A Non-Canonical Role for the Glycosyltransferase Enzyme UGT2B17 as a Novel Constituent of the B Cell Receptor Signalosome
Abstract
1. Introduction
2. Materials and Methods
2.1. CLL Patient Cell Samples
2.2. Gene Expression Analysis
2.3. Cell Models and Cell-Based Assays
2.4. Antibodies and Immunoblotting
2.5. Immunoprecipitation
2.6. Immunofluorescence
2.7. Protein Stability
3. Results
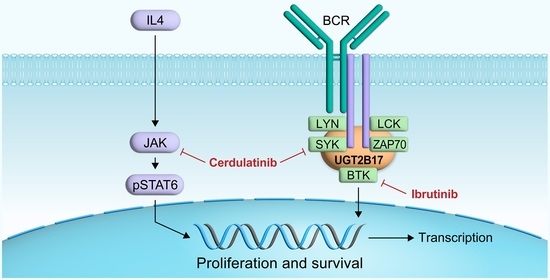
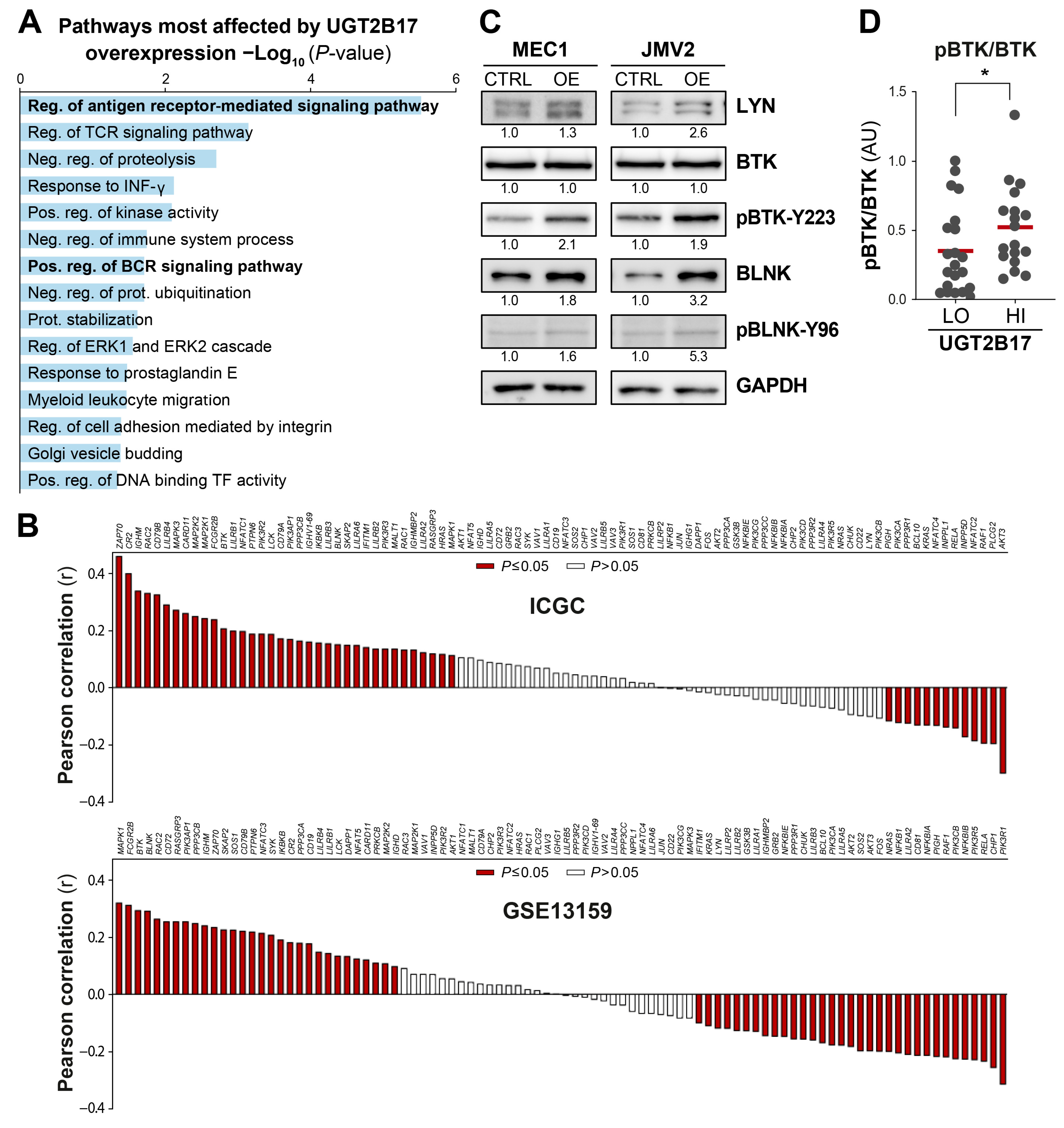
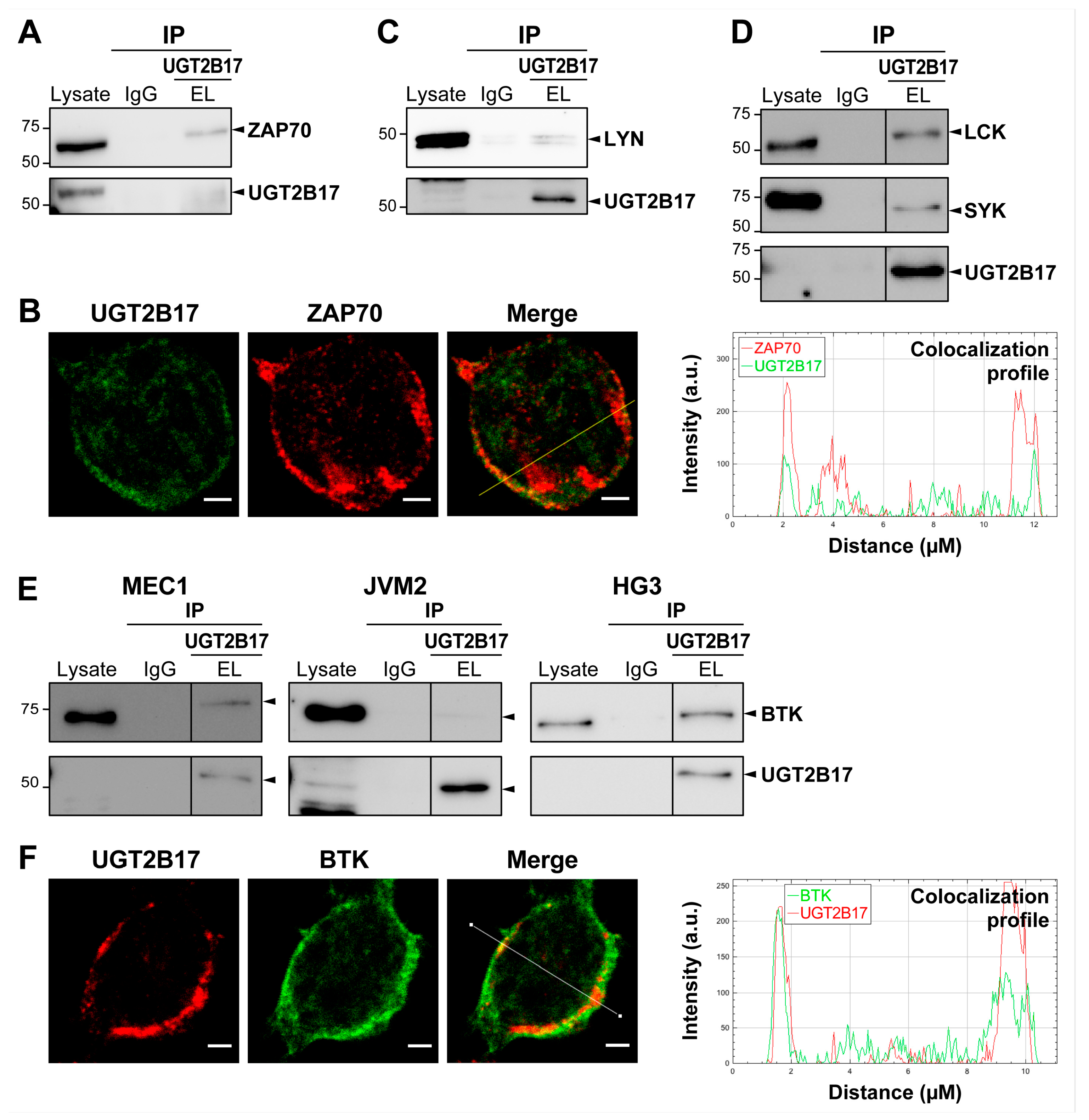
3.1. Interplay between UGT2B17 and the BCR Signalosome
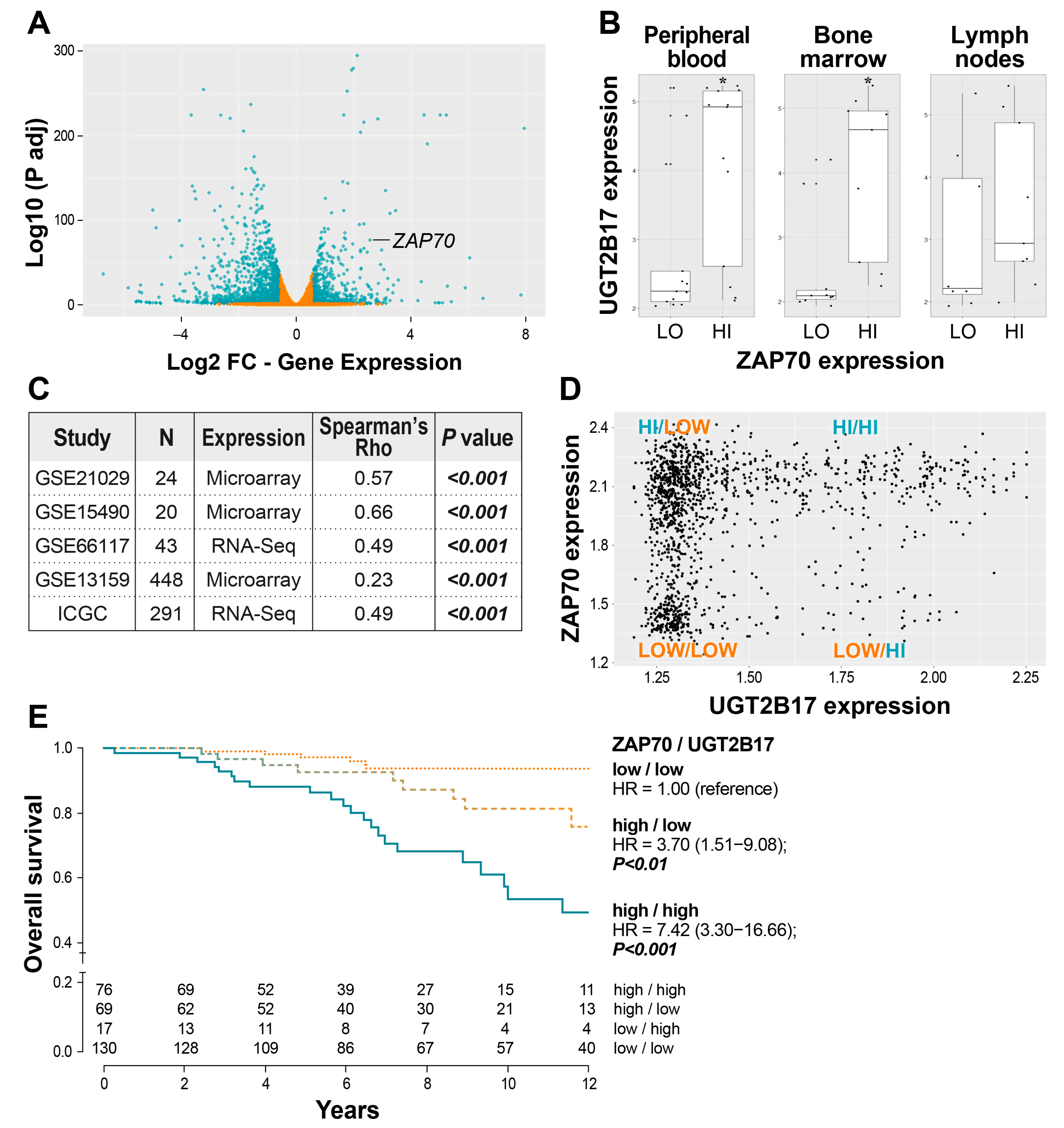
3.2. Correlation of UGT2B17 Expression with the Adverse Marker ZAP70 and the Influence of Their Co-Expression on the Overall Survival of CLL Patients
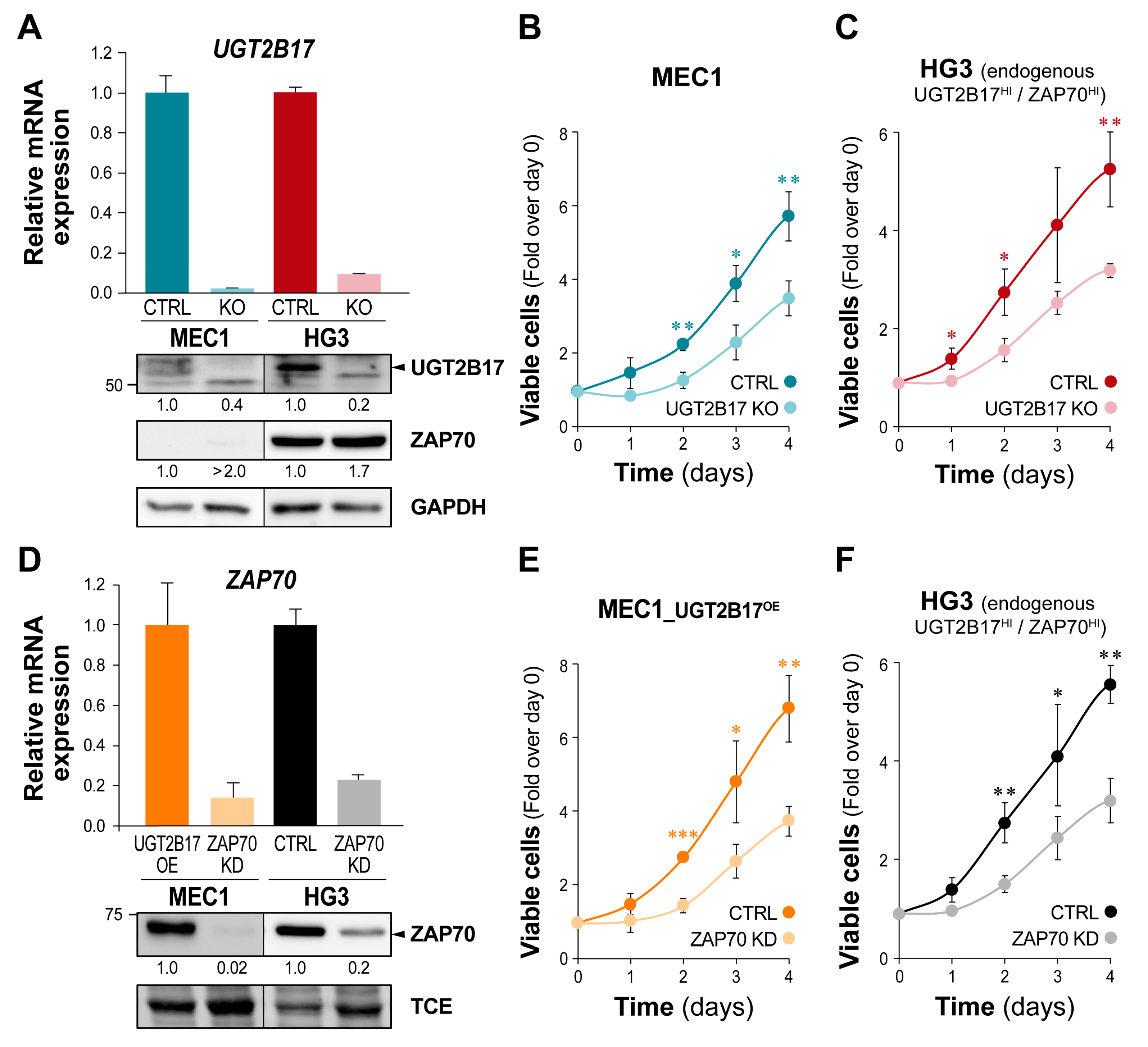
3.3. The UGT2B17-Dependent Enhanced Proliferation of Leukemic Cells Is Impaired by ZAP70 Depletion
3.4. UGT2B17 Interacts with Proximal BCR Effector Proteins in Leukemic Cells
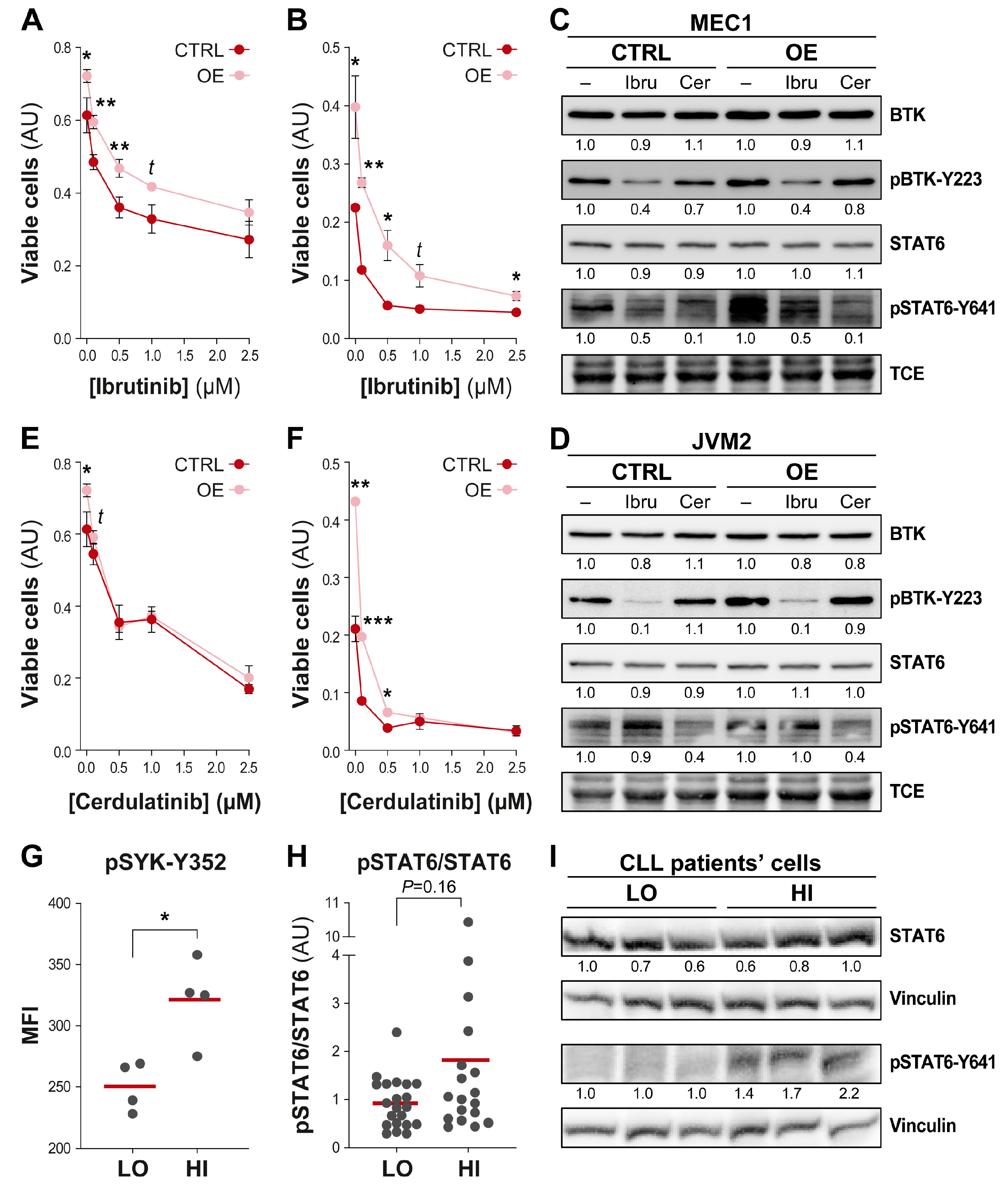
3.5. Efficient Targeting of the UGT2B17 Pro-Proliferative Effect by Dual Inhibition of SYK/JAK Kinases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seftel, M.D.; Demers, A.A.; Banerji, V.; Gibson, S.B.; Morales, C.; Musto, G.; Pitz, M.W.; Johnston, J.B. High incidence of chronic lymphocytic leukemia (CLL) diagnosed by immunophenotyping: A population-based Canadian cohort. Leuk. Res. 2009, 33, 1463–1468. [Google Scholar] [CrossRef] [PubMed]
- Hampel, P.J.; Parikh, S.A.; Call, T.G. Incorporating molecular biomarkers into the continuum of care in chronic lymphocytic leukemia. Leuk. Lymphoma 2021, 62, 1289–1301. [Google Scholar] [CrossRef]
