Using Microfluidic Hepatic Spheroid Cultures to Assess Liver Toxicity of T-2 Mycotoxin
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Fabrication of Microfluidic Devices
2.3. HepG2 Cell Cultures
2.3.1. Cell Culture in Standard Large Volume 3D Plates
2.3.2. Cell Culture in Microfluidic Devices
2.4. T-2 Toxicity in Microfluidic Devices
2.5. Live/Dead Assay and Analysis
2.6. Spheroid Formation Image Analysis
2.6.1. Assessing Area, Solidity, and Circularity of Hepatic Spheroids
2.6.2. HepG2 and HepG2-LX2 Spheroid Growth Analysis
2.7. Analysis of Hepatic Albumin Production by ELISA
2.8. RT-PCR Analysis of Hepatic Gene Expression
2.9. Secretome Analysis
2.10. Statistical Analysis
3. Results and Discussion
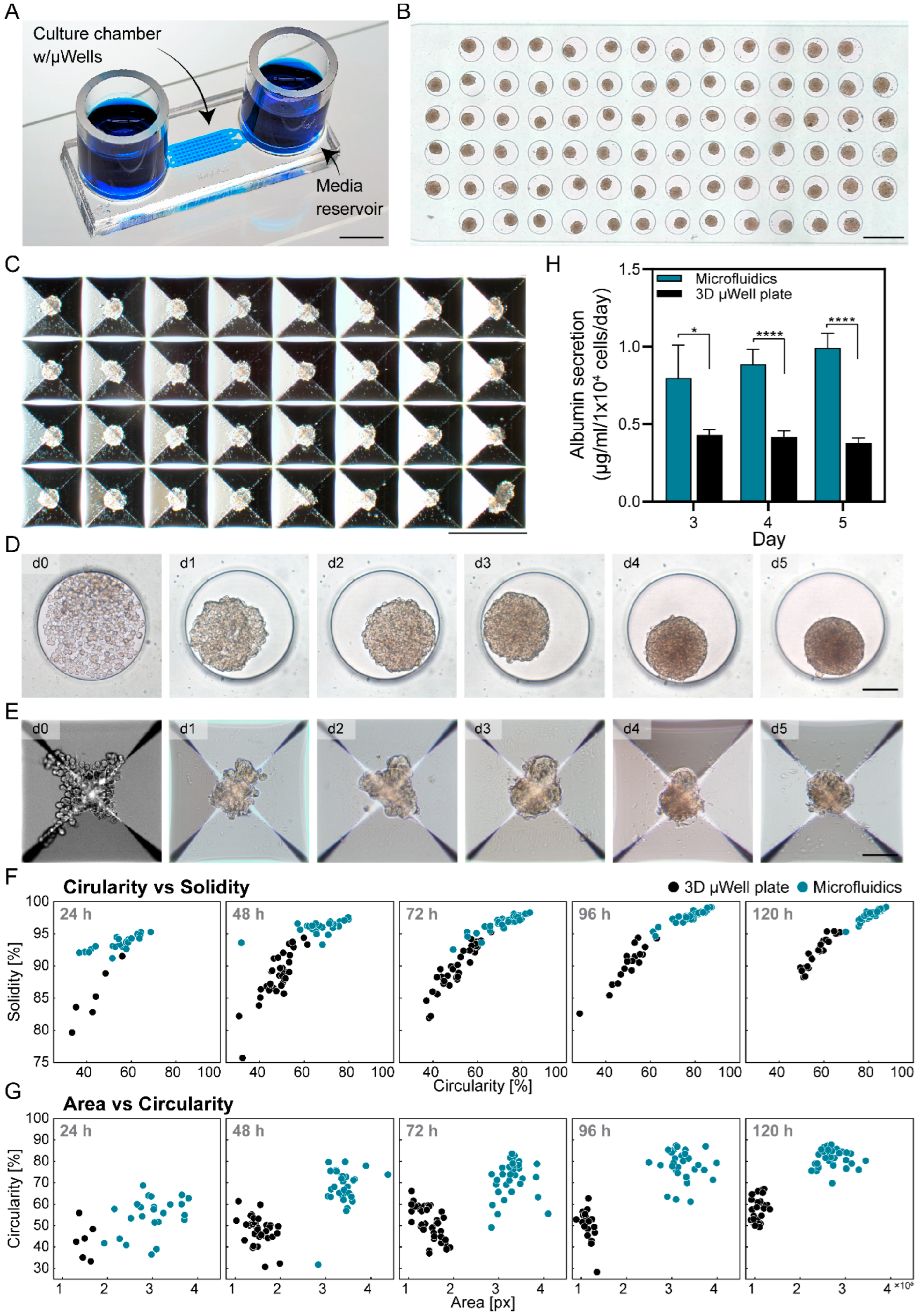
3.1. Cultivation of Hepatic Spheroids in Microfluidic Devices vs. Standard Large Volume 3D Plates
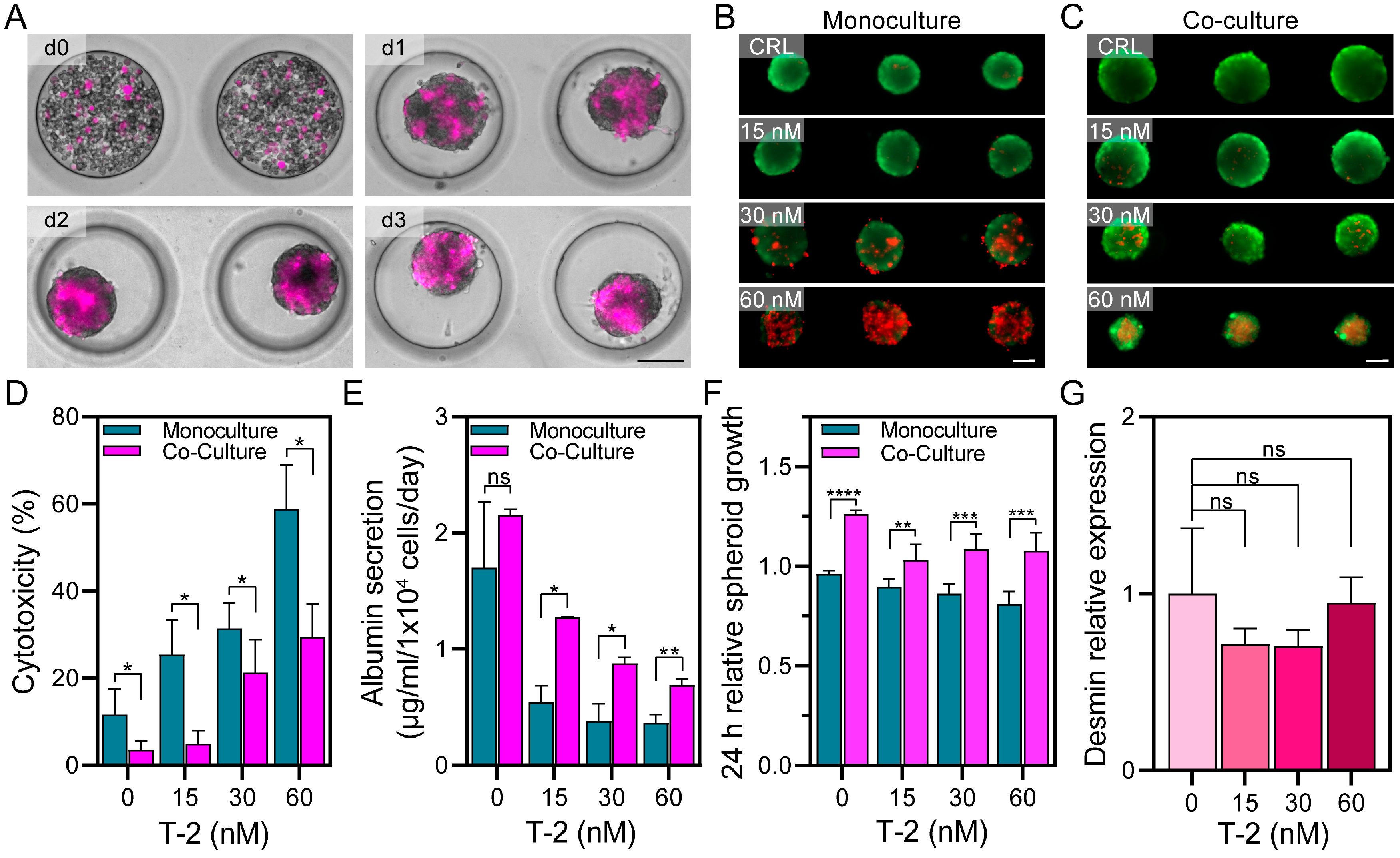
3.2. Creating Microfluidic 3D Co-Cultures and Assessing Viability upon T-2 Exposure
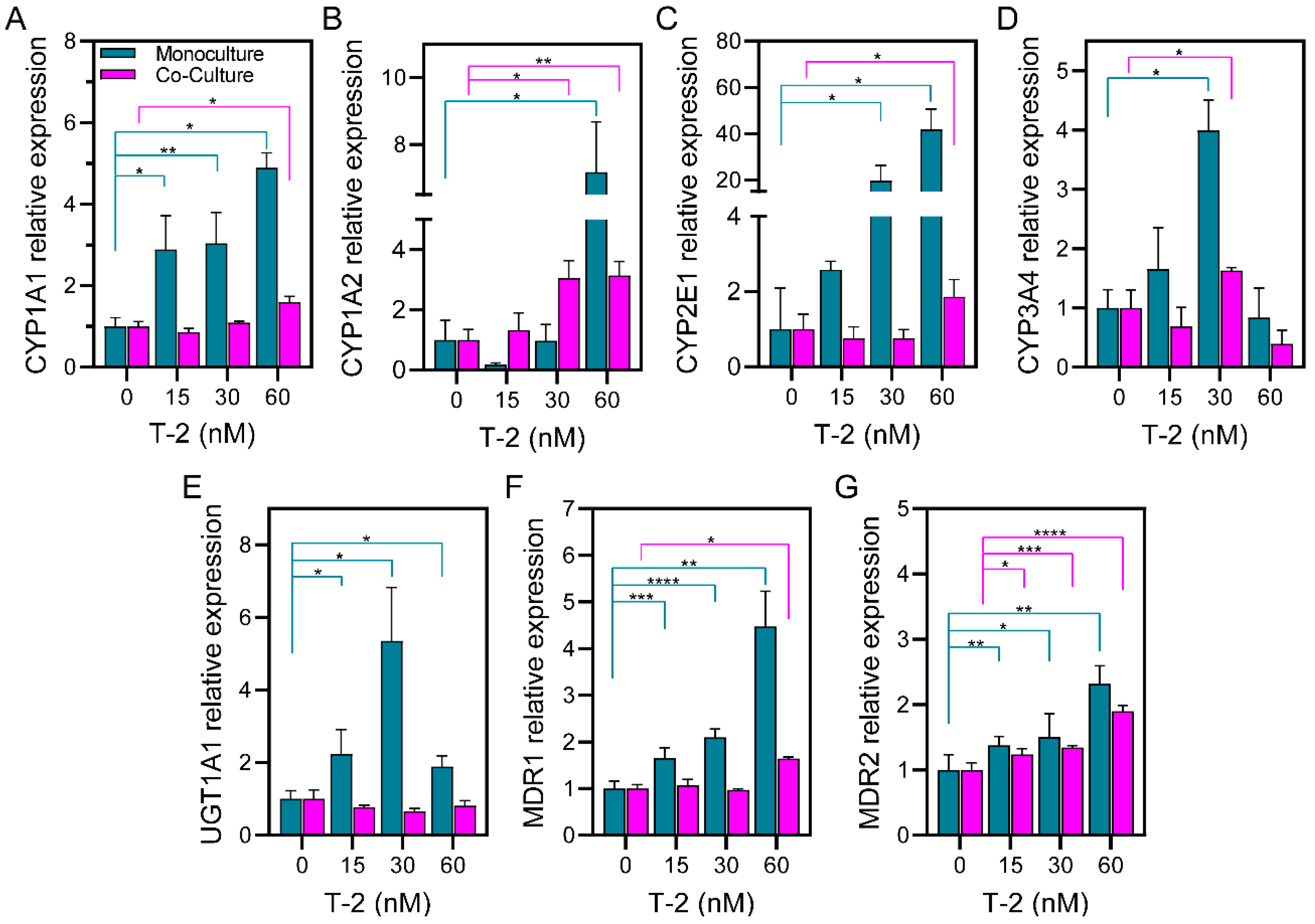
3.3. Evaluating the Expression of Hepatic Enzymes in Microfluidic Spheroid Cultures
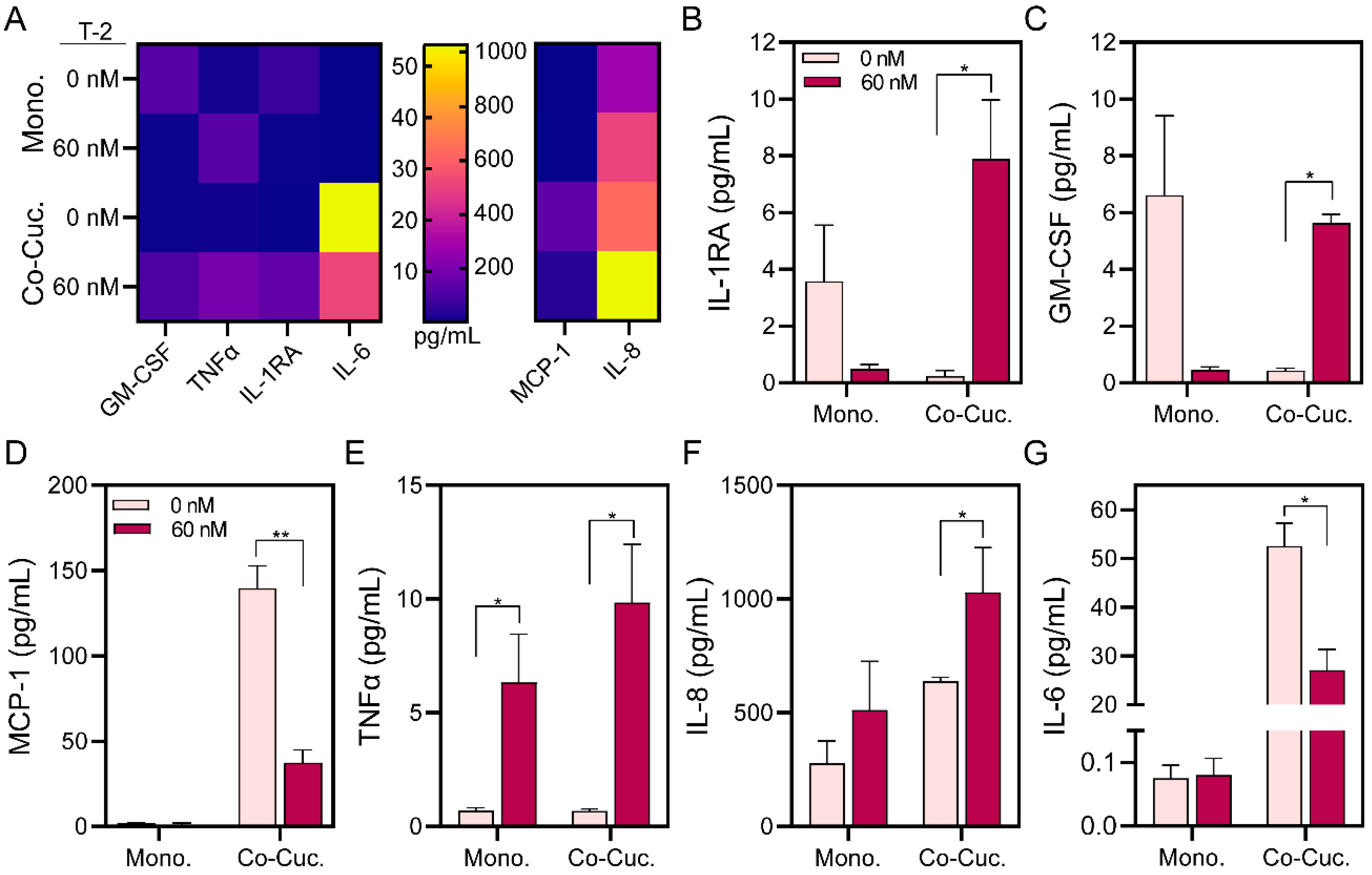
3.4. Assessing Inflammatory Markers in Microfluidic Hepatic Spheroid Cultures
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- EFSA. Human and Animal Dietary Exposure to T-2 and HT-2 Toxin. EFSA J. 2017, 15, e04972. [Google Scholar] [CrossRef] [PubMed]
- Pereira, V.L.; Fernandes, J.O.; Cunha, S.C. Mycotoxins in Cereals and Related Foodstuffs: A Review on Occurrence and Recent Methods of Analysis. Trends Food Sci. Technol. 2014, 36, 96–136. [Google Scholar] [CrossRef]
- Alshannaq, A.; Yu, J.H. Occurrence, Toxicity, and Analysis of Major Mycotoxins in Food. Int. J. Environ. Res. Public Health 2017, 14, 632. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Appropriateness to Set a Group Health Based Guidance Value for T2 and HT2 Toxin and Its Modified Forms. EFSA J. 2017, 15, e04655. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Scientific Opinion on the Risks for Animal and Public Health Related to the Presence of T-2 and HT-2 Toxin in Food and Feed. EFSA J. 2011, 9, 2481. [Google Scholar] [CrossRef]
- Steinkellner, H.; Binaglia, M.; Dall’Asta, C.; Gutleb, A.C.; Metzler, M.; Oswald, I.P.; Parent-Massin, D.; Alexander, J. Combined Hazard Assessment of Mycotoxins and Their Modified Forms Applying Relative Potency Factors: Zearalenone and T2/HT2 Toxin. Food Chem. Toxicol. 2019, 131, 110599. [Google Scholar] [CrossRef] [PubMed]
- EFSA. Assessment of Information as Regards the Toxicity of T-2 and HT-2 Toxin for Ruminants. EFSA J. 2022, 20, e07564. [Google Scholar] [CrossRef] [PubMed]
