Immunol. 2020, 11, 1030. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Edilova, M.I.; Wagar, L.E.; Mujib, S.; Singer, M.; Bernard, N.F.; Croughs, T.; Lederman, M.M.; Sereti, I.; Fischl, M.A.; et al. Effect of IL-7 Therapy on Phospho-Ribosomal Protein S6 and TRAF1 Expression in HIV-Specific CD8 T Cells in Patients Receiving Antiretroviral Therapy. J. Immunol. 2018, 200, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Cubero, E.; Subirá, D.; Sanz-de-Villalobos, E.; Parra-Cid, T.; Madejón, A.; Miquel, J.; Olveira, A.; González-Praetorius, A.; García-Samaniego, J.; Larrubia, J.R. According to Hepatitis C Virus (HCV) Infection Stage, Interleukin-7 Plus 4-1BB Triggering Alone or Combined with PD-1 Blockade Increases TRAF1low HCV-Specific CD8+ Cell Reactivity. J. Virol. 2018, 92, e01443-17. [Google Scholar] [CrossRef] [PubMed]
- Eliopoulos, A.G.; Waites, E.R.; Blake, S.M.; Davies, C.; Murray, P.; Young, L.S. TRAF1 is a critical regulator of JNK signaling by the TRAF-binding domain of the Epstein-Barr virus-encoded latent infection membrane protein 1 but not CD40. J. Virol. 2003, 77, 1316–1328. [Google Scholar] [CrossRef]
- Akiyama, T.; Shiraishi, T.; Qin, J.; Konno, H.; Akiyama, N.; Shinzawa, M.; Miyauchi, M.; Takizawa, N.; Yanai, H.; Ohashi, H.; et al. Mitochondria-nucleus shuttling FK506-binding protein 51 interacts with TRAF proteins and facilitates the RIG-I-like receptor-mediated expression of type I IFN. PLoS ONE 2014, 9, e95992. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Fujinaga, K.; Gross, J.D.; Frankel, A.D. Enhanced NF-κB activation via HIV-1 Tat-TRAF6 cross-talk. Sci. Adv. 2024, 10, eadi4162. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhu, C.; Wang, T.; Jiang, H.; Ren, Y.; Zhang, Q.; Wu, K.; Liu, F.; Liu, Y.; Wu, J. GP73 represses host innate immune response to promote virus replication by facilitating MAVS and TRAF6 degradation. PLoS Pathog. 2017, 13, e1006321. [Google Scholar] [CrossRef]



| Primers | Sequence (5′-3′) | Purpose |
|---|---|---|
| CSFV-F | GCCATGCCCATAGTAGGACT | detection of CSFV gRNA |
| CSFV-R | GCTTCTGCTCACGTCGAACT | |
| RIG-I-F | CACATTTGCGGATATACAAC | detection of RIG-I mRNA |
| RIG-I-R | TCGGGCATGTTCATTGATAA | |
| MAVS-F | CTGGAGATTCTGCCTTACCTGT | detection of MAVS mRNA |
| MAVS-R | CAGATCCTCAGTGCCCCGAT | |
| TRAF1-F | AATGCAAAGGCGACGACACTCC | detection of TRAF1 mRNA |
| TRAF1-R | CCAACACCAGCAAAAGGGCATC | |
| TRAF2-F | CGCTCTTCTGCCCCGTCT | detection of TRAF2 mRNA |
| TRAF2-R | TAGAGCCCCGTCAGGTCCAC | |
| TRAF3-F | ACAGCGAGTCATAGACAGCC | detection of TRAF3 mRNA |
| TRAF3-R | TCCACGCTGCTCTTCATGCT | |
| TRAF4-F | GCAGCTTCAATGTGGTTCCCT | detection of TRAF4 mRNA |
| TRAF4-R | AAGTCACAGCCACAGAACTCG | |
| TRAF5-F | TCTTCAGCCAGCCCTTCTAC | detection of TRAF5 mRNA |
| TRAF5-R | TCCCCATTCAGGTACGCTCT | |
| TRAF6-F | CAAAGCCTGCATCATCAAGTC | detection of TRAF6 mRNA |
| TRAF6-R | ATTTGGACACTTCACCGTCAG | |
| IRF1-F | AGTCCAAGTCCAGCCGAGAT | detection of IRF1 mRNA |
| IRF1-R | TACTGATGGCACACGGTGAC | |
| ISG15-F | GCCTTCCAGCAGCGTCT | detection of ISG15 mRNA |
| ISG15-R | GCGTTGCTGCGACCCT | |
| GAPDH-F | TGGTGAAGGTCGGAGTGAAC | detection of GAPDH mRNA |
| GAPDH-R | GGAAGATGGTGATGGGATTTC | |
| TRAF1 siRNA-F | GCCUGCGGCUCUACCUGAATT | Knockdown of TRAF1 |
| TRAF1 siRNA-R | UUCAGGUAGAGCCGCAGGCTT |
| Protein Name | Fold Changes to Protein Levels of PK-15 Cells Infection with CSFV (MOI = 1) | |||
|---|---|---|---|---|
| 24 hpi | 36 hpi | 48 hpi | 72 hpi | |
| CSFV E2 | 27.9 | 32.6 | 30.1 | 15.4 |
| TRAF1 | 4.7 | 6.0 | 6.7 | 6.4 |
| RIG-I | 3.7 | 4.8 | 4.8 | 5.0 |
| MAVS | 7.5 | 8.8 | 8.6 | 8.4 |
| IRF1 | 6.7 | 11.0 | 11.8 | 10.5 |
| ISG15 | 6.1 | 8.0 | 8.8 | 8.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Tang, R.; Liang, D.; Wang, W.; Min, K.; Luo, T.; Li, X. Uncovering the Interaction between TRAF1 and MAVS in the RIG-I Pathway to Enhance the Upregulation of IRF1/ISG15 during Classical Swine Fever Virus Infection. Cells 2024, 13, 1165. https://doi.org/10.3390/cells13131165
Zhang L, Tang R, Liang D, Wang W, Min K, Luo T, Li X. Uncovering the Interaction between TRAF1 and MAVS in the RIG-I Pathway to Enhance the Upregulation of IRF1/ISG15 during Classical Swine Fever Virus Infection. Cells. 2024; 13(13):1165. https://doi.org/10.3390/cells13131165
Chicago/Turabian StyleZhang, Liyuan, Rongze Tang, Dongli Liang, Wenfeng Wang, Kaijun Min, Tingrong Luo, and Xiaoning Li. 2024. "Uncovering the Interaction between TRAF1 and MAVS in the RIG-I Pathway to Enhance the Upregulation of IRF1/ISG15 during Classical Swine Fever Virus Infection" Cells 13, no. 13: 1165. https://doi.org/10.3390/cells13131165
APA StyleZhang, L., Tang, R., Liang, D., Wang, W., Min, K., Luo, T., & Li, X. (2024). Uncovering the Interaction between TRAF1 and MAVS in the RIG-I Pathway to Enhance the Upregulation of IRF1/ISG15 during Classical Swine Fever Virus Infection. Cells, 13(13), 1165. https://doi.org/10.3390/cells13131165