- Yun, X.; Zhang, Y.; Wang, X. Recent progress of prognostic biomarkers and risk scoring systems in chronic lymphocytic leukemia. Biomark. Res. 2020, 8, 40. [Google Scholar] [CrossRef]
- Hampel, P.J.; Parikh, S.A. Chronic lymphocytic leukemia treatment algorithm 2022. Blood Cancer J. 2022, 12, 161. [Google Scholar] [CrossRef] [PubMed]
- Owen, C.; Banerji, V.; Johnson, N.; Gerrie, A.; Aw, A.; Chen, C.; Robinson, S. Canadian evidence-based guideline for frontline treatment of chronic lymphocytic leukemia: 2022 update. Leuk. Res. 2023, 125, 107016. [Google Scholar] [CrossRef]
- Allain, E.P.; Rouleau, M.; Le, T.; Vanura, K.; Villeneuve, L.; Caron, P.; Turcotte, V.; Levesque, E.; Guillemette, C. Inactivation of Prostaglandin E(2) as a Mechanism for UGT2B17-Mediated Adverse Effects in Chronic Lymphocytic Leukemia. Front. Oncol. 2019, 9, 606. [Google Scholar] [CrossRef] [PubMed]
- Bhoi, S.; Baliakas, P.; Cortese, D.; Mattsson, M.; Engvall, M.; Smedby, K.E.; Juliusson, G.; Sutton, L.A.; Mansouri, L. UGT2B17 expression: A novel prognostic marker within IGHV-mutated chronic lymphocytic leukemia? Haematologica 2016, 101, e63–e65. [Google Scholar] [CrossRef]
- Gruber, M.; Bellemare, J.; Hoermann, G.; Gleiss, A.; Porpaczy, E.; Bilban, M.; Le, T.; Zehetmayer, S.; Mannhalter, C.; Gaiger, A.; et al. Overexpression of uridine diphospho glucuronosyltransferase 2B17 in high-risk chronic lymphocytic leukemia. Blood 2013, 121, 1175–1183. [Google Scholar] [CrossRef]