- Kimura, H.; Sakai, Y.; Fujii, T. Organ/Body-on-a-Chip Based on Microfluidic Technology for Drug Discovery. Drug Metab. Pharmacokinet. 2018, 33, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Goossens, J.; Pasmans, F.; Verbrugghe, E.; Vandenbroucke, V.; De Baere, S.; Meyer, E.; Haesebrouck, F.; De Backer, P.; Croubels, S. Porcine Intestinal Epithelial Barrier Disruption by the Fusarium Mycotoxins Deoxynivalenol and T-2 Toxin Promotes Transepithelial Passage of Doxycycline and Paromomycin. BMC Vet. Res. 2012, 8, 245. [Google Scholar] [CrossRef]
- Jia, Z.; Cheng, Y.; Jiang, X.; Zhang, C.; Wang, G.; Xu, J.; Li, Y.; Peng, Q.; Gao, Y. 3D Culture System for Liver Tissue Mimicking Hepatic Plates for Improvement of Human Hepatocyte (C3A) Function and Polarity. BioMed Res. Int. 2020, 2020, 6354183. [Google Scholar] [CrossRef]
- Martínez-Alonso, C.; Taroncher, M.; Rodríguez-Carrasco, Y.; Ruiz, M.J. Evaluation of the Bioaccessible Fraction of T-2 Toxin from Cereals and Its Effect on the Viability of Caco-2 Cells Exposed to Tyrosol. Toxins 2023, 15, 493. [Google Scholar] [CrossRef] [PubMed]
- Mulac, D.; Lepski, S.; Ebert, F.; Schwerdtle, T.; Humpf, H.U. Cytotoxicity and Fluorescence Visualization of Ergot Alkaloids in Human Cell Lines. J. Agric. Food Chem. 2013, 61, 462–471. [Google Scholar] [CrossRef]
- Yang, L.; Tu, D.; Wang, N.; Deng, Z.; Zhan, Y.; Liu, W.; Hu, Y.; Liu, T.; Tan, L.; Li, Y.; et al. The Protective Effects of DL-Selenomethionine against T-2/HT-2 Toxins-Induced Cytotoxicity and Oxidative Stress in Broiler Hepatocytes. Toxicol. Vitr. 2019, 54, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Taroncher, M.; Rodríguez-Carrasco, Y.; Ruiz, M.J. T-2 Toxin and Its Metabolites: Characterization, Cytotoxic Mechanisms and Adaptive Cellular Response in Human Hepatocarcinoma (HepG2) Cells. Food Chem. Toxicol. 2020, 145, 111654. [Google Scholar] [CrossRef] [PubMed]
- Taroncher, M.; Halbig, F.; Rodríguez-Carrasco, Y.; Ruiz, M.J. Stressful Effects of T-2 Metabolites and Defense Capability of HepG2 Cells. Toxins 2022, 14, 841. [Google Scholar] [CrossRef]
- Martínez-Alonso, C.; Taroncher, M.; Castaldo, L.; Izzo, L.; Rodríguez-Carrasco, Y.; Ritieni, A.; Ruiz, M.-J. Effect of Phenolic Extract from Red Beans (Phaseolus vulgaris L.) on T-2 Toxin-Induced Cytotoxicity in HepG2 Cells. Foods 2022, 11, 1033. [Google Scholar] [CrossRef] [PubMed]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning from 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef]
- de Hoyos-Vega, J.M.; Hong, H.J.; Stybayeva, G.; Revzin, A. Hepatocyte Cultures: From Collagen Gel Sandwiches to Microfluidic Devices with Integrated Biosensors. APL Bioeng. 2021, 5, 041504. [Google Scholar] [CrossRef]
