Conservation of cis-Regulatory Syntax Underlying Deuterostome Gastrulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Raw ATAC-Seq Data Retrieval
2.2. ATAC-Seq Analysis
2.3. Human Reactome Pathway Enrichment Analysis
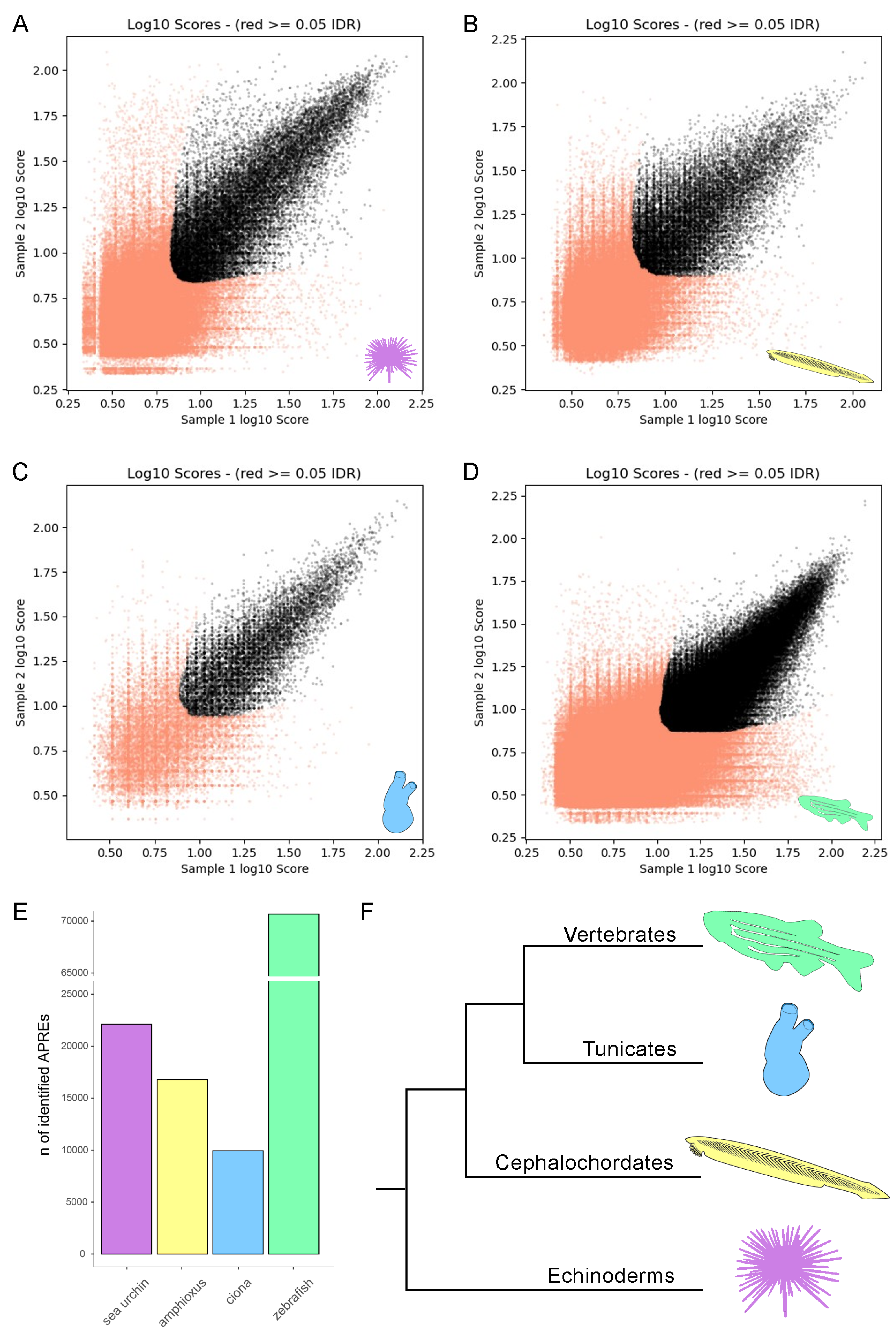
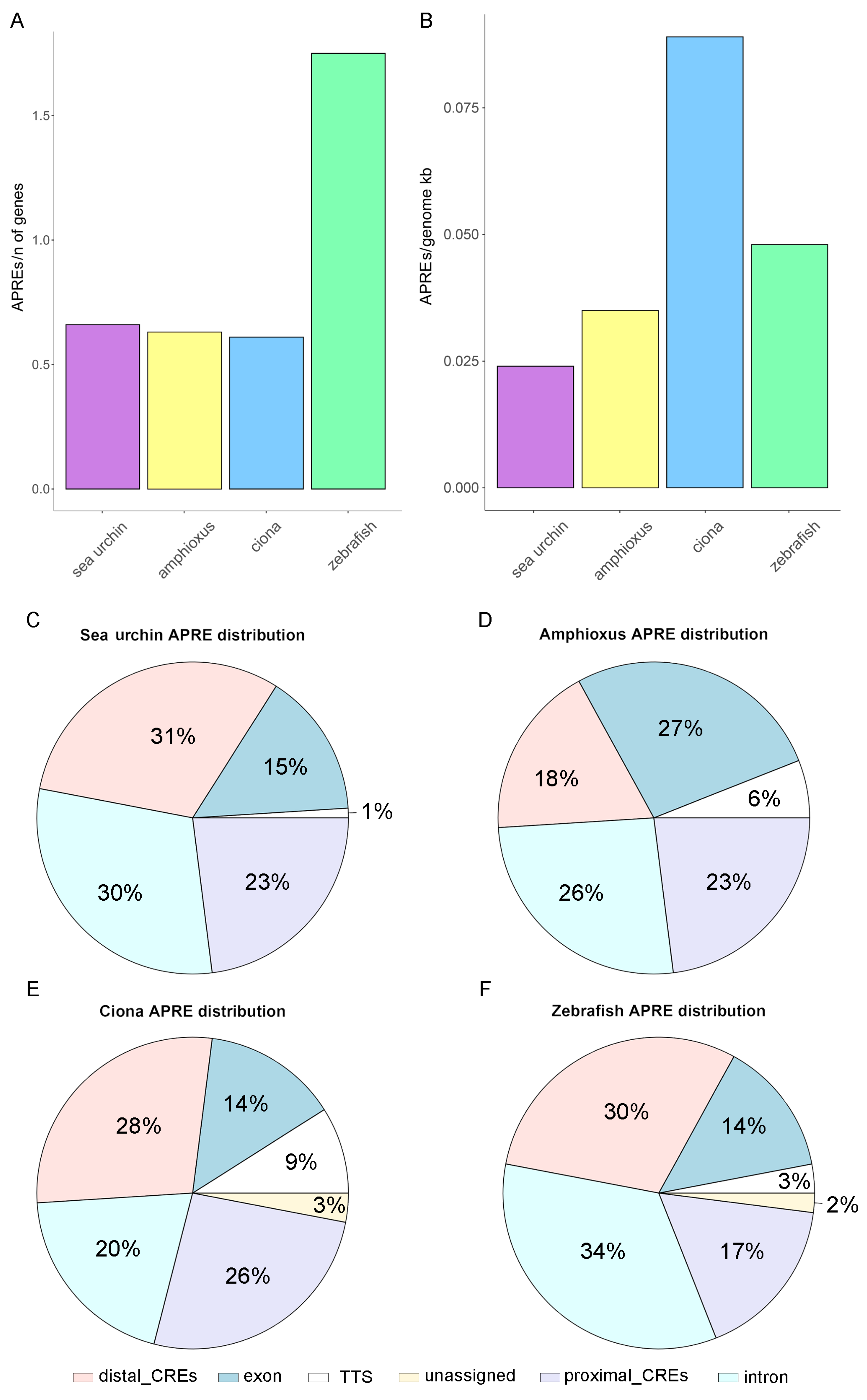
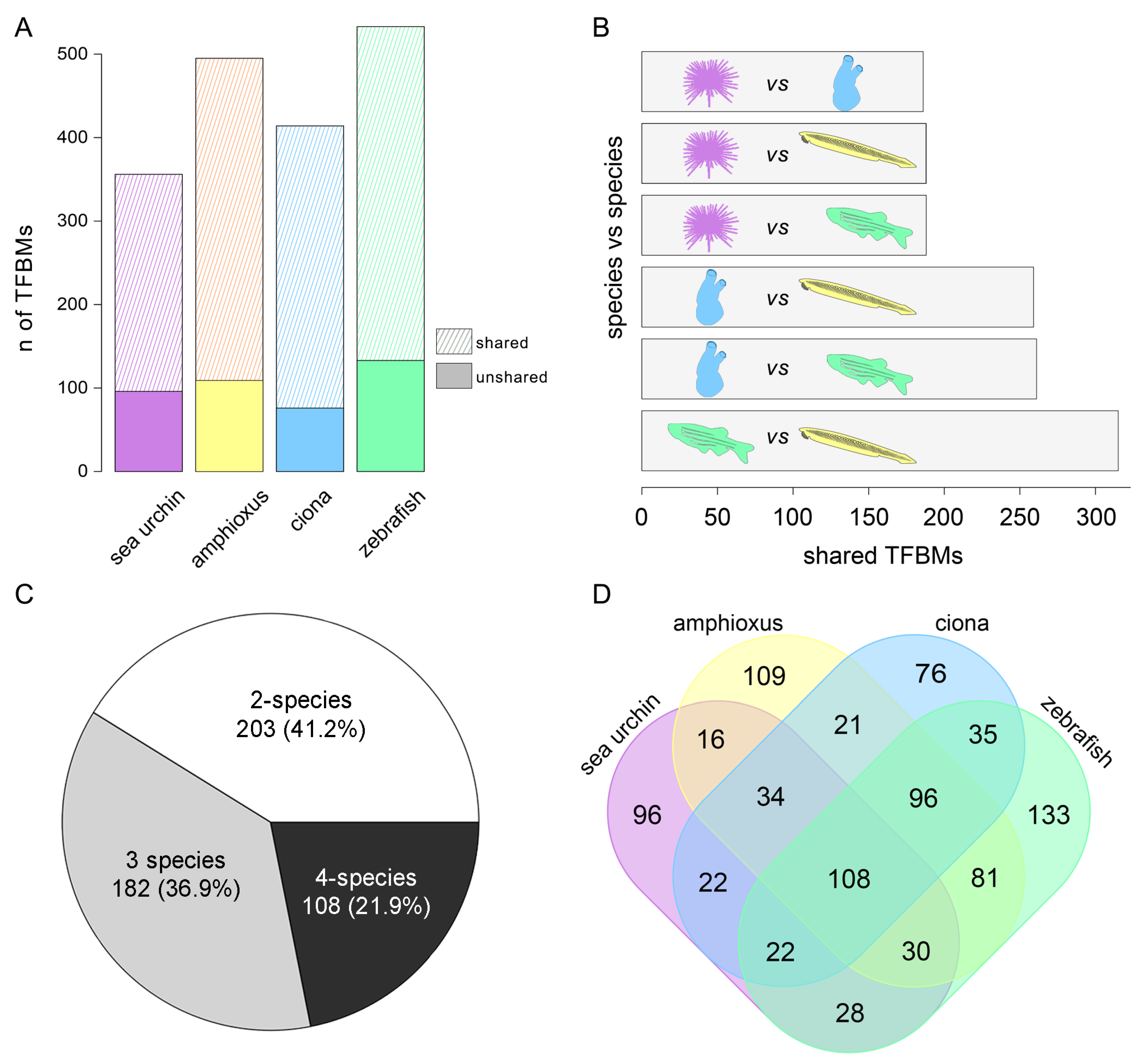
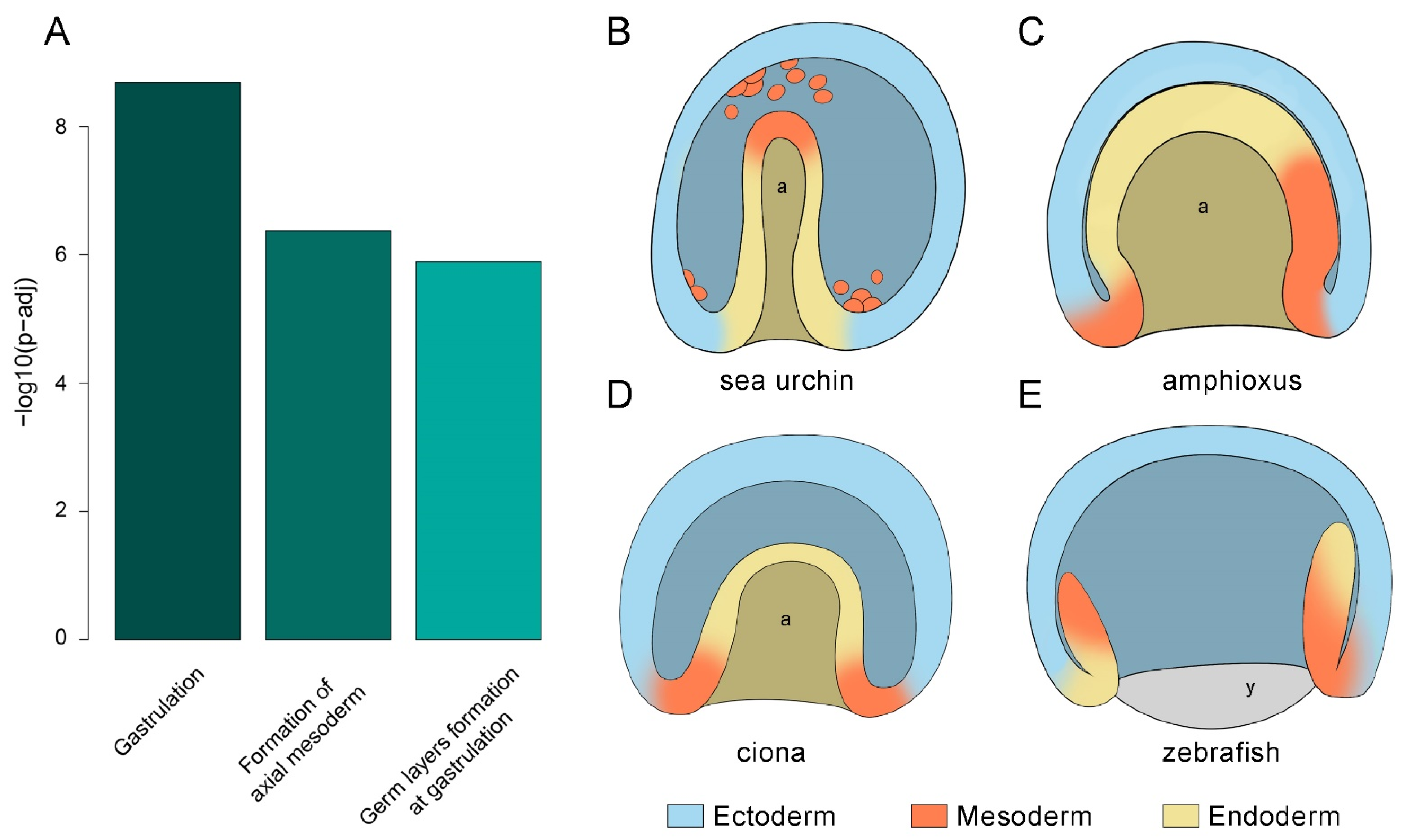
3. Results
3.1. Identification of Accessible Putative Regulatory Regions (APREs)
3.2. Characterization of the APRE Distribution
3.3. Transcription Factor Binding Motifs Analyses
3.4. Identification of a Conserved Core of Transcription Factor-Binding Motifs during Gastrulation
| Binding Motif Sequence | TF Family | TF | Germ Layer Pathway | Known Role in Gastrulation | Role in Organogenesis | Refs. |
|---|---|---|---|---|---|---|
| WAAGTAAACA | Forkhead | FoxA1 | Endoderm Mesoderm | Specification of anterior ectodermal program and repressor of posterior fates | Specification and differentiation of endodermal structures: gut, lungs, liver, kidneys, pancreas, prostate, notochord, nodes | [30,31,34,35,39,46,48,49,50,51,52,53] |
| CYTGTTTACWYW | FoxA2 | Ectoderm | ||||
| BSNTGTTTACWYWGN | FoxA3 | Endoderm Mesoderm | ||||
| NVWTGTTTAC | FoxK1 | Mesoderm | Myogenesis | |||
| SCHTGTTTACAT | FoxK2 | |||||
| WWTRTAAACAVG | FoxL2 | Mesoderm Ectoderm | Specification of ovaries and eyes | |||
| DGTAAACA | FoxO3 | Mesoderm | Vasculogenesis (blood vessels) | |||
| TRTTTACTTW | FoxM1 | Ectoderm | Activation of G2-M cell-cycle regulators | Neuronal differentiation | ||
| WWATRTAAACAN | FoxF1 | Mesoderm | Lung, liver, and gut development | |||
| KTGTTTGC | FoxJ2 | Angiogenesis, spermatogenesis, ciliogenesis | ||||
| CTGTTTAC | FoxO1 | Mesoderm | Vasculogenesis (blood vessels) | |||
| NYYTGTTTACHN | FoxP1 | Ectoderm | Specification of nervous system and heart | |||
| TTRAGTGSYK | Homeobox | Nkx3-2 | Endoderm Mesoderm | Neuroendoderm specification | Development of embryonic skeletal system and intestinal epithelium | [36,37,38,39,40,41,42,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68] |
| VNNGGATTADNN | Gcs | Endoderm Mesoderm | Head organizer, notochord formation, cell migration | Anterior brain induction, craniofacial development, head patterning | ||
| WACACGTAACTT | Irx3 | Ectoderm Mesoderm | Organizer, specification of neuronal progenitor cells of the spinal cord, anteroposterior patterning of neural axis | SHH-dependent neural patterning, development of inner ear, limbs, heart, kidneys, and facial/gill cartilage (regulated by the Wnt pathway) | ||
| NGTGTTCAVTSAAGCGKAAA | Pax6 | Ectoderm Mesoderm | Specification of neuronal progenitor cells of the spinal cord | Development of eyes, pancreas, nose, nervous system, and pituitary gland | ||
| RTGATTKATRGN | Pbx2 | Mesoderm | Development of hindbrain, tectum, retina, axial skeleton, and thyme | |||
| SCTGTCAVTCAV | Pknox1 | Mesoderm | Apoptosis, chick primitive streak formation, (novel) regulator of EMT | Adipogenesis, differentiation of hematopoietic precursor cells, hindbrain segmentation, head cartilage development | ||
| NYTAATCCYB | Otx2 | Ectoderm Endoderm | Specification of neuroectoderm and endoderm | Development of eyes, nose, ears, nervous system, first pharyngeal arch formation, and midbrain–hindbrain border (MHB) induction (regulated by retinoic acid signaling) | ||
| KACACGTCTCTY | bHLH | Hey2 | Mesoderm | Cardiovascular development (Notch signaling) | [44,45,69,70,71] | |
| VVCCACGTGG | c-Myc | Mesoderm Extraembryonic tissues | Cellular plasticity maintenance | |||
| VRCCACGTGG | n-Myc | |||||
| BAACAGCTGT | Myf5 | Mesoderm | Myogenesis | |||
| AACAGCTG | MyoG | |||||
| DGCACACGTG | Mnt | |||||
| WNBCACGTGA | Arntl1 | Chick primitive streak, mediates hypoxia-induced IGFBP-1 expression | Development of nervous system, optic vesicles, notochord, foregut, and somites | |||
| GHCACGTG | Clock | Chick primitive streak | Development of nervous system, optic vesicles, notochord, foregut, somites, and heart | |||
| KCCACGTGAC | Npas2 | |||||
| KCACGTGMCN | Dec2 | Prechordal plate | Neural crest cells | |||
| VTTACGTAAYNNNNN | bZip | Nfil3 | Immune system | [72,73] | ||
| RTTATGYAAB | Hlf | Clock-controlled gene, never associated with gastrulation | ||||
| VTGACTCATC | AP-1 | |||||
| GATGASTCATCN RATGASTCAT | JunB | Innate and adaptive immune responses, tumorigenesis | ||||
| NNATGASTCATH | Fra1 | Differentiation of adipocytes, chondrocytes, and osteoblasts; placental vascularization | ||||
| GGATGACTCATC NATGASTCABNN | FosL2 | Cell proliferation and differentiation | ||||
| DATGASTCATHN | Atf3 | Mesoderm | ||||
| DATGASTCAT | Batf | Differentiation of immune system cells | ||||
| NKGMCACGTGDCMNN | Creb3L2 | Chondrogenesis, regulation of the secretory pathway | ||||
| RRTSACGTSD | Usf2 | |||||
| BCCATTGTTC | HMG | Sox2 | Ectoderm Endoderm | Development of nervous system, progenitor state maintenance | Embryonic stem cell pluripotency; specification of nervous system and eyes | [43,47,74,75,76,77,78,79,80,81,82,83] |
| CCWTTGTY | Sox3 | Ectoderm Endoderm | Nervous system development, progenitor state maintenance | Formation of hypothalamic–pituitary axis, suppression of neuronal differentiation, craniofacial morphogenesis, sex determination | ||
| YCTTTGTTCC | Sox4 | Regulator of epithelial–mesenchymal transition (EMT) | Development of eyes, pancreas, and skeletal system; differentiation of noradrenergic neurons | |||
| CCATTGTTNY | Sox6 | Ectoderm Endoderm Mesoderm | Development of nervous system, chondrogenesis, maintenance of cardiac and skeletal muscle cells | |||
| CCWTTGTYYB | Sox10 | Specification of neural crest | Development of neural crest and peripheral nervous system; glia and melanocyte development | |||
| RAACAATGGN | Sox15 | Inhibition of myoblast differentiation, skeletal muscle regeneration, regulation of stem cell pluripotency, germline development | ||||
| AGGVNCCTTTGT | Sox9 | Endoderm | Specification of neural crest | Chondrocyte, otic placode and glial differentiation, development of skeletal system, inner ear, craniofacial, male sex determination | ||
| ASWTCAAAGG | Tcf3 | Endoderm | Axis specification, regulator of pluripotency | Neuronal differentiation, mesenchymal–epithelial transition (MET), eye development | ||
| ASATCAAAGGVA | Tcf4 | Mesoderm | Neuronal differentiation | |||
| CCTTTGATST | Lef1 | Ectoderm | ||||
| CTTTGATGTGSB | Tcf7 | Mesoderm | T-cell lymphocyte differentiation | |||
| CTGTCTGG | MAD | Smad2 | Endoderm | Specification of the anterior primitive streak, dorsoventral axis specification | [84,85,86,87] | |
| VBSYGTCTGG | Smad4 | Endoderm | Specification of the anterior primitive streak | Heart and skeletal muscle development | ||
| BGTTGACTWH | NR | Nr2E1 | Anterior brain differentiation, eye (retinal) development | [23] | ||
| CGTTGACTWW NCGTTGACTT | Nr2F1 | Neurogenesis (regulated by retinoic acid signaling) | ||||
| NNNTTGACYWNNNNN | Nr4A1 | Endoderm Mesoderm | Regeneration, immune response | |||
| YACGTMAY | Zinc Finger | Atf1 | Role in correct gastrulation | (regulated by Notch signaling) | [88,89,90] | |
| CCTGCTGAGH | Zic | Ectoderm | Left–right axis formation | Early neurogenesis (BMP signaling) | ||
| MAATCACTGC | Gfi1b | Mesoderm | Hematopoiesis | |||
| GGCGGCTG | Znf460 | |||||
| Several BMs | Yy1 Yy2 | Embryonic and extraembryonic tissues | Morphogenetic movements | Cardiac morphogenesis (regulated by nodal signaling) | ||
| HNACGCTCCT | Klf13 | Heart development |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Davidson, E.H. The Regulatory Genome; Elsevier: Amsterdam, The Netherlands, 2006; ISBN 9780120885633. [Google Scholar]
- Stern, D.L.; Orgogozo, V. The Loci of Evolution: How Predictable Is Genetic Evolution? Evolution 2008, 62, 2155–2177. [Google Scholar] [CrossRef]
- Hill, M.S.; Vande Zande, P.; Wittkopp, P.J. Molecular and Evolutionary Processes Generating Variation in Gene Expression. Nat. Rev. Genet. 2021, 22, 203–215. [Google Scholar] [CrossRef]
- Royo, J.L.; Maeso, I.; Irimia, M.; Gao, F.; Peter, I.S.; Lopes, C.S.; D’Aniello, S.; Casares, F.; Davidson, E.H.; Garcia-Fernández, J.; et al. Transphyletic Conservation of Developmental Regulatory State in Animal Evolution. Proc. Natl. Acad. Sci. USA 2011, 108, 14186–14191. [Google Scholar] [CrossRef]
- Peter, I.S.; Davidson, E.H. Evolution of Gene Regulatory Networks Controlling Body Plan Development. Cell 2011, 144, 970–985. [Google Scholar] [CrossRef]
- Hinman, V.F.; Yankura, K.A.; McCauley, B.S. Evolution of Gene Regulatory Network Architectures: Examples of Subcircuit Conservation and Plasticity between Classes of Echinoderms. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2009, 1789, 326–332. [Google Scholar] [CrossRef]
- Wolpert, L.; Skinner, D. The Triumph of the Embryo; Dover Publications, Inc.: Mineola, NY, USA, 1991; p. 211. [Google Scholar]
- Wanninger, A. Hox, Homology, and Parsimony: An Organismal Perspective. Semin. Cell Dev. Biol. 2024, 152–153, 16–23. [Google Scholar] [CrossRef]
- Moreau, C.; Caldarelli, P.; Rocancourt, D.; Roussel, J.; Denans, N.; Pourquie, O.; Gros, J. Timed Collinear Activation of Hox Genes during Gastrulation Controls the Avian Forelimb Position. Curr. Biol. 2019, 29, 35–50.e4. [Google Scholar] [CrossRef]
- Chen, K.; Rajewsky, N. The Evolution of Gene Regulation by Transcription Factors and MicroRNAs. Nat. Rev. Genet. 2007, 8, 93–103. [Google Scholar] [CrossRef]
- Marlétaz, F.; Firbas, P.N.; Maeso, I.; Tena, J.J.; Bogdanovic, O.; Perry, M.; Wyatt, C.D.R.; de la Calle-Mustienes, E.; Bertrand, S.; Burguera, D.; et al. Amphioxus Functional Genomics and the Origins of Vertebrate Gene Regulation. Nature 2018, 564, 64–70. [Google Scholar] [CrossRef]
- Madgwick, A.; Magri, M.S.; Dantec, C.; Gailly, D.; Fiuza, U.-M.; Guignard, L.; Hettinger, S.; Gomez-Skarmeta, J.L.; Lemaire, P. Evolution of Embryonic Cis-Regulatory Landscapes between Divergent Phallusia and Ciona Ascidians. Dev. Biol. 2019, 448, 71–87. [Google Scholar] [CrossRef]
- Skvortsova, K.; Bertrand, S.; Voronov, D.; Duckett, P.E.; Ross, S.E.; Magri, M.S.; Maeso, I.; Weatheritt, R.J.; Gómez Skarmeta, J.L.; Arnone, M.I.; et al. Active DNA Demethylation of Developmental Cis-Regulatory Regions Predates Vertebrate Origins. Sci. Adv. 2022, 8, 2258. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Brozovic, M.; Martin, C.; Dantec, C.; Dauga, D.; Mendez, M.; Simion, P.; Percher, M.; Laporte, B.; Scornavacca, C.; Di Gregorio, A.; et al. ANISEED 2015: A Digital Framework for the Comparative Developmental Biology of Ascidians. Nucleic Acids Res. 2016, 44, D808–D818. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Magri, M.S.; Jiménez-Gancedo, S.; Bertrand, S.; Madgwick, A.; Escrivà, H.; Lemaire, P.; Gómez-Skarmeta, J.L. Assaying Chromatin Accessibility Using ATAC-Seq in Invertebrate Chordate Embryos. Front. Cell Dev. Biol. 2020, 7, 500146. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-Based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Brown, J.B.; Huang, H.; Bickel, P.J. Measuring Reproducibility of High-Throughput Experiments. Ann. Appl. Stat. 2011, 5, 1752–1779. [Google Scholar] [CrossRef]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime Cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W. Quantifying Similarity between Motifs. Genome Biol. 2007, 8, R24. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. G:Profiler—Interoperable Web Service for Functional Enrichment Analysis and Gene Identifier Mapping (2023 Update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Love, C.E.; Prince, V.E. Expression and Retinoic Acid Regulation of the Zebrafish Nr2f Orphan Nuclear Receptor Genes. Dev. Dyn. 2012, 241, 1603–1615. [Google Scholar] [CrossRef] [PubMed]
- Satou, Y.; Nakamura, R.; Yu, D.; Yoshida, R.; Hamada, M.; Fujie, M.; Hisata, K.; Takeda, H.; Satoh, N. A Nearly Complete Genome of Ciona Intestinalis Type A (C. robusta) Reveals the Contribution of Inversion to Chromosomal Evolution in the Genus Ciona. Genome Biol. Evol. 2019, 11, 3144–3157. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, G.J.; Tao, J.; Westin, E.M.; Duttke, S.H.; Spann, N.J.; Strid, T.; Shen, Z.; Stender, J.D.; Sakai, M.; Link, V.M.; et al. Diverse Motif Ensembles Specify Non-Redundant DNA Binding Activities of AP-1 Family Members in Macrophages. Nat. Commun. 2019, 10, 414. [Google Scholar] [CrossRef] [PubMed]
- Rosanova, A.; Colliva, A.; Osella, M.; Caselle, M. Modelling the Evolution of Transcription Factor Binding Preferences in Complex Eukaryotes. Sci. Rep. 2017, 7, 7596. [Google Scholar] [CrossRef] [PubMed]
- Letelier, J.; Buono, L.; Almuedo-Castillo, M.; Zang, J.; Mounieres, C.; González-Díaz, S.; Polvillo, R.; Sanabria-Reinoso, E.; Corbacho, J.; Sousa-Ortega, A.; et al. Mutation of vsx Genes in Zebrafish Highlights the Robustness of the Retinal Specification Network. eLife 2023, 12, e85594. [Google Scholar] [CrossRef]
- Buono, L.; Corbacho, J.; Naranjo, S.; Almuedo-Castillo, M.; Moreno-Marmol, T.; de la Cerda, B.; Sanbria-Reinoso, E.; Polvillo, R.; Díaz-Corrales, F.J.; Bogdanovic, O.; et al. Analysis of Gene Network Bifurcation during Optic Cup Morphogenesis in Zebrafish. Nat. Commun. 2021, 12, 3866. [Google Scholar] [CrossRef] [PubMed]
- Milacic, M.; Beavers, D.; Conley, P.; Gong, C.; Gillespie, M.; Griss, J.; Haw, R.; Jassal, B.; Matthews, L.; May, B.; et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res. 2024, 52, D672–D678. [Google Scholar] [CrossRef]
- Ang, S.-L.; Rossant, J. HNF-3β Is Essential for Node and Notochord Formation in Mouse Development. Cell 1994, 78, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Lamy, C.; Rothbächer, U.; Caillol, D.; Lemaire, P. Ci-FoxA-a Is the Earliest Zygotic Determinant of the Ascidian Anterior Ectoderm and Directly Activates Ci-SFRP1/5. Development 2006, 133, 2835–2844. [Google Scholar] [CrossRef]
- Pei, W.; Noushmehr, H.; Costa, J.; Ouspenskaia, M.V.; Elkahloun, A.G.; Feldman, B. An Early Requirement for Maternal FoxH1 during Zebrafish Gastrulation. Dev. Biol. 2007, 310, 10–22. [Google Scholar] [CrossRef]
- Tu, Q.; Brown, C.T.; Davidson, E.H.; Oliveri, P. Sea Urchin Forkhead Gene Family: Phylogeny and Embryonic Expression. Dev. Biol. 2006, 300, 49–62. [Google Scholar] [CrossRef] [PubMed]
- Aldea, D.; Leon, A.; Bertrand, S.; Escriva, H. Expression of Fox Genes in the Cephalochordate Branchiostoma lanceolatum. Front. Ecol. Evol. 2015, 3, 153908. [Google Scholar] [CrossRef]
- Feuda, R.; Peter, I.S. Homologous Gene Regulatory Networks Control Development of Apical Organs and Brains in Bilateria. Sci. Adv. 2022, 8, 2416. [Google Scholar] [CrossRef] [PubMed]
- Roure, A.; Lemaire, P.; Darras, S. An Otx/Nodal Regulatory Signature for Posterior Neural Development in Ascidians. PLoS Genet. 2014, 10, e1004548. [Google Scholar] [CrossRef] [PubMed]
- Glardon, S.; Callaerts, P.; Halder, G.; Gehring, W.J. Conservation of Pax-6 in a Lower Chordate, the Ascidian Phallusia mammillata. Development 1997, 124, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Howard-Ashby, M.; Materna, S.C.; Brown, C.T.; Chen, L.; Cameron, R.A.; Davidson, E.H. Identification and Characterization of Homeobox Transcription Factor Genes in Strongylocentrotus purpuratus, and Their Expression in Embryonic Development. Dev. Biol. 2006, 300, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Saudemont, A.; Haillot, E.; Mekpoh, F.; Bessodes, N.; Quirin, M.; Lapraz, F.; Duboc, V.; Röttinger, E.; Range, R.; Oisel, A.; et al. Ancestral Regulatory Circuits Governing Ectoderm Patterning Downstream of Nodal and BMP2/4 Revealed by Gene Regulatory Network Analysis in an Echinoderm. PLoS Genet. 2010, 6, e1001259. [Google Scholar] [CrossRef] [PubMed]
- Yasuoka, Y.; Tando, Y.; Kubokawa, K.; Taira, M. Evolution of Cis-Regulatory Modules for the Head Organizer Gene Goosecoid in Chordates: Comparisons between Branchiostoma and Xenopus. Zool. Lett. 2019, 5, 27. [Google Scholar] [CrossRef] [PubMed]
- Onai, T.; Aramaki, T.; Inomata, H.; Hirai, T.; Kuratani, S. Ancestral Mesodermal Reorganization and Evolution of the Vertebrate Head. Zool. Lett. 2015, 1, 29. [Google Scholar] [CrossRef]
- Vlachakis, N.; Ellstrom, D.R.; Sagerström, C.G. A Novel Pbx Family Member Expressed during Early Zebrafish Embryogenesis Forms Trimeric Complexes with Meis3 and Hoxb1b. Dev. Dyn. 2000, 217, 109–119. [Google Scholar] [CrossRef]
- Poustka, A.J.; Kühn, A.; Groth, D.; Weise, V.; Yaguchi, S.; Burke, R.D.; Herwig, R.; Lehrach, H.; Panopoulou, G. A Global View of Gene Expression in Lithium and Zinc Treated Sea Urchin Embryos: New Components of Gene Regulatory Networks. Genome Biol. 2007, 8, R85. [Google Scholar] [CrossRef] [PubMed]
- Bellmeyer, A.; Krase, J.; Lindgren, J.; LaBonne, C. The Protooncogene C-Myc Is an Essential Regulator of Neural Crest Formation in Xenopus. Dev. Cell 2003, 4, 827–839. [Google Scholar] [CrossRef] [PubMed]
- Osborn, D.P.S.; Li, K.; Cutty, S.J.; Nelson, A.C.; Wardle, F.C.; Hinits, Y.; Hughes, S.M. Fgf-Driven Tbx Protein Activities Directly Induce Myf5 and Myod to Initiate Zebrafish Myogenesis. Development 2020, 147, dev184689. [Google Scholar] [CrossRef] [PubMed]
- Golson, M.L.; Kaestner, K.H. Fox Transcription Factors: From Development to Disease. Development 2016, 143, 4558–4570. [Google Scholar] [CrossRef]
- Lefebvre, V.; Dumitriu, B.; Penzo-Méndez, A.; Han, Y.; Pallavi, B. Control of Cell Fate and Differentiation by Sry-Related High-Mobility-Group Box (Sox) Transcription Factors. Int. J. Biochem. Cell Biol. 2007, 39, 2195–2214. [Google Scholar] [CrossRef]
- Friedman, J.R.; Kaestner, K.H. The Foxa Family of Transcription Factors in Development and Metabolism. Cell. Mol. Life Sci. 2006, 63, 2317–2328. [Google Scholar] [CrossRef]
- Pohl, B.S.; Rossner, A.; Knochel, W. The Fox Gene Family in Xenopus Laevis:FoxI2, FoxM1 and FoxP1 in Early Development. Int. J. Dev. Biol. 2005, 49, 53–58. [Google Scholar] [CrossRef]
- Pohl, B.S.; Schön, C.; Rößner, A.; Knöchel, W. The FoxO-Subclass in Xenopus laevis Development. Gene Expr. Patterns 2004, 5, 187–192. [Google Scholar] [CrossRef]
- Ueno, H.; Nakajo, N.; Watanabe, M.; Isoda, M.; Sagata, N. FoxM1-Driven Cell Division Is Required for Neuronal Differentiation in Early Xenopus Embryos. Development 2008, 135, 2023–2030. [Google Scholar] [CrossRef]
- Xie, X.; Liu, J.-X.; Hu, B.; Xiao, W. Zebrafish foxo3b Negatively Regulates Canonical Wnt Signaling to Affect Early Embryogenesis. PLoS ONE 2011, 6, e24469. [Google Scholar] [CrossRef]
- Seiliez, I.; Thisse, B.; Thisse, C. FoxA3 and Goosecoid Promote Anterior Neural Fate through Inhibition of Wnt8a Activity before the Onset of Gastrulation. Dev. Biol. 2006, 290, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Glardon, S.; Holland, L.Z.; Gehring, W.J.; Holland, N.D. Isolation and Developmental Expression of the Amphioxus Pax-6 Gene (AmphiPax-6): Insights into Eye and Photoreceptor Evolution. Development 1998, 125, 2701–2710. [Google Scholar] [CrossRef]
- Kumar, V.; Umair, Z.; Lee, U.; Kim, J. Two Homeobox Transcription Factors, Goosecoid and Ventx1.1, Oppositely Regulate Chordin Transcription in Xenopus Gastrula Embryos. Cells 2023, 12, 874. [Google Scholar] [CrossRef] [PubMed]
- Neidert, A.H.; Panopoulou, G.; Langeland, J.A. Amphioxus goosecoid and the Evolution of the Head Organizer and Prechordal Plate. Evol. Dev. 2000, 2, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Stachel, S.E.; Grunwald, D.J.; Myers, P.Z. Lithium Perturbation and goosecoid Expression Identify a Dorsal Specification Pathway in the Pregastrula Zebrafish. Development 1993, 117, 1261–1274. [Google Scholar] [CrossRef] [PubMed]
- Yasuo, H.; Lemaire, P. Role of Goosecoid, Xnot and Wnt Antagonists in the Maintenance of the Notochord Genetic Programme in Xenopus Gastrulae. Development 2001, 128, 3783–3793. [Google Scholar] [CrossRef] [PubMed]
- Bosse, A.; Zülch, A.; Becker, M.-B.; Torres, M.; Gómez-Skarmeta, J.L.; Modolell, J.; Gruss, P. Identification of the Vertebrate Iroquois Homeobox Gene Family with Overlapping Expression during Early Development of the Nervous System. Mech. Dev. 1997, 69, 169–181. [Google Scholar] [CrossRef]
- Kudoh, T.; Tsang, M.; Hukriede, N.A.; Chen, X.; Dedekian, M.; Clarke, C.J.; Kiang, A.; Schultz, S.; Epstein, J.A.; Toyama, R.; et al. A Gene Expression Screen in Zebrafish Embryogenesis. Genome Res. 2001, 11, 1979–1987. [Google Scholar] [CrossRef]
- Wei, Z.; Angerer, R.C.; Angerer, L.M. Direct Development of Neurons within Foregut Endoderm of Sea Urchin Embryos. Proc. Natl. Acad. Sci. USA 2011, 108, 9143–9147. [Google Scholar] [CrossRef]
- Acampora, D.; Mazan, S.; Lallemand, Y.; Avantaggiato, V.; Maury, M.; Simeone, A.; Brûlet, P. Forebrain and Midbrain Regions Are Deleted in Otx2−/− Mutants Due to a Defective Anterior Neuroectoderm Specification during Gastrulation. Development 1995, 121, 3279–3290. [Google Scholar] [CrossRef]
- Oda-Ishii, I.; Bertrand, V.; Matsuo, I.; Lemaire, P.; Saiga, H. Making Very Similar Embryos with Divergent Genomes: Conservation of Regulatory Mechanisms of Otx between the Ascidians Halocynthia roretzi and Ciona intestinalis. Development 2005, 132, 1663–1674. [Google Scholar] [CrossRef]
- Tour, E.; Pillemer, G.; Gruenbaum, Y.; Fainsod, A. Otx2 Can Activate the Isthmic Organizer Genetic Network in the Xenopus Embryo. Mech. Dev. 2002, 110, 3–13. [Google Scholar] [CrossRef]
- Yankura, K.A.; Martik, M.L.; Jennings, C.K.; Hinman, V.F. Uncoupling of Complex Regulatory Patterning during Evolution of Larval Development in Echinoderms. BMC Biol. 2010, 8, 143. [Google Scholar] [CrossRef]
- Deflorian, G.; Tiso, N.; Ferretti, E.; Meyer, D.; Blasi, F.; Bortolussi, M.; Argenton, F. Prep1.1 Has Essential Genetic Functions in Hindbrain Development and Cranial Neural Crest Cell Differentiation. Development 2004, 131, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Diaz, L.C.; Laurent, A.; Girasoli, S.; Turco, M.; Longobardi, E.; Iotti, G.; Jenkins, N.A.; Fiorenza, M.T.; Copeland, N.G.; Blasi, F. The Absence of Prep1 Causes P53-Dependent Apoptosis of Mouse Pluripotent Epiblast Cells. Development 2010, 137, 3393–3403. [Google Scholar] [CrossRef] [PubMed]
- Risolino, M.; Mandia, N.; Iavarone, F.; Dardaei, L.; Longobardi, E.; Fernandez, S.; Talotta, F.; Bianchi, F.; Pisati, F.; Spaggiari, L.; et al. Transcription Factor PREP1 Induces EMT and Metastasis by Controlling the TGF-β–SMAD3 Pathway in Non-Small Cell Lung Adenocarcinoma. Proc. Natl. Acad. Sci. USA 2014, 111, E3775–E3784. [Google Scholar] [CrossRef]
- Downs, K.M.; Martin, G.R.; Bishop, J.M. Contrasting Patterns of Myc and N-Myc Expression during Gastrulation of the Mouse Embryo. Genes. Dev. 1989, 3, 860–869. [Google Scholar] [CrossRef]
- Gonçalves, L.; Vinhas, M.; Pereira, R.; Pais De Azevedo, T.; Bajanca, F.; Palmeirim, I. Circadian Clock Genes Bmal1 and Clock during Early Chick Development. Dev. Dyn. 2012, 241, 1365–1373. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhou, J.; Xu, H.; Xu, G.; Xue, J. Identification and Developmental Expression of Dec2 in Zebrafish. Fish. Physiol. Biochem. 2010, 36, 667–675. [Google Scholar] [CrossRef]
- Gaitán-Espitia, J.D.; Hofmann, G.E. Gene Expression Profiling during the Embryo-to-larva Transition in the Giant Red Sea Urchin Mesocentrotus franciscanus. Ecol. Evol. 2017, 7, 2798–2811. [Google Scholar] [CrossRef]
- Imai, K.S.; Hino, K.; Yagi, K.; Satoh, N.; Satou, Y. Gene Expression Profiles of Transcription Factors and Signaling Molecules in the Ascidian Embryo: Towards a Comprehensive Understanding of Gene Networks. Development 2004, 131, 4047–4058. [Google Scholar] [CrossRef]
- Meulemans, D.; Bronner-Fraser, M. The Amphioxus SoxB Family: Implications for the Evolution of Vertebrate Placodes. Int. J. Biol. Sci. 2007, 3, 356–364. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cizelsky, W.; Hempel, A.; Metzig, M.; Tao, S.; Hollemann, T.; Kühl, M.; Kühl, S.J. Sox4 and Sox11 Function during Xenopus laevis Eye Development. PLoS ONE 2013, 8, e69372. [Google Scholar] [CrossRef] [PubMed]
- Saint-Germain, N.; Lee, Y.-H.; Zhang, Y.; Sargent, T.D.; Saint-Jeannet, J.-P. Specification of the Otic Placode Depends on Sox9 Function in Xenopus. Development 2004, 131, 1755–1763. [Google Scholar] [CrossRef] [PubMed]
- Aoki, Y.; Saint-Germain, N.; Gyda, M.; Magner-Fink, E.; Lee, Y.-H.; Credidio, C.; Saint-Jeannet, J.-P. Sox10 Regulates the Development of Neural Crest-Derived Melanocytes in Xenopus. Dev. Biol. 2003, 259, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Vonica, A.; Weng, W.; Gumbiner, B.M.; Venuti, J.M. TCF Is the Nuclear Effector of the β-Catenin Signal That Patterns the Sea Urchin Animal–Vegetal Axis. Dev. Biol. 2000, 217, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Esposito, R.; Racioppi, C.; Pezzotti, M.R.; Branno, M.; Locascio, A.; Ristoratore, F.; Spagnuolo, A. The Ascidian Pigmented Sensory Organs: Structures and Developmental Programs. Genesis 2015, 53, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Merrill, B.J.; Pasolli, H.A.; Polak, L.; Rendl, M.; García-García, M.J.; Anderson, K.V.; Fuchs, E. Tcf3: A Transcriptional Regulator of Axis Induction in the Early Embryo. Development 2004, 131, 263–274. [Google Scholar] [CrossRef]
- Gregorieff, A.; Grosschedl, R.; Clevers, H. Hindgut Defects and Transformation of the Gastro-Intestinal Tract in Tcf4−/−/Tcf1−/− Embryos. EMBO J. 2004, 23, 1825–1833. [Google Scholar] [CrossRef]
- Onai, T.; Lin, H.-C.; Schubert, M.; Koop, D.; Osborne, P.W.; Alvarez, S.; Alvarez, R.; Holland, N.D.; Holland, L.Z. Retinoic Acid and Wnt/β-Catenin Have Complementary Roles in Anterior/Posterior Patterning Embryos of the Basal Chordate Amphioxus. Dev. Biol. 2009, 332, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Wood, H.B.; Episkopou, V. Comparative Expression of the Mouse Sox1, Sox2 and Sox3 Genes from Pre-Gastrulation to Early Somite Stages. Mech. Dev. 1999, 86, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Chu, G.C.; Dunn, N.R.; Anderson, D.C.; Oxburgh, L.; Robertson, E.J. Differential Requirements for Smad4 in TGFβ-Dependent Patterning of the Early Mouse Embryo. Development 2004, 131, 3501–3512. [Google Scholar] [CrossRef] [PubMed]
- Müller, F.; Blader, P.; Rastegar, S.; Fischer, N.; Knöchel, W.; Strähle, U. Characterization of Zebrafish Smad1, Smad2 and Smad5: The Amino-Terminus of Smad1 and Smad5 Is Required for Specific Function in the Embryo. Mech. Dev. 1999, 88, 73–88. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, J.; Zeng, Z.; Qiao, L.; Zhuang, L.; Jiang, L.; Wei, J.; Ma, Q.; Wu, M.; Ye, S.; et al. Smad4 Is Required for the Development of Cardiac and Skeletal Muscle in Zebrafish. Differentiation 2016, 92, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Dunn, N.R.; Vincent, S.D.; Oxburgh, L.; Robertson, E.J.; Bikoff, E.K. Combinatorial Activities of Smad2 and Smad3 Regulate Mesoderm Formation and Patterning in the Mouse Embryo. Development 2004, 131, 1717–1728. [Google Scholar] [CrossRef] [PubMed]
- Grinblat, Y.; Sive, H. Zic Gene Expression Marks Anteroposterior Pattern in the Presumptive Neurectoderm of the Zebrafish Gastrula. Dev. Dyn. 2001, 222, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Merzdorf, C.S. Emerging Roles for Zic Genes in Early Development. Dev. Dyn. 2007, 236, 922–940. [Google Scholar] [CrossRef] [PubMed]
- Trask, M.C.; Tremblay, K.D.; Mager, J. Yin-Yang1 Is Required for Epithelial-to-Mesenchymal Transition and Regulation of Nodal Signaling during Mammalian Gastrulation. Dev. Biol. 2012, 368, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Berná, L.; Alvarez-Valin, F. Evolutionary Genomics of Fast Evolving Tunicates. Genome Biol. Evol. 2014, 6, 1724–1738. [Google Scholar] [CrossRef]
- Brasó-Vives, M.; Marlétaz, F.; Echchiki, A.; Mantica, F.; Acemel, R.D.; Gómez-Skarmeta, J.L.; Hartasánchez, D.A.; Le Targa, L.; Pontarotti, P.; Tena, J.J.; et al. Parallel Evolution of Amphioxus and Vertebrate Small-Scale Gene Duplications. Genome Biol. 2022, 23, 243. [Google Scholar] [CrossRef]
- Delsuc, F.; Philippe, H.; Tsagkogeorga, G.; Simion, P.; Tilak, M.-K.; Turon, X.; López-Legentil, S.; Piette, J.; Lemaire, P.; Douzery, E.J.P. A Phylogenomic Framework and Timescale for Comparative Studies of Tunicates. BMC Biol. 2018, 16, 39. [Google Scholar] [CrossRef] [PubMed]
- Erwin, D.H.; Laflamme, M.; Tweedt, S.M.; Sperling, E.A.; Pisani, D.; Peterson, K.J. The Cambrian Conundrum: Early Divergence and Later Ecological Success in the Early History of Animals. Science 2011, 334, 1091–1097. [Google Scholar] [CrossRef]
- Bonifer, C.; Cockerill, P.N. Chromatin Priming of Genes in Development: Concepts, Mechanisms and Consequences. Exp. Hematol. 2017, 49, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vallone, D.; Lahiri, K.; Dickmeis, T.; Foulkes, N.S. Start the Clock! Circadian Rhythms and Development. Dev. Dyn. 2007, 236, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Green, C.B.; Durston, A.J.; Morgan, R. The Circadian Gene Clock Is Restricted to the Anterior Neural Plate Early in Development and Is Regulated by the Neural Inducer Noggin and the Transcription Factor Otx2. Mech. Dev. 2001, 101, 105–110. [Google Scholar] [CrossRef]
- Wu, F.; Olson, B.G.; Yao, J. DamID-Seq: Genome-Wide Mapping of Protein-DNA Interactions by High Throughput Sequencing of Adenine-Methylated DNA Fragments. J. Vis. Exp. 2016, 2016, e53620. [Google Scholar] [CrossRef]
| Species | Genome Assembly | Genome Size |
|---|---|---|
| Strongylocentrotus purpuratus | GCF_000002235.5 | 9.21 × 108 |
| Branchiostoma lanceolatum | GCA_927797965.1 | 4.74 × 108 |
| Ciona intestinalis | GCA_000224145.1 | 1.12 × 108 |
| Danio rerio | GCF_000002035.5 | 1.46 × 109 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buono, L.; Annona, G.; Magri, M.S.; Negueruela, S.; Sepe, R.M.; Caccavale, F.; Maeso, I.; Arnone, M.I.; D’Aniello, S. Conservation of cis-Regulatory Syntax Underlying Deuterostome Gastrulation. Cells 2024, 13, 1121. https://doi.org/10.3390/cells13131121
Buono L, Annona G, Magri MS, Negueruela S, Sepe RM, Caccavale F, Maeso I, Arnone MI, D’Aniello S. Conservation of cis-Regulatory Syntax Underlying Deuterostome Gastrulation. Cells. 2024; 13(13):1121. https://doi.org/10.3390/cells13131121
Chicago/Turabian StyleBuono, Lorena, Giovanni Annona, Marta Silvia Magri, Santiago Negueruela, Rosa Maria Sepe, Filomena Caccavale, Ignacio Maeso, Maria Ina Arnone, and Salvatore D’Aniello. 2024. "Conservation of cis-Regulatory Syntax Underlying Deuterostome Gastrulation" Cells 13, no. 13: 1121. https://doi.org/10.3390/cells13131121
APA StyleBuono, L., Annona, G., Magri, M. S., Negueruela, S., Sepe, R. M., Caccavale, F., Maeso, I., Arnone, M. I., & D’Aniello, S. (2024). Conservation of cis-Regulatory Syntax Underlying Deuterostome Gastrulation. Cells, 13(13), 1121. https://doi.org/10.3390/cells13131121







