1. Introduction
Mast cells (MCs) are hematopoietic cells that are positioned at the interphase between tissue and the environment, such as the skin, the tongue, the intestinal mucosa, and the lung [
1]. They are highly specialized cells filled with electron-dense granules that can act on external threats to initiate an inflammatory response. A number of physiologically highly potent substances, such as histamine and heparin, and a number of abundant granule proteases are stored in these granules. The granule-stored proteases can constitute up to 35% of the total cellular protein and the transcript levels for these proteases are among the highest in the entire cell [
1,
2]. The levels can reach several % of the total transcriptome [
1,
3]. Heparin and the related chondroitin sulfate are highly sulfated and negatively charged proteoglycans expressed by MCs. They play important roles as anticoagulants, as stabilizing agents for MC proteases, and as charge-compensating molecules in the granule storage of positively charged mediators, including histamine. The enzymes involved in the synthesis of these proteoglycans and of histamine are therefore important markers for MCs.
MCs can rapidly release their granule material upon stimulation of the cell by the crosslinking of receptors for immunoglobulin E (IgE) or by stimulation by complement components like the anaphylatoxins C3a, C4a, and C5a. MCs can also be activated by positively charged low-molecular-weight compounds, like substance P and compound 48/80. This response is mediated by the MRGPRX2 receptor, which is almost exclusively expressed by connective tissue-type MCs (CTMCs) [
4,
5,
6]. Upon activation, MCs also produce a number of lipid mediators, including leukotriene C4 (LTC4) and prostaglandin D2 (PGD2). The enzymes involved in their synthesis, such as phospholipase A2 (PLA2), the hematopoietic prostaglandin D synthase (HPGDS), and leukotriene C4 synthase (LTC4S), are also important markers for MCs.
MC development and migration are regulated by cytokines and chemokines that deliver their information by binding to specific cell surface receptors. The growth and differentiation of MCs are primarily regulated by stem cell factor (SCF), interleukin-3 (IL-3) (especially in the mouse), IL-4, and IL-33, and the receptors for these cytokines are important characteristics of MCs [
7,
8,
9,
10,
11,
12,
13,
14,
15,
16,
17,
18,
19,
20,
21,
22].
Transcription factors play a key role in cell growth and differentiation and several of them are important characteristics of MCs, including GATA1, GATA2, and MITF [
1,
5,
23,
24,
25]. Analyzing the transcript levels for these transcription factors can therefore reveal important features of MCs and how their phenotype changes upon in vitro culture conditions. These transcription factors are also important for the physiological role of MCs and can thereby give information on differences in MC biology between species. The MC-related transcription factors were therefore used in the present study, together with a panel of other proteins, as markers for the comparison between MCs in mouse and man and for the characterization of how MCs change upon culture. A selection of results from published studies on mouse peritoneal MCs and in vitro expanded mouse bone marrow-derived MCs (BMMCs), the two most commonly used models in mouse MC research, using the Ampliseq technology are summarized in
Table 1 [
1,
26] for comparison with the human MC data presented in this article.
The majority of tissue MCs seem to originate from an early wave of cells coming from the yolk sac [
27,
28]. However, during inflammatory conditions, MC precursors can also be recruited from the bone marrow to increase in number in the inflamed tissue [
29]. The fate of these cells upon returning to steady state conditions of the tissue is not fully known but most likely involves apoptosis of the majority of the cells. This relatively recent information concerning the origin of the majority of tissue MCs and macrophages has changed the view of the role of the bone marrow for both MCs and other hematopoietic cells like macrophages. Such information is of major importance for our view of the characteristics of in vitro-developed cells originating from the bone marrow compared to tissue-resident MCs and macrophages [
30,
31,
32,
33,
34,
35,
36]. The most commonly used in vitro model for mouse MCs is namely bone marrow-derived MCs (BMMCs). These cells most likely represent cells of a different origin compared to the majority of tissue MCs, which appear to be yolk sac-derived. We have also recently shown that these BMMCs represent relatively immature cells that differ markedly in phenotype from tissue-resident MCs and should therefore be used with care as in vitro models of MC physiology [
26] (see also
Table 1). An interesting in vitro model for studies of MC physiology is therefore human skin MCs that can be expanded in vitro by a medium containing serum and recombinant SCF and IL-4, as described in this article. We can show here that they resemble tissue-resident MCs much better than mouse BMMCs and are therefore superior as an in vitro model, in particular for research of human MC biology.