- Mukundan, S.; Bell, J.; Teryek, M.; Hernandez, C.; Love, A.C.; Parekkadan, B.; Chan, L.L.Y. Automated Assessment of Cancer Drug Efficacy on Breast Tumor Spheroids in AggrewellTM400 Plates Using Image Cytometry. J. Fluoresc. 2022, 32, 521–531. [Google Scholar] [CrossRef]
- Dunn, J.C.Y.; Yarmush, M.L.; Koebe, H.G.; Tompkins, R.G. Hepatocyte Function and Extracellular Matrix Geometry: Long-Term Culture in a Sandwich Configuration. FASEB J. 1989, 3, 174–177. [Google Scholar] [CrossRef]
- Foster, E.; You, J.; Siltanen, C.; Patel, D.; Haque, A.; Anderson, L.; Revzin, A. Heparin Hydrogel Sandwich Cultures of Primary Hepatocytes. Eur. Polym. J. 2015, 72, 726–735. [Google Scholar] [CrossRef]
- Vongchan, P.; Warda, M.; Toyoda, H.; Toida, T.; Marks, R.M.; Linhardt, R.J. Structural Characterization of Human Liver Heparan Sulfate. Biochim. Biophys. Acta Gen. Subj. 2005, 1721, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Berthiaume, F.; Moghe, P.V.; Toner, M.; Yarmush, M.L. Effect of Extracellular Matrix Topology on Cell Structure, Function, and Physiological Responsiveness: Hepatocytes Cultured in a Sandwich Configuration. FASEB J. 1996, 10, 1471–1484. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.L.; Hughes, A.; Soonawalla, Z.; Mukherjee, S.; O’neill, E. Dynamic Physiological Culture of Ex Vivo Human Tissue: A Systematic Review. Cancers 2021, 13, 2870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Zhang, Y.N.; Zhang, W. Cancer-on-a-Chip Systems at the Frontier of Nanomedicine. Drug Discov. Today 2017, 22, 1392–1399. [Google Scholar] [CrossRef]
- Trujillo-de Santiago, G.; Flores-Garza, B.G.; Tavares-Negrete, J.A.; Lara-Mayorga, I.M.; González-Gamboa, I.; Zhang, Y.S.; Rojas-Martínez, A.; Ortiz-López, R.; Álvarez, M.M. The Tumor-on-Chip: Recent Advances in the Development of Microfluidic Systems to Recapitulate the Physiology of Solid Tumors. Materials 2019, 12, 2945. [Google Scholar] [CrossRef]
- Hoon Choi, J.; Loarca, L.; De Hoyos-Vega, J.M.; Neda Dadgar, X.; Loutherback, K.; Vijay Shah, X.H.; Stybayeva, G.; Revzin, A.; Hoyos-Vega, D.J. Microfluidic Confinement Enhances Phenotype and Function of Hepatocyte Spheroids Microfluidic Confine-Ment Enhances Phenotype and Function of Hepatocyte Spheroids. Am. J. Physiol. Cell Physiol. 2020, 319, C552–C560. [Google Scholar]
- Dadgar, N.; Gonzalez-Suarez, A.M.; Fattahi, P.; Hou, X.; Weroha, J.S.; Gaspar-Maia, A.; Stybayeva, G.; Revzin, A. A Microfluidic Platform for Cultivating Ovarian Cancer Spheroids and Testing Their Responses to Chemotherapies. Microsyst. Nanoeng. 2020, 6, 93. [Google Scholar] [CrossRef] [PubMed]