- Knisbacher, B.A.; Lin, Z.; Hahn, C.K.; Nadeu, F.; Duran-Ferrer, M.; Stevenson, K.E.; Tausch, E.; Delgado, J.; Barbera-Mourelle, A.; Taylor-Weiner, A.; et al. Molecular map of chronic lymphocytic leukemia and its impact on outcome. Nat. Genet. 2022, 54, 1664–1674. [Google Scholar] [CrossRef]
- Allain, E.P.; Rouleau, M.; Vanura, K.; Tremblay, S.; Vaillancourt, J.; Bat, V.; Caron, P.; Villeneuve, L.; Labriet, A.; Turcotte, V.; et al. UGT2B17 modifies drug response in chronic lymphocytic leukaemia. Br. J. Cancer 2020, 123, 240–251. [Google Scholar] [CrossRef]
- Allain, E.P.; Rouleau, M.; Levesque, E.; Guillemette, C. Emerging roles for UDP-glucuronosyltransferases in drug resistance and cancer progression. Br. J. Cancer 2020, 122, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Allain, E.P.; Venzl, K.; Caron, P.; Turcotte, V.; Simonyan, D.; Gruber, M.; Le, T.; Levesque, E.; Guillemette, C.; Vanura, K. Sex-dependent association of circulating sex steroids and pituitary hormones with treatment-free survival in chronic lymphocytic leukemia patients. Ann. Hematol. 2018, 97, 1649–1661. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Baran, J.; Cros, A.; Guberman, J.M.; Haider, S.; Hsu, J.; Liang, Y.; Rivkin, E.; Wang, J.; Whitty, B.; et al. International Cancer Genome Consortium Data Portal--a one-stop shop for cancer genomics data. Database 2011, 2011, bar026. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, G.; Dozmorov, M.; Wren, J.D.; Qiu, J.; Shi, H.; Xu, D. Hypomethylation coordinates antagonistically with hypermethylation in cancer development: A case study of leukemia. Hum. Genom. 2016, 10 (Suppl. 2), 18. [Google Scholar] [CrossRef] [PubMed]