2. Materials and Methods
2.1. Purification of Human Skin MCs
MC purification was performed as previously described [
37,
38] with several modifications specified in more recent work [
21]. Skin was cut into strips and treated with dispase (24.5 mL per preparation, activity: 50 U/mL; Corning, Kaiserslautern, Germany) at 4 °C overnight. After removal of the epidermis, the dermis was chopped into small pieces and digested with 2.29 mg/mL collagenase (Worthington, Lakewood, NJ, USA), 0.75 mg/mL hyaluronidase (Sigma, Deisenhofen, Germany), DNase I at 10 µg/mL (both from Roche, Basel, Switzerland), and 5 mM MgSO
4 for 1 h at 37 °C.
The cell suspensions were separated from remaining tissue by three steps of filtration. In the case of breast skin, the undigested tissue still remaining after the first digestion was subjected to a second digestion step of 1 h at 37 °C after the first filtration. MC purification from the dispersates was achieved by positive selection with anti-human c-Kit microbeads and an Auto-MACS separation device (both from Miltenyi Biotec, Bergisch Gladbach, Germany). MC purity always exceeded 98%, as assessed by acidic toluidine-blue staining (0.1% in 0.5 N HCl). Viability by trypan blue exclusion exceeded 99%. We used between 4.8 and 6.2 × 106 MCs for one RNA isolation (ex vivo samples). Cultures were started from 1.5–6 × 106 MCs; around 3 × 106 were eventually used for one RNA preparation.
2.2. In Vitro Culture of Human Skin MCs
The isolated skin MCs were expanded in vitro for 2–3 weeks in cell culture medium (basal Iscove’s medium; BioSell, Feucht, Germany) containing 10% fetal calf serum (Biochrom, Berlin, Germany), 1% P/S (Fisher Scientific, Berlin, Germany), 1% non-essential amino acid solution (Roth, Karlsruhe, Germany), 100 ng/mL recombinant human stem cell factor (SCF) (Peprotech, Rocky Hill, NJ, USA), and 20 ng/mL of recombinant human IL-4 (Peprotech, Rocky Hill, NJ, USA). The medium was changed every three days to ensure there was a sufficient quantity of the recombinant cytokines.
2.3. RNA Isolation and Heparinase Treatment
Total RNA was prepared from freshly isolated MCs and in vitro-cultured MCs, following an established protocol for each preparation. Briefly, MCs were lysed in 700 µL QIAzol®lysis reagent (Qiagen, Hilden, Germany), mixed with 140 µL chloroform (Sigma) and 60 µL DEPC-treated water, and transferred to a 2 mL gel tube (Quanta bio/VWR, Dresden, Germany). After centrifugation, the supernatant was transferred to a NucleoSpin® filter and RNA was isolated using the NucleoSpin RNA kit from Machery-Nagel (Düren, Germany) following the manufacturer’s instructions. For heparinase (BioLab, Braunschweig, Germany) treatment, the resulting RNA solution was mixed with RNAse inhibitor (Thermo Fisher Scientific) and heparinase buffer (BioLab, Braunschweig, Germany) and incubated for 3 h at 25 °C. Another RNA isolation procedure was followed, using the NucleoSpin RNA kit from Machery-Nagel (Düren, Germany) according to the manufacturer’s protocol. To further concentrate preparations, RNA was precipitated overnight at −80 °C using 100% ethanol and sodium acetate (Merck, Darmstadt, Germany). The RNA of each preparation was eventually solved in 20 µL DEPC-treated water. After each treatment step, RNA concentration was determined by using a Nanodrop ND-1000 (Nano Drop Technologies, Wilmington, DE, USA).
2.4. Ampliseq Analysis of the Total Transcriptome
The transcriptome of freshly isolated mast cells and the different cultures were analyzed for their total transcriptome by the Thermo-Fisher chip-based Ampliseq transcriptomic platform at the SciLife lab in Uppsala, Sweden (Ion-Torrent next-generation sequencing system—Thermofisher, Waltham, MA, USA). The sequence results were delivered in the form of Excel files with normalized expression levels for an easy comparison between samples. In the Ampliseq analysis, all transcripts are read only once; this is why no normalization is needed.