- de Hoyos-Vega, J.M.; Hong, H.J.; Loutherback, K.; Stybayeva, G.; Revzin, A. A Microfluidic Device for Long-Term Maintenance of Organotypic Liver Cultures. Adv. Mater. Technol. 2023, 8, 2201121. [Google Scholar] [CrossRef]
- Fattahi, P.; de Hoyos-Vega, J.M.; Choi, J.H.; Duffy, C.D.; Gonzalez-Suarez, A.M.; Ishida, Y.; Nguyen, K.M.; Gwon, K.; Peterson, Q.P.; Saito, T.; et al. Guiding Hepatic Differentiation of Pluripotent Stem Cells Using 3D Microfluidic Co-Cultures with Human Hepatocytes. Cells 2023, 12, 1982. [Google Scholar] [CrossRef]
- Kane, B.J.; Zinner, M.J.; Yarmush, M.L.; Toner, M. Liver-Specific Functional Studies in a Microfluidic Array of Primary Mammalian Hepatocytes. Anal. Chem. 2006, 78, 4291–4298. [Google Scholar] [CrossRef] [PubMed]
- Clement, B.; Guguen-guillouzo, C.; Campion, J.; Glaise, D.; Bourel, M.; Guillouzo, A.; Guguen-Guillouzo, C. Long-Term Co-Cultures of Adult Human Hepatocytes with Rat Liver Epithelial Cells: Modulation of Albumin Secretion and Accumulation of Extracellular Material. Hepatology 1984, 4, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Khetani, S.R.; Bhatia, S.N. Microscale Culture of Human Liver Cells for Drug Development. Nat. Biotechnol. 2008, 26, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.-J.; Otieno, M.A.; Ronxhi, J.; Lim, H.-K.; Ewart, L.; Kodella, K.R.; Petropolis, D.B.; Kulkarni, G.; Rubins, J.E.; Conegliano, D.; et al. Reproducing Human and Cross-Species Drug Toxicities Using a Liver-Chip. Sci. Transl. Med. 2019, 11, eaax5516. [Google Scholar] [CrossRef] [PubMed]
- Ukairo, O.; Kanchagar, C.; Moore, A.; Shi, J.; Gaffney, J.; Aoyama, S.; Rose, K.; Krzyzewski, S.; Mcgeehan, J.; Andersen, M.E.; et al. Long-Term Stability of Primary Rat Hepatocytes in Micropatterned Cocultures. J. Biochem. Mol. Toxicol. 2013, 27, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Patel, D.; Kwa, T.; Haque, A.; Matharu, Z.; Stybayeva, G.; Gao, Y.; Diehl, A.M.; Revzin, A. Liver Injury-on-a-Chip: Microfluidic Co-Cultures with Integrated Biosensors for Monitoring Liver Cell Signaling during Injury. Lab. Chip 2015, 15, 4467–4478. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L. Hepatic Stellate Cells: Protean, Multifunctional, and Enigmatic Cells of the Liver. Physiol. Rev. 2008, 88, 125–172. [Google Scholar]
- Arzt, M.; Deschamps, J.; Schmied, C.; Pietzsch, T.; Schmidt, D.; Tomancak, P.; Haase, R.; Jug, F. LABKIT: Labeling and Segmentation Toolkit for Big Image Data. Front. Comput. Sci. 2022, 4, 777728. [Google Scholar] [CrossRef]
- Amaral, R.L.F.; Miranda, M.; Marcato, P.D.; Swiech, K. Comparative Analysis of 3D Bladder Tumor Spheroids Obtained by Forced Floating and Hanging Drop Methods for Drug Screening. Front. Physiol. 2017, 8, 284532. [Google Scholar] [CrossRef] [PubMed]