- Kohlmann, A.; Kipps, T.J.; Rassenti, L.Z.; Downing, J.R.; Shurtleff, S.A.; Mills, K.I.; Gilkes, A.F.; Hofmann, W.K.; Basso, G.; Dell’orto, M.C.; et al. An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: The Microarray Innovations in LEukemia study prephase. Br. J. Haematol. 2008, 142, 802–807. [Google Scholar] [CrossRef]
- Shehata, M.; Demirtas, D.; Schnabl, S.; Hilgarth, M.; Hubmann, R.; Fonatsch, C.; Schwarzinger, I.; Hopfinger, G.; Eigenberger, K.; Heintel, D.; et al. Sequential gene expression profiling during treatment for identification of predictive markers and novel therapeutic targets in chronic lymphocytic leukemia. Leukemia 2010, 24, 2122–2127. [Google Scholar] [CrossRef]
- Herishanu, Y.; Perez-Galan, P.; Liu, D.; Biancotto, A.; Pittaluga, S.; Vire, B.; Gibellini, F.; Njuguna, N.; Lee, E.; Stennett, L.; et al. The lymph node microenvironment promotes B-cell receptor signaling, NF-kappaB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood 2011, 117, 563–574. [Google Scholar] [CrossRef]
- Gutierrez, A., Jr.; Tschumper, R.C.; Wu, X.; Shanafelt, T.D.; Eckel-Passow, J.; Huddleston, P.M., 3rd; Slager, S.L.; Kay, N.E.; Jelinek, D.F. LEF-1 is a prosurvival factor in chronic lymphocytic leukemia and is expressed in the preleukemic state of monoclonal B-cell lymphocytosis. Blood 2010, 116, 2975–2983. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Claus, R.; Lucas, D.M.; Ruppert, A.S.; Williams, K.E.; Weng, D.; Patterson, K.; Zucknick, M.; Oakes, C.C.; Rassenti, L.Z.; Greaves, A.W.; et al. Validation of ZAP-70 methylation and its relative significance in predicting outcome in chronic lymphocytic leukemia. Blood 2014, 124, 42–48. [Google Scholar] [CrossRef]
- Nguyen Van Long, F.; Valcout-Gendron, D.; Caron, P.; Rouleau, M.; Villeneuve, L.; Sergerie, R.; Laverdière, I.; Vanura, K.; Guillemette, C. Untargeted metabolomics identifies metabolic dysregulation of sphingolipids associated with aggressive chronic lymphocytic leukemia and poor survival. Submitted 2023. [Google Scholar]
- Emond, J.P.; Labriet, A.; Desjardins, S.; Rouleau, M.; Villeneuve, L.; Hovington, H.; Brisson, H.; Lacombe, L.; Simonyan, D.; Caron, P.; et al. Factors Affecting Interindividual Variability of Hepatic UGT2B17 Protein Expression Examined Using a Novel Specific Monoclonal Antibody. Drug. Metab. Dispos. 2019, 47, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, L.; Hovington, H.; Brisson, H.; Mehdi, S.; Beillevaire, D.; Emond, J.P.; Wagner, A.; Villeneuve, L.; Simonyan, D.; Ouellet, V.; et al. UGT2B28 accelerates prostate cancer progression through stabilization of the endocytic adaptor protein HIP1 regulating AR and EGFR pathways. Cancer Lett. 2023, 553, 215994. [Google Scholar] [CrossRef] [PubMed]
- Tsang, M.; Gantchev, J.; Ghazawi, F.M.; Litvinov, I.V. Protocol for adhesion and immunostaining of lymphocytes and other non-adherent cells in culture. Biotechniques 2017, 63, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Guillemette, C.; Levesque, E.; Beaulieu, M.; Turgeon, D.; Hum, D.W.; Belanger, A. Differential regulation of two uridine diphospho-glucuronosyltransferases, UGT2B15 and UGT2B17, in human prostate LNCaP cells. Endocrinology 1997, 138, 2998–3005. [Google Scholar] [CrossRef]