2.5. Validations and RT-qPCR
For the first approach, cultured skin MCs were stimulated with an anti-FcεRI-Ab, AER-37 (0.5 µg/mL) (Abcam, Cambridge, UK), for 30 min, 90 min, and 24 h and compared to cultured unstimulated skin MCs. In the second approach, skin MCs were harvested 1 and 2 d after isolation, as well as after 3 weeks of cultivation. For the final part, cultured skin MCs were exposed to enzymes identically to what is used for skin MC isolation (2.29 mg/mL collagenase, 0.75 mg/mL hyaluronidase, DNase I at 10 µg/mL, and 5 mM MgSO
4) and kept at 37 °C with or without shaking prior to harvest. RNA was extracted with the NucleoSpin RNA kit (Macherey-Nagel, Düren, Germany) and reverse transcribed with TaqMan Reverse Transcription reagent (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s recommendations. Quantitative PCR was performed with the LightCycler
® FastStart DNA Master SYBR Green I reaction mix (Roche Diagnostics, Mannheim, Germany) in a QIAquant 96 5plex real-time PCR cycler (Qiagen, Hilden, Germany). The primers for target genes were designed with NCBI primer blast software and synthesized by TIB Molbiol, Berlin, Germany. The 2
−ΔΔCT method was used to quantify the relative expression levels of target genes to several reference genes (appearing at the end of the table, i.e., β-actin, GAPDH, HPRT, and Cyclophilin B = Peptidylprolyl Isomerase B, gene name PPIB), as described (
Table 2) [
39,
40]. Statistical analysis and visualization were performed with GraphPad Prism software (La Jolla, CA, USA, version 10.0.3.)
4. Discussion
A detailed analysis of the total transcriptome of freshly isolated and cultured human skin MCs was performed to validate the quality of the in vitro-expanded MCs concerning their potential use as models of human MC biology. The cultured cells were found to be remarkably stable in their transcriptome compared to what has been observed for mouse BMMCs (
Table 1) [
26]. Mouse BMMCs represent a very immature version of mouse MCs where the expression levels of several important MC markers are very low compared to freshly isolated mouse peritoneal MCs, including the granule-stored chymase, Mcpt4 (mMCP-4), the tryptase Mcpt6 (mMCP-6), and the drug/neuropeptide receptor Mrgprb2 (
Table 1) [
26]. One additional important difference between these two model systems for studies of MC biology is that they represent cells of very different developmental origin. The mouse BMMCs originate from adult bone marrow, whereas the majority of human skin MCs have their origin primarily in an early wave of cells from the yolk sac [
27,
28]. A recent study of MCs from different mouse organs has shown that, also in the mouse, the organ of origin is a decisive factor for the resulting MC phenotype even though cells from different sources were exposed to identical microenvironments during their recent history [
61].
The skin MCs used herein are human, which makes them better models of human biology than mouse MCs of any origin. The presented data harmonize with a previous analysis of skin MCs using another technology for transcriptome analysis, namely, deep-CAGE sequencing [
5]. We have expanded this analysis here to include additional samples and another sequencing technology to verify the findings, and, most importantly, to extend to foreskin MCs, which had not been analyzed previously. Concerning granzymes, we did not detect expression of any of the granzymes under steady state conditions. However, some of them may be induced by LPS or IgE receptor crosslinking, as has been observed for GZMB, which was found to be highly inducible following FcεRI aggregation in human skin MCs in the Motakis study [
5]. A fivefold increase in GZMB levels was also seen in mouse BMMCs upon LPS stimulation [
1]. Also, mouse GZMC was actually upregulated from 0.1 reads to 100 reads, which corresponds to a 1000-fold increase but from very low starting levels [
1]. However, during normal steady state conditions, granzymes seem to be almost totally absent from both mouse and human MCs.
In this study, the multiagonist receptor MRGPRX2 remained largely stable, although a slightly lower expression was detected in cultured MCs. This was in contrast to previous studies, demonstrating a prominent downregulation of this receptor at the transcript, protein, and functional levels in cultured skin MCs [
5,
62]. Even so, skin MCs still retain decent levels of MRGPRX2 in culture and can be used for MRGPRX2-related research, as substantiated by the findings herein. This is in marked contrast to mouse BMMCs that almost totally lack expression of the corresponding mouse receptor Mrgprb2 (
Table 1). Expression of the dopamine receptor DRD2 was also observed in this dataset at similar expression levels as in previous studies [
49].
Skin is a heterogeneous organ with different micromilieus prevailing in distinct locations. In the skin, MCs interact chiefly with endothelial cells of arterioles and fibroblasts, which shape the MC phenotype [
63,
64]. MCs interact with fibroblasts by different receptors, including but not limited to KIT, different integrins, and still unidentified receptor–receptor pairs [
65,
66,
67]. The known receptors were also found to be expressed in our current study. Here, we analyzed the transcriptome in MCs derived from both foreskin and breast skin. We previously compared these two skin MC subsets by low-throughput techniques and found that the differences were rather small [
68]. For example, breast skin MCs displayed slightly higher tryptase activity but lower histamine and chymase activity; however, there were huge variations within groups [
68]. The very minor differences observed here, in global gene expression, between female and male samples further confirmed this concept. These findings suggest that despite their different sources (female, adult versus male, juvenile), major characteristics of skin MCs are surprisingly stable and are not strongly influenced by age, sex, or the precise location.
Concerning the cultured human skin MCs, one additional factor is the total number of cells. Upon in vitro culture, the cells start to proliferate, and we found that the expression of genes related to cell growth were increased (
Table 13). After three weeks of culturing, the number of cells had increased by 6–10 times, resulting in more cells available for experiments. However, it is advantageous to use ex vivo skin MCs at least in a few experiments, to verify findings obtained with the precultured counterparts. The notion that expanded cells will resemble their respective prototype cell in the original tissue, even after culture, was recently reported also for mouse fetal skin or fetal liver-derived MCs [
61]. However, our findings demonstrate a number of slightly unusual features that should be taken into consideration when using the in vitro-expanded human skin MCs in studies of MC biology (
Figure 4 and
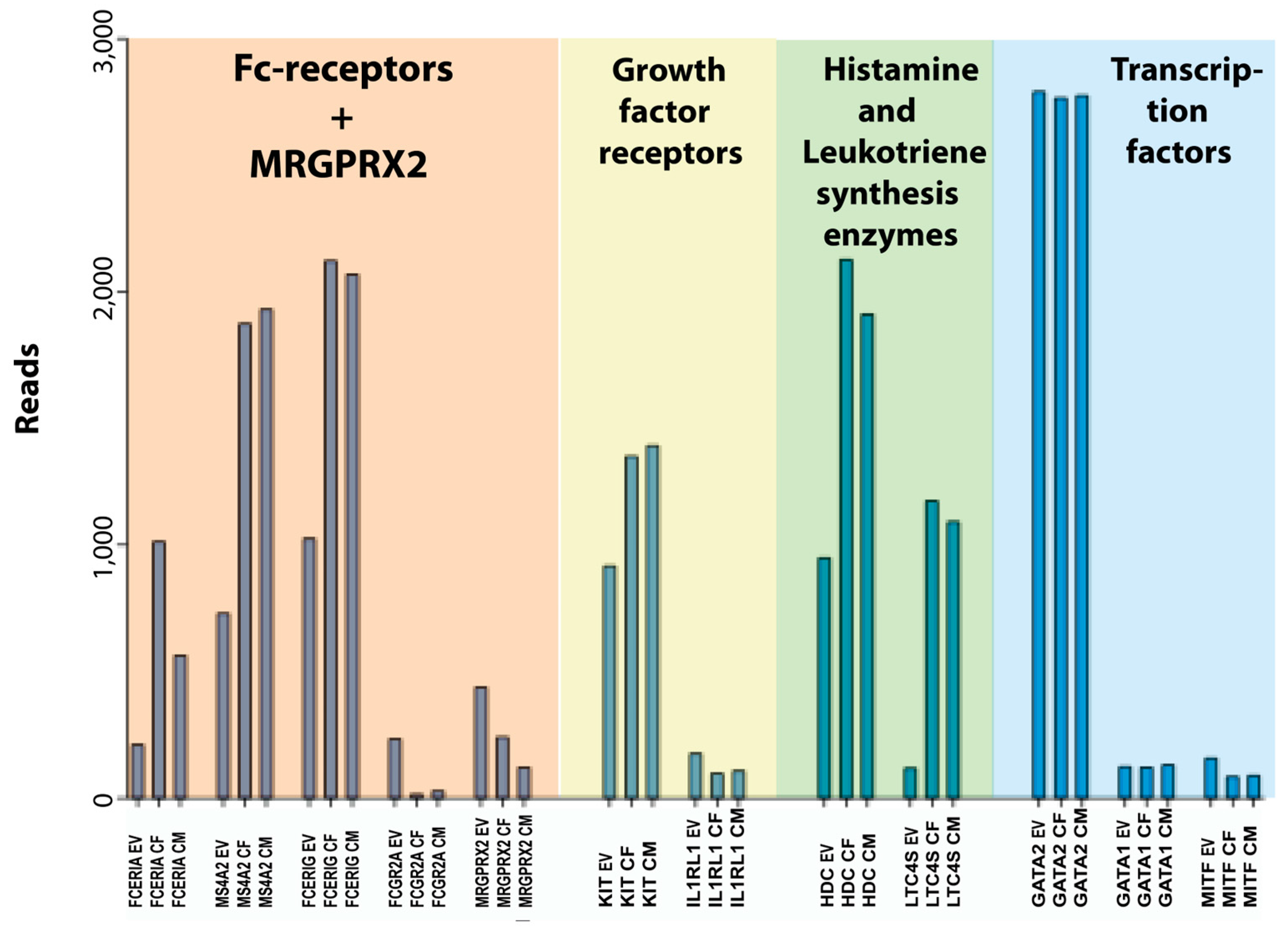
Figure 5). Notably, the expression of the three genes for the high-affinity IgE receptor was increased by 2–5 times and there was an almost 10-fold reduction in the transcript level for the low-affinity IgG receptor FCGR2A. Moreover, there was an almost 10-fold upregulation of the leukotriene synthesis enzyme, leukotriene C4 synthase (LTC4S) (
Figure 5). The demonstrated upregulation of high-affinity IgE receptor subunits and the leukotriene synthesis enzymes harmonizes with previous studies, and it is most likely explained by an induced response to the added SCF and IL-4 in the culture medium [
18,
38,
62,
69]. This also expands to IL-5 [
70], GATA3 [
71,
72], and FCER1A [
73]. There were also changes in cell adhesion molecules with a shift from integrin alpha 5 and 9 to beta 2A (
Table 11). We found no or very minor changes in the enzymes involved in proteoglycan synthesis but a clear upregulation of some enzymes involved in leukotriene and prostaglandin synthesis (
Table 10). In spite of these changes, the in vitro-expanded human skin MCs seem to be a very interesting model for studies of human MC biology. We can also say from this study that there are no or very minor differences between cells isolated from men or females. The only major sex-related differences we found were in the non-coding transcript XIST for X chromosome inactivation, which was found essentially only in the two female samples, and in the male-specific gene RPS4Y1, which is expressed from the Y chromosome (
Table 3). XIST acts to cover one copy of the X chromosome, rendering it inactive [
74].
For future studies including the design and evaluation of novel treatment strategies based on targeting MCs, there are some findings that need to be taken into serious consideration. One example is the very limited complexity in Fc receptor expression in human skin MCs. Attempts are made to use antibodies against the inhibitory IgG receptor FCGR2B for downregulating MC activation through the high-affinity IgE receptor. The possibility of inhibiting MC activation by FCGR2B crosslinking is probably very limited in view of the almost total absence of the transcripts for this receptor both in freshly isolated and cultured skin MCs (
Table 6). However, this does not exclude that FCGR2B can be induced in skin MCs under disease conditions or that FCGR2B is expressed by human MCs in other tissues. Moreover, we did not detect the expression of any of the granzymes, which is in contrast to published work. Here, we found that the levels of all five granzymes in the freshly isolated skin MCs were very low, close to zero, indicating that granzymes have no or only a very limited role in human MC biology unless cells are stimulated.
The high expression of selective IEGs and HSPs was a hallmark of ex vivo MCs, while this quality was lost in culture, as reported previously [
53]. Because the high expression of IEGs was likewise detected in monocytes, basophils, neutrophils, and other cells [
53], we hypothesized that this was an inherent property of fully mature leukocytes, also encompassing ex vivo MCs, to be gradually lost outside of the body. Alternatively, it could have resulted from the isolation process.
Using several strategies, we found that the first hypothesis was most likely correct accounting for most of the effect. Firstly, we showed that mimicking the steps of isolation did not lead to a general upregulation of HSPs and IEGs in cultured MCs (only to a 20-fold increase in one gene), while the expression in cultured versus ex vivo cells was >10,000-fold lower (by RT-qPCR; up to ≈1000-fold by sequencing). This is in accordance with findings in the mouse, where the exposure of peritoneal MCs to digestive enzymes led to a maximally fivefold difference in a limited number of genes [
75]. Interestingly, several IEGs were found among the regulated genes in this mouse study and FOSB was third in their list, in line with what we found here for FOSB (but not for FOS). However, this moderate increase can by no means explain the differences between ex vivo and cultured MCs, but it does indicate that a slight modulation during purification can occur in a gene-dependent manner. This upregulation is most likely transient, though. In fact, FOSB was down more strongly after 1 and 2 d compared to FOS (the latter not enhanced by digestive enzymes/shaking).
Additional evidence for an inherently high expression of HSPs and IEGs in skin MCs came from the fact that expression following MC purification did not diminish as rapidly as expression induced by MC stimulation. Indeed, the levels in ex vivo MCs were still increased at 1 or even 2 d vis-à-vis cultured MCs. In contrast, when IEGs were induced by FcεRI aggregation, the levels were back to their initial values after 1 d or less.
Our data are in line with the comprehensive FANTOM5 atlas. For example, granulocytes and monocytes expressed very high levels of certain IEGs, including FOS and JUNB. Both of the HSPs uncovered as high in skin MCs were also expressed in brain regions and reproductive organs (which were not exposed to digestive enzymes). Moreover, only some HSPs were highly expressed in skin MCs, while others were not; and the same applied to IEGs. Therefore, these genes are not expressed across the board in ex vivo cells/tissues but in clearly defined, cell- and tissue-specific patterns. For example, monocytes expressed substantial FOS but only little FOSB, while FOSB reached similar levels as in skin MCs also in basophils, eosinophils, Langerhans cells, and CD8
+ cells [
53]. Expression of HSPA1A/B was pronounced in the newborn brain, adult reproductive tissues (breast and vagina), heart, aorta, and adipocytes (in addition to influenza-infected macrophages). It is not entirely clear why primary cells accumulate such high levels of these transcripts. However, abundant expression seems to extend to the protein level in MCs and monocytes, whereby FOS (transcript and protein) abundance in primary monocytes was particularly striking [
25]. In that paper, we compared the expression in primary myelocytes with corresponding cell lines (THP-1 and HMC-1), detecting much higher levels in primary cells, while reduction was a hallmark of actively cycling cells [
25]. This was confirmed herein and extended to HSPs in MCs (yet not monocytes), emphasizing that each ex vivo cell expresses a unique combination of IEGs/HSPs. Regarding FOS specifically, it is described as indispensable in monocyte differentiation, increasing during maturation [
76]. We assume that FOS and other factors play similar roles in MCs in vivo where they maintain the fully differentiated state and long-term survival within the cutaneous environment. In this regard, certain IEGs, HSPs, and several TFs like KLF2 and KLF4 (all much higher in freshly extracted MCs) may all be part of the same program.
In summary, this study provides a detailed transcriptomic characterization of in vivo matured human skin MCs in comparison with their in vitro-expanded counterparts. The findings confirm and extend our understanding of the phenotype of skin MCs in their normal tissue environment. Such information is also essential for the design and evaluation of novel potential treatment strategies aimed at targeting MCs in vivo. Based on previous and current findings, in vitro-expanded human skin MCs seem to be one of the best alternatives presently available for in vitro studies of the general in vivo function of human skin MCs. However, it should be noted that the large amounts of human skin needed to obtain a sufficient number of freshly isolated MCs is still an obstacle for the wider use of this model. To fully see the potential of these cells as in vitro models of human MC biology, future studies of the activation of these cells, both freshly isolated and cultured, by IgE receptor crosslinking and by activation of the MRGPRX2 receptor are needed. Some of these experiments are currently underway in our laboratories.