- Mora, C.F.; Kwan, A.K.H. Sphericity, Shape Factor, and Convexity Measurement of Coarse Aggregate for Concrete Using Digital Image Processing. Cem. Concr. Res. 2000, 30, 351–358. [Google Scholar] [CrossRef]
- Kurosawa, H.; Yasumoto, K.; Kimura, T.; Amano, Y. Polyurethane Membrane as an Efficient Immobilization Carrier for High-Density Culture of Rat Hepatocytes in the Fixed-Bed Reactor. Biotechnol. Bioeng. 2000, 70, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Nitou, M.; Ishikawa, K.; Shiojiri, N. Immunohistochemical analysis of development of desmin-positive hepatic stellate cells in mouse liver. J Anat. 2000, 197 Pt 4, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Bechmann, L.P.; Hannivoort, R.A.; Gerken, G.; Hotamisligil, G.S.; Trauner, M.; Canbay, A. The Interaction of Hepatic Lipid and Glucose Metabolism in Liver Diseases. J. Hepatol. 2012, 56, 952–964. [Google Scholar] [CrossRef] [PubMed]
- Crettol, S.; Petrovic, N.; Murray, M. Pharmacogenetics of Phase I and Phase II Drug Metabolism. Curr. Pharm. Des. 2010, 16, 204–219. [Google Scholar] [CrossRef] [PubMed]
- Guengerich, F.P. Cytochrome P450s and Other Enzymes in Drug Metabolism and Toxicity. AAPS J. 2006, 8, E101–E111. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.A.; Hyland, R.; Jones, B.C.; Smith, D.A.; Hurst, S.; Goosen, T.C.; Peterkin, V.; Koup, J.R.; Ball, S.E. Drug-Drug Interactions for UDP-Glucuronosyltransferase Substrates: A Pharmacokinetic Explanation for Typically Observed Low Exposure (AUC 1/AUC) Ratios. Drug Metab. Dispos. 2004, 32, 1201–1208. [Google Scholar] [CrossRef] [PubMed]
- Taroncher, M.; Rodríguez-Carrasco, Y.; Ruiz, M.J. Interactions between T-2 Toxin and Its Metabolites in HepG2 Cells and in Silico Approach. Food Chem. Toxicol. 2021, 148, 111942. [Google Scholar] [CrossRef] [PubMed]
- Lootens, O.; Vermeulen, A.; Croubels, S.; De Saeger, S.; Van Bocxlaer, J.; De Boevre, M. Possible Mechanisms of the Interplay between Drugs and Mycotoxins—Is There a Possible Impact? Toxins 2022, 14, 873. [Google Scholar] [CrossRef]
- Song, W.; Wang, Y.; Huang, T.; Liu, Y.; Chen, F.; Chen, Y.; Jiang, Y.; Zhang, C.; Yang, X. T-2 Toxin Metabolism and Its Hepatotoxicity: New Insights on the Molecular Mechanism and Detoxification. Environ. Pollut. 2023, 330, 121784. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Mu, P.; Deng, Y. Mycotoxins: Cytotoxicity and Biotransformation in Animal Cells. Toxicol. Res. 2016, 5, 377–387. [Google Scholar] [CrossRef]
- Lin, N.N.; Chen, J.; Xu, B.; Wei, X.; Guo, L.; Xie, J.W. The Roles of Carboxylesterase and CYP Isozymes on the In Vitro Metabolism of T-2 Toxin. Mil. Med. Res. 2015, 2, 13. [Google Scholar] [CrossRef][Green Version]
