Structured Illumination Microscopy Improves Spot Detection Performance in Spatial Transcriptomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Histology
2.3. In Situ Sequencing
2.4. Imaging
2.5. Deconvolution
2.6. Data Analysis
3. Results
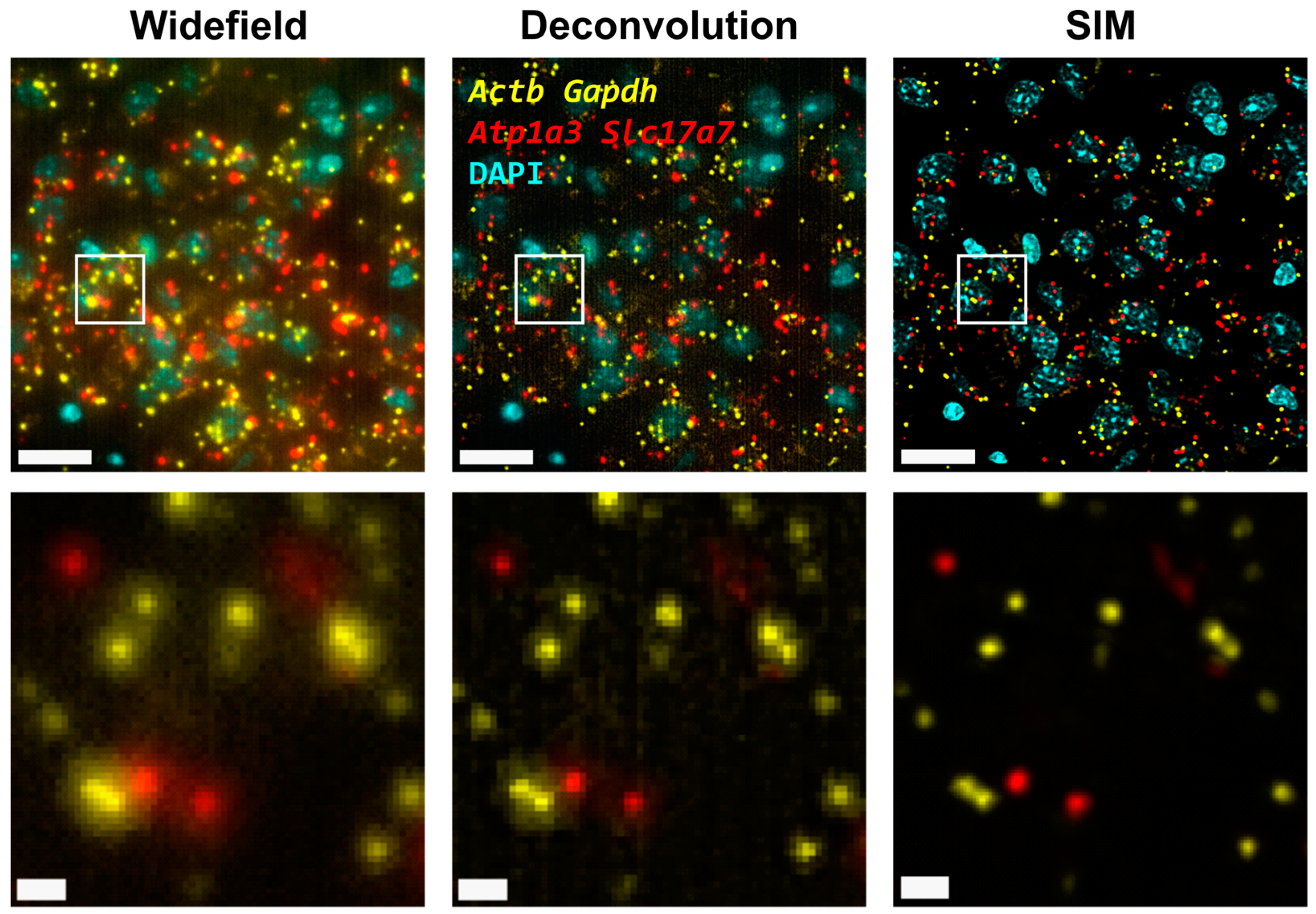
3.1. Structured-Illumination Microscopy Enhances Contrast and Improves Resolution of Amplified Individual Transcripts
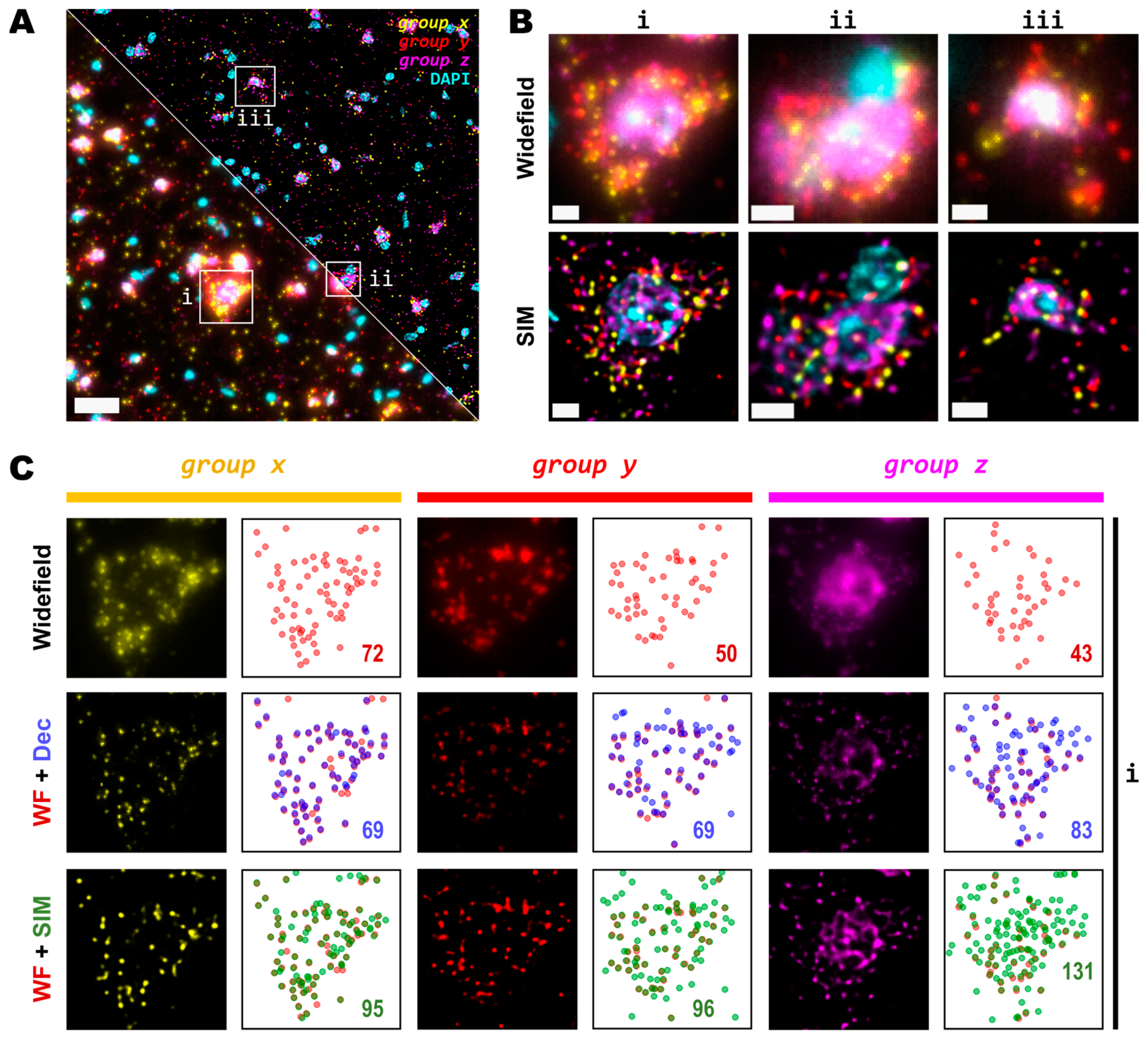
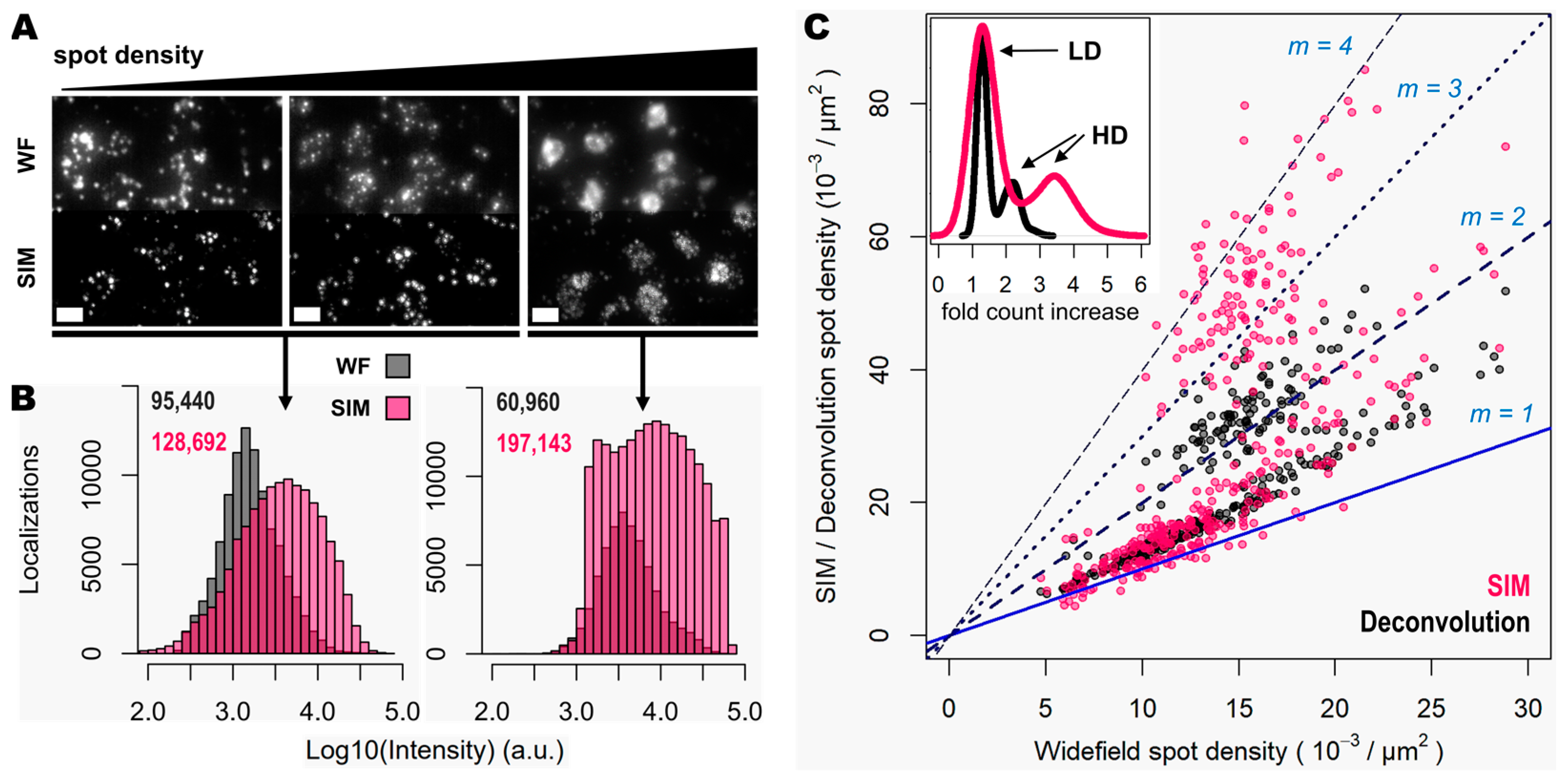
3.2. Spot Detection Performance Is Enhanced for Relatively Low- and Highly Expressed Genes with Moderate Magnification and NA
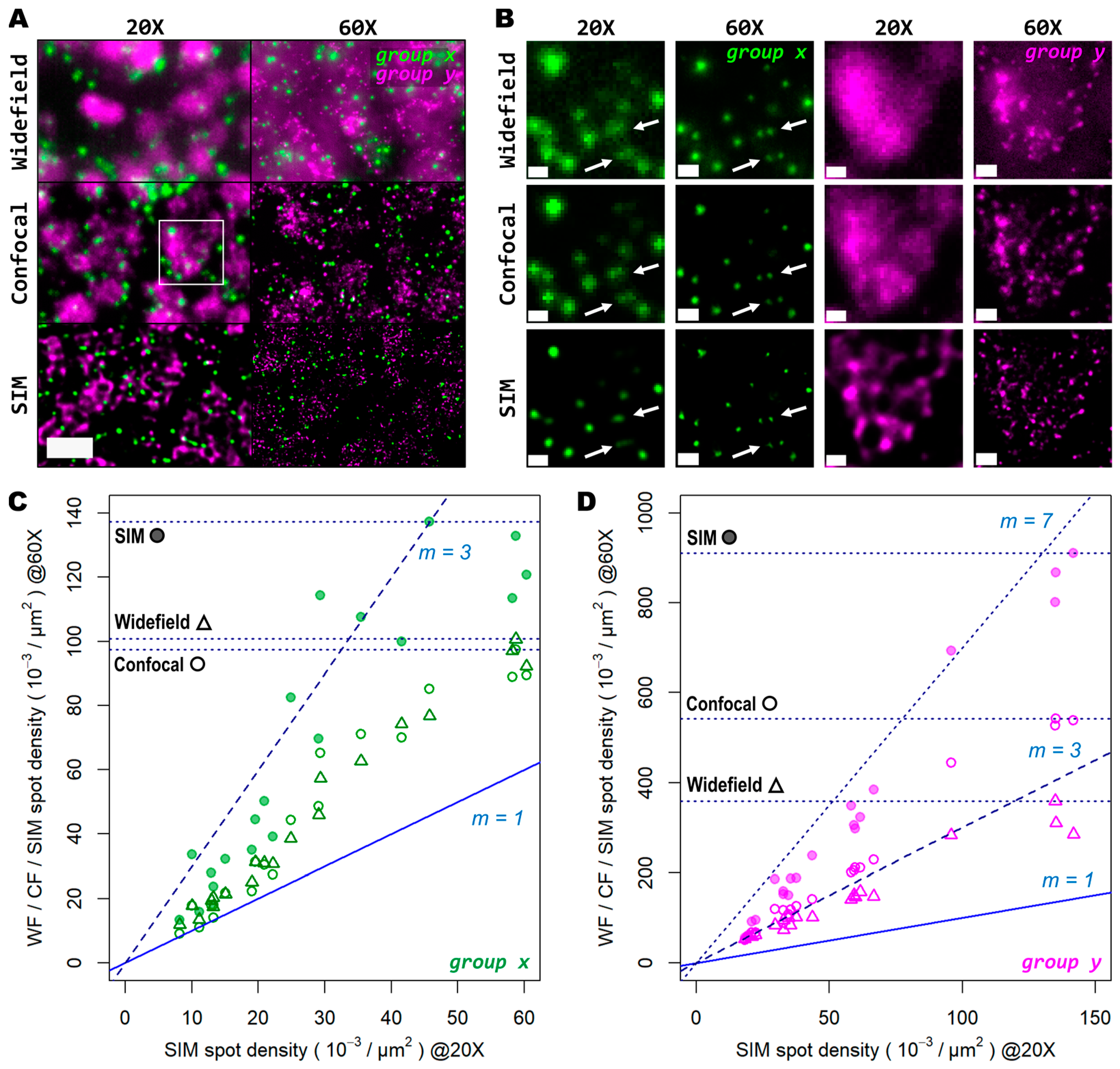
3.3. High Magnification, High NA and Super Resolution Is Needed for Precise Transcript Localisation in Crowded Regions
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| 20× (Figure 3 and Figure 4) | 25× (Figure 1 and Figure 2) | |||||||||
| Sigma | Thres. | S.R.R. | Inlier R. | Max E. | Sigma | Thres. | S.R.R. | Inlier R. | Max E. | |
| WF | 1.75 | 0.002 | 2 | 0.9 | 1.25 | 2.5 | 0.0015 | 2.5 | 0.75 | 1.25 |
| DEC | 2.25 | 0.0015 | 2 | 0.5 | 1.5 | 2.5 | 0.0015 | 2.5 | 0.75 | 1.25 |
| SIM | 1.5 | 0.0015 | 2 | 0.5 | 1.25 | 2 | 0.0015 | 2 | 0.9 | 1 |
| 20× (Figure 5 and Figure S1) | 60× (Figure 5, Figures S1 and S2) | |||||||||