- Mockridge, C.I.; Potter, K.N.; Wheatley, I.; Neville, L.A.; Packham, G.; Stevenson, F.K. Reversible anergy of sIgM-mediated signaling in the two subsets of CLL defined by VH-gene mutational status. Blood 2007, 109, 4424–4431. [Google Scholar] [CrossRef]
- Chen, L.; Widhopf, G.; Huynh, L.; Rassenti, L.; Rai, K.R.; Weiss, A.; Kipps, T.J. Expression of ZAP-70 is associated with increased B-cell receptor signaling in chronic lymphocytic leukemia. Blood 2002, 100, 4609–4614. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Huynh, L.; Apgar, J.; Tang, L.; Rassenti, L.; Weiss, A.; Kipps, T.J. ZAP-70 enhances IgM signaling independent of its kinase activity in chronic lymphocytic leukemia. Blood 2008, 111, 2685–2692. [Google Scholar] [CrossRef]
- Gobessi, S.; Laurenti, L.; Longo, P.G.; Sica, S.; Leone, G.; Efremov, D.G. ZAP-70 enhances B-cell-receptor signaling despite absent or inefficient tyrosine kinase activation in chronic lymphocytic leukemia and lymphoma B cells. Blood 2007, 109, 2032–2039. [Google Scholar] [CrossRef]
- Claus, R.; Lucas, D.M.; Stilgenbauer, S.; Ruppert, A.S.; Yu, L.; Zucknick, M.; Mertens, D.; Buhler, A.; Oakes, C.C.; Larson, R.A.; et al. Quantitative DNA methylation analysis identifies a single CpG dinucleotide important for ZAP-70 expression and predictive of prognosis in chronic lymphocytic leukemia. J. Clin. Oncol. 2012, 30, 2483–2491. [Google Scholar] [CrossRef] [PubMed]