- Trusal, L.R. Metabolism of T-2 mycotoxin by cultured cells. Toxicon 1986, 24, 597–603. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, L.A.; Gröhn, Y.; Karppanen, E. Massive Lipid Accumulation in Mink Liver Stellate Cells May Be Caused by Fusarium Mycotoxins in the Feed. Acta Vet. Scand. 1985, 26, 423–424. [Google Scholar] [CrossRef]
- Stanley, L.A.; Wolf, C.R. Through a Glass, Darkly? HepaRG and HepG2 Cells as Models of Human Phase I Drug Metabolism. Drug Metab. Rev. 2022, 54, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Artursson, P.; Palm, K.; Luthman, K. Caco-2 Monolayers in Experimental and Theoretical Predictions q of Drug Transport. Adv. Drug Deliv. Rev. 2001, 46, 27–43. [Google Scholar] [CrossRef]
- Kadota, T.; Furusawa, H.; Hirano, S.; Tajima, O.; Kamata, Y.; Sugita-Konishi, Y. Comparative Study of Deoxynivalenol, 3-Acetyldeoxynivalenol, and 15-Acetyldeoxynivalenol on Intestinal Transport and IL-8 Secretion in the Human Cell Line Caco-2. Toxicol. Vitr. 2013, 27, 1888–1895. [Google Scholar] [CrossRef]
- Sergent, T.; Parys, M.; Garsou, S.; Pussemier, L.; Schneider, Y.J.; Larondelle, Y. Deoxynivalenol Transport across Human Intestinal Caco-2 Cells and Its Effects on Cellular Metabolism at Realistic Intestinal Concentrations. Toxicol. Lett. 2006, 164, 167–176. [Google Scholar] [CrossRef]
- Kim, Y.S.; Ho, S.B. Intestinal Goblet Cells and Mucins in Health and Disease: Recent Insights and Progress. Curr. Gastroenterol. Rep. 2010, 12, 319–330. [Google Scholar] [CrossRef]
- Döll, S.; Schrickx, J.A.; Dänicke, S.; Fink-Gremmels, J. Deoxynivalenol-Induced Cytotoxicity, Cytokines and Related Genes in Unstimulated or Lipopolysaccharide Stimulated Primary Porcine Macrophages. Toxicol. Lett. 2009, 184, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Königs, M.; Schwerdt, G.; Gekle, M.; Humpf, H.U. Effects of the Mycotoxin Deoxynivalenol on Human Primary Hepatocytes. Mol. Nutr. Food Res. 2008, 52, 830–839. [Google Scholar] [CrossRef]
- Volarevic, V.; Al-Qahtani, A.; Arsenijevic, N.; Pajovic, S.; Lukic, M.L. Interleukin-1 Receptor Antagonist (IL-1Ra) and IL-1Ra Producing Mesenchymal Stem Cells as Modulators of Diabetogenesis. Autoimmunity 2010, 43, 255–263. [Google Scholar] [CrossRef]
- Karmacharya, M.B.; Hada, B.; Park, S.R.; Kim, K.H.; Choi, B.H. Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF) Shows Therapeutic Effect on Dimethylnitrosamine (DMN)-Induced Liver Fibrosis in Rats. PLoS ONE 2022, 17, e0274126. [Google Scholar] [CrossRef]
- Maher, J.J. Interactions between Hepatic Stellate Cells and the Immune System. Semin. Liver Dis. 2001, 21, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Marra, F.; Romanelli, R.G.; Giannini, C.; Failli, P.; Pastacaldi, S.; Arrighi, M.C.; Pinzani, M.; Laffi, G.; Montalto, P.; Gentilini, P. Monocyte Chemotactic Protein-1 as a Chemoattractant for Human Hepatic Stellate Cells. Hepatology 1999, 29, 140–148. [Google Scholar] [CrossRef]
- Pei, X.; Jiang, H.; Liu, X.; Li, L.; Li, C.; Xiao, X.; Li, D.; Tang, S. Targeting HMGB1 Inhibits T-2 Toxin-Induced Neurotoxicity via Regulation of Oxidative Stress, Neuroinflammation and Neuronal Apoptosis. Food Chem. Toxicol. 2021, 151, 112134. [Google Scholar] [CrossRef]
- Norris, C.A.; He, M.; Kang, L.I.; Ding, M.Q.; Radder, J.E.; Haynes, M.M.; Yang, Y.; Paranjpe, S.; Bowen, W.C.; Orr, A.; et al. Synthesis of IL-6 by Hepatocytes Is a Normal Response to Common Hepatic Stimuli. PLoS ONE 2014, 9, e96053. [Google Scholar] [CrossRef]
- Kim, Y.; Fiel, M.I.; Albanis, E.; Chou, H.I.; Zhang, W.; Khitrov, G.; Friedman, S.L. Anti-Fibrotic Activity and Enhanced Interleukin-6 Production by Hepatic Stellate Cells in Response to Imatinib Mesylate. Liver Int. 2012, 32, 1008–1017. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Pan, Z.; Wang, Y. Ultrasound-Assisted Turbine Bead Milling for Disintegration of Nannochloropsis oculata Cells. J. Appl. Phycol. 2019, 31, 1651–1659. [Google Scholar] [CrossRef]
- Döll, S.; Schrickx, J.A.; Dänicke, S.; Fink-Gremmels, J. Interactions of Deoxynivalenol and Lipopolysaccharides on Cytokine Excretion and MRNA Expression in Porcine Hepatocytes and Kupffer Cell Enriched Hepatocyte Cultures. Toxicol. Lett. 2009, 190, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Sugita-Konishi, Y.; Pestka, J.J. Differential Upregulation of TNF-α, IL-6, and IL-8 Production by Deoxynivalenol (Vomitoxin) and Other 8-Ketotrichothecenes in a Human Macrophage Model. J. Toxicol. Environ. Health A 2001, 64, 619–636. [Google Scholar] [CrossRef]
- Nagashima, H.; Nakamura, K.; Goto, T. Rubratoxin B Induced the Secretion of Hepatic Injury-Related Colony Stimulating Factors in Human Hepatoma Cells. Toxicol. Lett. 2003, 145, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Zhang, X.; Yue, L.; Hu, H.; Wei, X.; Guo, Q.; Zhang, B.; Fan, X.; Xin, Y.; Oh, Y.; et al. Thiamethoxam Induces Nonalcoholic Fatty Liver Disease in Mice via Methionine Metabolism Disturb via Nicotinamide N-Methyltransferase Overexpression. Chemosphere 2021, 273, 129727. [Google Scholar] [CrossRef] [PubMed]
- Liu, A.; Guo, M.; He, L.; Martínez, M.A.; Martínez, M.; Lopez-Torres, B.; Martínez-Larrañaga, M.R.; Wang, X.; Anadón, A.; Ares, I. Nicotinamide N-Methyltransferase Protects against Deoxynivalenol-Induced Growth Inhibition by Suppressing pro-Inflammatory Cytokine Expression. Food Chem. Toxicol. 2022, 163, 112969. [Google Scholar] [CrossRef]
- Xu, J.; Moatamed, F.; Caldwell, J.S.; Walker, J.R.; Kraiem, Z.; Taki, K.; Brent, G.A.; Hershman, J.M. Enhanced Expression of Nicotinamide N-Methyltransferase in Human Papillary Thyroid Carcinoma Cells. J. Clin. Endocrinol. Metab. 2003, 88, 4990–4996. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Gene Symbol | Gene Name | Tm (°C) Forward Primer/Revers Primer | Forward Primer/Revers Primer |
|---|---|---|---|
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | 57.2/57.7 | TGTTGCCATCAATGACCCCTT/ CTCCACGACGTACTCAGCG |
| CYP1A1 | Cytochrome P450 1A1 | 59.4/57.4 | CATTAACATCGTCTTGGACC/ TCTTGGATCTTTCTCTGTACC |
| CYP1A2 | Cytochrome P450 1A2 | 52.5/56.4 | GCTTCTACATCCCCAAGAAAT/ ACCACTTGGCCAGGACT |
| CYP2E1 | Cytochrome P450 2E1 | 57.5/57.8 | GACACCATTTTCAGAGGATAC/ TTCATTCAGGAAGTGTTCTG |
| CYP3A4 | Cytochrome P450 3A4 | 50.1/51.8 | ACTGCCTTTTTTGGGAAATA/ GGCTGTTGACCATCATAAAAG |
| UGT1A1 | UDP Glucuronosyltransferase 1A1 | 54.8/56.7 | TTGATCCCAGTGGATGGC/ ATGCTCCGTCTCTGATGTACAAC |
| MDR1 | Multidrug resistance mutation 1 | 56.3/56.1 | GTCATCGCTGGTTTCGATGATG/ ATTTCCTGCTGTCTGCATTGTG |
| MDR2 | Multidrug resistance protein 2 | 53.1/53.1 | TCCAACTGTGCTTCAAGC/ GGCATCCACAGACATCAG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Taroncher, M.; Gonzalez-Suarez, A.M.; Gwon, K.; Romero, S.; Reyes-Figueroa, A.D.; Rodríguez-Carrasco, Y.; Ruiz, M.-J.; Stybayeva, G.; Revzin, A.; de Hoyos-Vega, J.M. Using Microfluidic Hepatic Spheroid Cultures to Assess Liver Toxicity of T-2 Mycotoxin. Cells 2024, 13, 900. https://doi.org/10.3390/cells13110900
Taroncher M, Gonzalez-Suarez AM, Gwon K, Romero S, Reyes-Figueroa AD, Rodríguez-Carrasco Y, Ruiz M-J, Stybayeva G, Revzin A, de Hoyos-Vega JM. Using Microfluidic Hepatic Spheroid Cultures to Assess Liver Toxicity of T-2 Mycotoxin. Cells. 2024; 13(11):900. https://doi.org/10.3390/cells13110900
Chicago/Turabian StyleTaroncher, Mercedes, Alan M. Gonzalez-Suarez, Kihak Gwon, Samuel Romero, Angel D. Reyes-Figueroa, Yelko Rodríguez-Carrasco, María-José Ruiz, Gulnaz Stybayeva, Alexander Revzin, and Jose M. de Hoyos-Vega. 2024. "Using Microfluidic Hepatic Spheroid Cultures to Assess Liver Toxicity of T-2 Mycotoxin" Cells 13, no. 11: 900. https://doi.org/10.3390/cells13110900
APA StyleTaroncher, M., Gonzalez-Suarez, A. M., Gwon, K., Romero, S., Reyes-Figueroa, A. D., Rodríguez-Carrasco, Y., Ruiz, M.-J., Stybayeva, G., Revzin, A., & de Hoyos-Vega, J. M. (2024). Using Microfluidic Hepatic Spheroid Cultures to Assess Liver Toxicity of T-2 Mycotoxin. Cells, 13(11), 900. https://doi.org/10.3390/cells13110900