| Sigma | Thres. | S.R.R. | Inlier R. | Max E. | Sigma | Thres. | S.R.R. | Inlier R. | Max E. | |
| WF | 1.5 | 0.002 | 2 | 0.85 | 1.25 | 1.5 | 0.002 | 2 | 0.85 | 1.25 |
| CF | 1.5 | 0.002 | 2 | 0.85 | 1.25 | 2 | 0.002 | 2 | 0.8 | 1.5 |
| SIM | 2 | 0.002 | 2 | 0.8 | 1.5 | 1.5 | 0.002 | 2 | 0.85 | 1.25 |
References
- Asp, M.; Bergenstrahle, J.; Lundeberg, J. Spatially Resolved Transcriptomes-Next Generation Tools for Tissue Exploration. Bioessays 2020, 42, e1900221. [Google Scholar] [CrossRef]
- Marx, V. Method of the Year: Spatially resolved transcriptomics. Nat. Methods 2021, 18, 9–14. [Google Scholar] [CrossRef]
- Rao, A.; Barkley, D.; Franca, G.S.; Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 2021, 596, 211–220. [Google Scholar] [CrossRef]
- Moses, L.; Pachter, L. Museum of spatial transcriptomics. Nat. Methods 2022, 19, 534–546. [Google Scholar] [CrossRef]
- Williams, C.G.; Lee, H.J.; Asatsuma, T.; Vento-Tormo, R.; Haque, A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 2022, 14, 68. [Google Scholar] [CrossRef]
- Rodriques, S.G.; Stickels, R.R.; Goeva, A.; Martin, C.A.; Murray, E.; Vanderburg, C.R.; Welch, J.; Chen, L.M.; Chen, F.; Macosko, E.Z. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science 2019, 363, 1463–1467. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, M.; Deng, Y.; Su, G.; Enninful, A.; Guo, C.C.; Tebaldi, T.; Zhang, D.; Kim, D.; Bai, Z.; et al. High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue. Cell 2020, 183, 1665–1681.e18. [Google Scholar] [CrossRef]