- Amin, N.A.; Balasubramanian, S.; Saiya-Cork, K.; Shedden, K.; Hu, N.; Malek, S.N. Cell-Intrinsic Determinants of Ibrutinib-Induced Apoptosis in Chronic Lymphocytic Leukemia. Clin. Cancer Res. 2017, 23, 1049–1059. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Blunt, M.D.; Koehrer, S.; Dobson, R.C.; Larrayoz, M.; Wilmore, S.; Hayman, A.; Parnell, J.; Smith, L.D.; Davies, A.; Johnson, P.W.M.; et al. The Dual Syk/JAK Inhibitor Cerdulatinib Antagonizes B-cell Receptor and Microenvironmental Signaling in Chronic Lymphocytic Leukemia. Clin. Cancer Res. 2017, 23, 2313–2324. [Google Scholar] [CrossRef] [PubMed]
- Spaner, D.E.; Luo, Y.; Wang, G.; Gallagher, J.; Tsui, H.; Shi, Y. Janus kinases restrain chronic lymphocytic leukemia cells in patients on ibrutinib: Results of a phase II trial. Cancer Med. 2021, 10, 8789–8798. [Google Scholar] [CrossRef]
- Talab, F.; Allen, J.C.; Thompson, V.; Lin, K.; Slupsky, J.R. LCK is an important mediator of B-cell receptor signaling in chronic lymphocytic leukemia cells. Mol. Cancer Res. 2013, 11, 541–554. [Google Scholar] [CrossRef]
- Chen, J.; Sathiaseelan, V.; Moore, A.; Tan, S.; Chilamakuri, C.S.R.; Roamio Franklin, V.N.; Shahsavari, A.; Jakwerth, C.A.; Hake, S.B.; Warren, A.J.; et al. ZAP-70 constitutively regulates gene expression and protein synthesis in chronic lymphocytic leukemia. Blood 2021, 137, 3629–3640. [Google Scholar] [CrossRef]
- Audet-Delage, Y.; Rouleau, M.; Rouleau, M.; Roberge, J.; Miard, S.; Picard, F.; Tetu, B.; Guillemette, C. Cross-Talk between Alternatively Spliced UGT1A Isoforms and Colon Cancer Cell Metabolism. Mol. Pharmacol. 2017, 91, 167–177. [Google Scholar] [CrossRef]
- Ishii, Y.; Takeda, S.; Yamada, H.; Oguri, K. Functional protein-protein interaction of drug metabolizing enzymes. Front. Biosci. 2005, 10, 887–895. [Google Scholar] [CrossRef][Green Version]
- Rouleau, M.; Audet-Delage, Y.; Desjardins, S.; Rouleau, M.; Girard-Bock, C.; Guillemette, C. Endogenous Protein Interactome of Human UDP-Glucuronosyltransferases Exposed by Untargeted Proteomics. Front. Pharmacol. 2017, 8, 23. [Google Scholar] [CrossRef]
- Rouleau, M.; Roberge, J.; Bellemare, J.; Guillemette, C. Dual roles for splice variants of the glucuronidation pathway as regulators of cellular metabolism. Mol. Pharmacol. 2014, 85, 29–36. [Google Scholar] [CrossRef]
- Liu, T.M.; Woyach, J.A.; Zhong, Y.; Lozanski, A.; Lozanski, G.; Dong, S.; Strattan, E.; Lehman, A.; Zhang, X.; Jones, J.A.; et al. Hypermorphic mutation of phospholipase C, gamma2 acquired in ibrutinib-resistant CLL confers BTK independency upon B-cell receptor activation. Blood 2015, 126, 61–68. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagner, A.; Rouleau, M.; Villeneuve, L.; Le, T.; Peltier, C.; Allain, É.P.; Beaudoin, C.; Tremblay, S.; Courtier, F.; Nguyen Van Long, F.; et al. A Non-Canonical Role for the Glycosyltransferase Enzyme UGT2B17 as a Novel Constituent of the B Cell Receptor Signalosome. Cells 2023, 12, 1295. https://doi.org/10.3390/cells12091295
Wagner A, Rouleau M, Villeneuve L, Le T, Peltier C, Allain ÉP, Beaudoin C, Tremblay S, Courtier F, