Analysis of Virus-Specific B Cell Epitopes Reveals Extensive Antigen Degradation Prior to Recognition
Abstract
1. Introduction
2. Materials and Methods
2.1. B Cell Epitope Data Collection and Processing
2.2. Antigen Annotation
2.3. Determining the Solvent Accessibility of B Cell Epitopes
2.4. Graphics and Statistics
3. Results
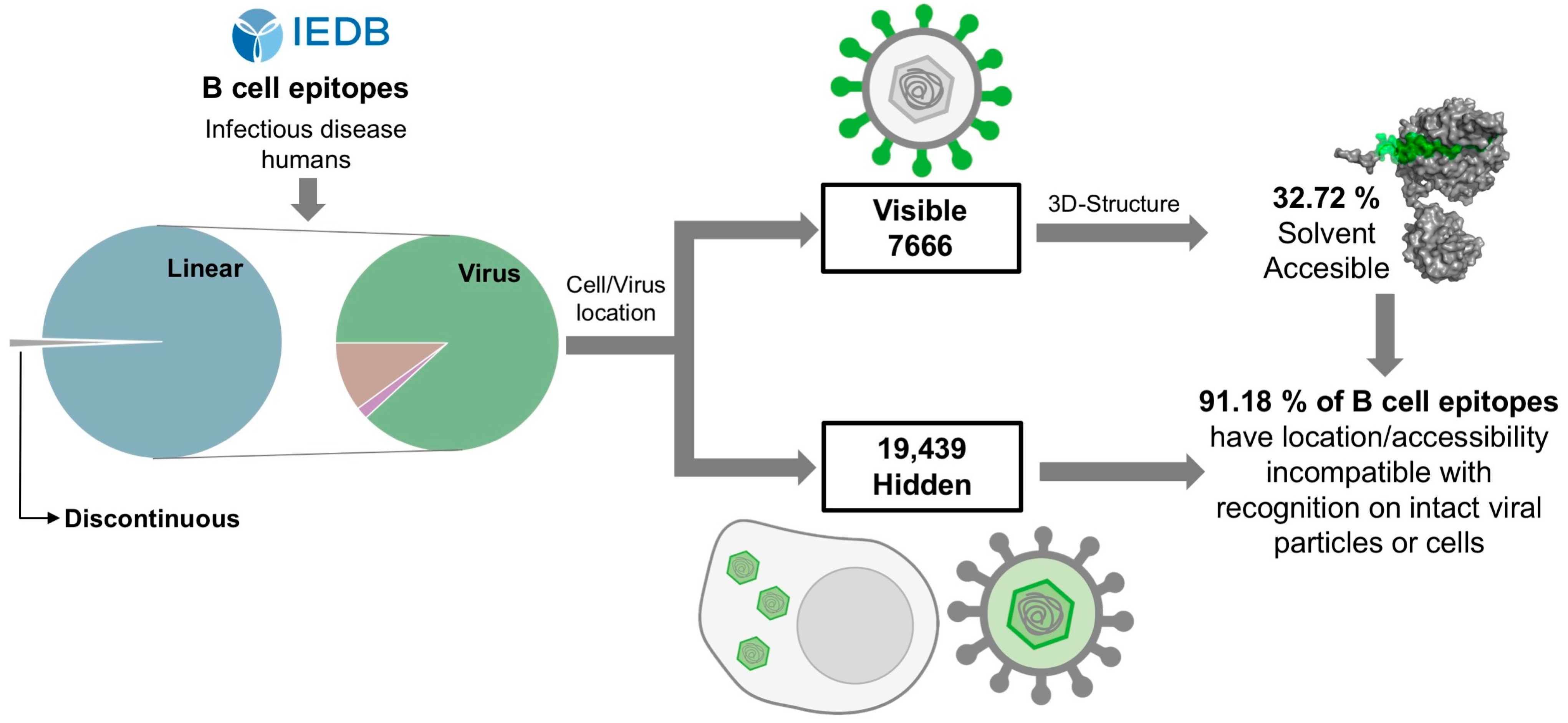
3.1. Characterization of B Cell Epitopes Targeted during the Course of a Natural Infection
3.2. Analysis of the Visibility of Viral Antigens Encompassing Linear B Cell Epitopes
3.3. Most Linear B Cell Epitopes Are Not Solvent Accessible in Native Antigens
4. Discussion
5. Conclusions and Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abbas, A.K.; Lichtman, A.H.; Pillai, S.; Baker, D.L.; Baker, A. Cellular and Molecular Immunology; Elsevier: Amsterdam, The Netherlands, 2018; ISBN 9780323479783/0323479782/9780323523240/0323523242. [Google Scholar]
- Sanchez-Trincado, J.L.; Gomez-Perosanz, M.; Reche, P.A. Fundamentals and Methods for T- and B-Cell Epitope Prediction. J. Immunol. Res. 2017, 2017, 2680160. [Google Scholar] [CrossRef] [PubMed]
- Van Regenmortel, M.H.V. What is a B-cell epitope. In Methods in Molecular Biology; Schutkowski, M., Reineke, U., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 524, pp. 3–20. ISBN 9781934115176. [Google Scholar]
- Ferdous, S.; Kelm, S.; Baker, T.S.; Shi, J.; Martin, A.C.R. B-cell epitopes: Discontinuity and conformational analysis. Mol. Immunol. 2019, 114, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Nilvebrant, J.; Rockberg, J. An Introduction to Epitope Mapping. Methods Mol. Biol. 2018, 1785, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.B.; Buus, S.; Schafer-Nielsen, C. Identification and mapping of linear antibody epitopes in human serum albumin using high-density Peptide arrays. PLoS ONE 2013, 8, e68902. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, L.C.; Kuo, H.-Y.; Mrksich, M. Peptide Arrays: Development and Application. Anal. Chem. 2018, 90, 266–282. [Google Scholar] [CrossRef] [PubMed]
- Salimi, N.; Fleri, W.; Peters, B.; Sette, A. Design and utilization of epitope-based databases and predictive tools. Immunogenetics 2010, 62, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Overton, J.A.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A.; et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015, 43, D405–D412. [Google Scholar] [CrossRef] [PubMed]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef]
- Ras-Carmona, A.; Gomez-Perosanz, M.; Reche, P.A. Prediction of unconventional protein secretion by exosomes. BMC Bioinform. 2021, 22, 333. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Ras-Carmona, A.; Pelaez-Prestel, H.F.; Lafuente, E.M.; Reche, P.A. BCEPS: A Web Server to Predict Linear B Cell Epitopes with Enhanced Immunogenicity and Cross-Reactivity. Cells 2021, 10, 2744. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, S.; Martin, A.C.R. AbDb: Antibody structure database-a database of PDB-derived antibody structures. Database 2018, 2018, bay040. [Google Scholar] [CrossRef] [PubMed]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.; Building, M.S. NACCESS, Computer Program; University College London: London, UK, 1993. [Google Scholar]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Yang, C.; Xu, X.; Xu, W.; Liu, S. Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020, 41, 1141–1149. [Google Scholar] [CrossRef] [PubMed]
- McAuley, J.L.; Gilbertson, B.P.; Trifkovic, S.; Brown, L.E.; McKimm-Breschkin, J.L. Influenza Virus Neuraminidase Structure and Functions. Front. Microbiol. 2019, 10, 432609. [Google Scholar] [CrossRef]
- Abbadi, N.; Mousa, J.J. Broadly Protective Neuraminidase-Based Influenza Vaccines and Monoclonal Antibodies: Target Epitopes and Mechanisms of Action. Viruses 2023, 15, 200. [Google Scholar] [CrossRef]
- Gordon, S. Phagocytosis: An Immunobiologic Process. Immunity 2016, 44, 463–475. [Google Scholar] [CrossRef]
- Underhill, D.M.; Ozinsky, A. Phagocytosis of Microbes: Complexity in Action. Annu. Rev. Immunol. 2002, 20, 825–852. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Querol, E.; Rosales, C. Phagocytosis: Our Current Understanding of a Universal Biological Process. Front. Immunol. 2020, 11, 1066. [Google Scholar] [CrossRef] [PubMed]
- Catz, S.D.; McLeish, K.R. Therapeutic targeting of neutrophil exocytosis. J. Leukoc. Biol. 2020, 107, 393–408. [Google Scholar] [CrossRef]
- Young, D.; Das, N.; Anowai, A.; Dufour, A. Matrix Metalloproteases as Influencers of the Cells’ Social Media. Int. J. Mol. Sci. 2019, 20, 3847. [Google Scholar] [CrossRef] [PubMed]
- Nissinen, L.; Kähäri, V.-M. Matrix metalloproteinases in inflammation. Biochim. Biophys. Acta-Gen. Subj. 2014, 1840, 2571–2580. [Google Scholar] [CrossRef] [PubMed]
- Reche, P.A. Potential Cross-Reactive Immunity to SARS-CoV-2 From Common Human Pathogens and Vaccines. Front. Immunol. 2020, 11, 586984. [Google Scholar] [CrossRef] [PubMed]
- Bodas-Pinedo, A.; Lafuente, E.M.; Pelaez-Prestel, H.F.; Ras-Carmona, A.; Subiza, J.L.; Reche, P.A. Combining different bacteria in vaccine formulations enhances the chance for antiviral cross-reactive immunity: A detailed in silico analysis for influenza A virus. Front. Immunol. 2023, 14, 1235053. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ras-Carmona, A.; Reche, P.A. Analysis of Virus-Specific B Cell Epitopes Reveals Extensive Antigen Degradation Prior to Recognition. Cells 2024, 13, 1076. https://doi.org/10.3390/cells13131076
Ras-Carmona A, Reche PA. Analysis of Virus-Specific B Cell Epitopes Reveals Extensive Antigen Degradation Prior to Recognition. Cells. 2024; 13(13):1076. https://doi.org/10.3390/cells13131076
Chicago/Turabian StyleRas-Carmona, Alvaro, and Pedro A. Reche. 2024. "Analysis of Virus-Specific B Cell Epitopes Reveals Extensive Antigen Degradation Prior to Recognition" Cells 13, no. 13: 1076. https://doi.org/10.3390/cells13131076
APA StyleRas-Carmona, A., & Reche, P. A. (2024). Analysis of Virus-Specific B Cell Epitopes Reveals Extensive Antigen Degradation Prior to Recognition. Cells, 13(13), 1076. https://doi.org/10.3390/cells13131076