- Ke, R.; Mignardi, M.; Pacureanu, A.; Svedlund, J.; Botling, J.; Wahlby, C.; Nilsson, M. In situ sequencing for RNA analysis in preserved tissue and cells. Nat. Methods 2013, 10, 857–860. [Google Scholar] [CrossRef]
- Chen, K.H.; Boettiger, A.N.; Moffitt, J.R.; Wang, S.; Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 2015, 348, aaa6090. [Google Scholar] [CrossRef]
- Eng, C.L.; Lawson, M.; Zhu, Q.; Dries, R.; Koulena, N.; Takei, Y.; Yun, J.; Cronin, C.; Karp, C.; Yuan, G.C.; et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 2019, 568, 235–239. [Google Scholar] [CrossRef]
- Kishi, J.Y.; Lapan, S.W.; Beliveau, B.J.; West, E.R.; Zhu, A.; Sasaki, H.M.; Saka, S.K.; Wang, Y.; Cepko, C.L.; Yin, P. SABER amplifies FISH: Enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Methods 2019, 16, 533–544. [Google Scholar] [CrossRef]
- Wang, S.; Su, J.H.; Beliveau, B.J.; Bintu, B.; Moffitt, J.R.; Wu, C.T.; Zhuang, X. Spatial organization of chromatin domains and compartments in single chromosomes. Science 2016, 353, 598–602. [Google Scholar] [CrossRef]
- Gyllborg, D.; Langseth, C.M.; Qian, X.; Choi, E.; Salas, S.M.; Hilscher, M.M.; Lein, E.S.; Nilsson, M. Hybridization-based in situ sequencing (HybISS) for spatially resolved transcriptomics in human and mouse brain tissue. Nucleic Acids Res. 2020, 48, e112. [Google Scholar] [CrossRef]
- Hirano, Y.; Matsuda, A.; Hiraoka, Y. Recent advancements in structured-illumination microscopy toward live-cell imaging. Microscopy 2015, 64, 237–249. [Google Scholar] [CrossRef]
- Wu, Y.; Shroff, H. Faster, sharper, and deeper: Structured illumination microscopy for biological imaging. Nat. Methods 2018, 15, 1011–1019. [Google Scholar] [CrossRef]
- Miron, E.; Oldenkamp, R.; Brown, J.M.; Pinto, D.M.S.; Xu, C.S.; Faria, A.R.; Shaban, H.A.; Rhodes, J.D.P.; Innocent, C.; de Ornellas, S.; et al. Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci. Adv. 2020, 6, eaba8811. [Google Scholar] [CrossRef]
- Li, D.; Shao, L.; Chen, B.-C.; Zhang, X.; Zhang, M.; Moses, B.; Milkie, D.E.; Beach, J.R.; Hammer, J.A.; Pasham, M.; et al. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science 2015, 349, aab3500. [Google Scholar] [CrossRef]
- Hilscher, M.M.; Gyllborg, D.; Yokota, C.; Nilsson, M. In Situ Sequencing: A High-Throughput, Multi-Targeted Gene Expression Profiling Technique for Cell Typing in Tissue Sections. Methods Mol. Biol. 2020, 2148, 313–329. [Google Scholar]
- Sountoulidis, A.; Liontos, A.; Nguyen, H.P.; Firsova, A.B.; Fysikopoulos, A.; Qian, X.; Seeger, W.; Sundstrom, E.; Nilsson, M.; Samakovlis, C. SCRINSHOT enables spatial mapping of cell states in tissue sections with single-cell resolution. PLoS Biol. 2020, 18, e3000675. [Google Scholar] [CrossRef]
- Wernersson, E.; Gelali, E.; Girelli, G.; Wang, S.; Castillo, D.; Langseth, C.M.; Nguyen, H.; Chattoraj, S.; Casals, A.M.; Lundberg, E.; et al. Deconwolf enables high-performance deconvolution of widefield fluorescence microscopy images. 2022; preprint. [Google Scholar] [CrossRef]
- Bahry, E.; Breimann, L.; Zouinkhi, M.; Epstein, L.; Kolyvanov, K.; Mamrak, N.; King, B.; Long, X.; Harrington, K.I.S.; Lionnet, T.; et al. RS-FISH: Precise, interactive, fast, and scalable FISH spot detection. Nat. Methods 2022, 19, 1563–1567. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 1 April 2023).
- Lee, H.; Marco Salas, S.; Gyllborg, D.; Nilsson, M. Direct RNA targeted in situ sequencing for transcriptomic profiling in tissue. Sci. Rep. 2022, 12, 7976. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Chang, B.J.; Roudot, P.; Zhou, F.; Sapoznik, E.; Marlar-Pavey, M.; Hayes, J.B.; Brown, P.T.; Zeng, C.W.; Lambert, T.; et al. Resolution doubling in light-sheet microscopy via oblique plane structured illumination. Nat. Methods 2022, 19, 1419–1426. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Han, X.; Su, Y.; Glidewell, M.; Daniels, J.S.; Liu, J.; Sengupta, T.; Rey-Suarez, I.; Fischer, R.; Patel, A.; et al. Multiview confocal super-resolution microscopy. Nature 2021, 600, 279–284. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Linares, A.; Brighi, C.; Espinola, S.; Bacchi, F.; Crevenna, Á.H. Structured Illumination Microscopy Improves Spot Detection Performance in Spatial Transcriptomics. Cells 2023, 12, 1310. https://doi.org/10.3390/cells12091310
Linares A, Brighi C, Espinola S, Bacchi F, Crevenna ÁH. Structured Illumination Microscopy Improves Spot Detection Performance in Spatial Transcriptomics. Cells. 2023; 12(9):1310. https://doi.org/10.3390/cells12091310
Chicago/Turabian StyleLinares, Alejandro, Carlo Brighi, Sergio Espinola, Francesco Bacchi, and Álvaro H. Crevenna. 2023. "Structured Illumination Microscopy Improves Spot Detection Performance in Spatial Transcriptomics" Cells 12, no. 9: 1310. https://doi.org/10.3390/cells12091310
APA StyleLinares, A., Brighi, C., Espinola, S., Bacchi, F., & Crevenna, Á. H. (2023). Structured Illumination Microscopy Improves Spot Detection Performance in Spatial Transcriptomics. Cells, 12(9), 1310. https://doi.org/10.3390/cells12091310







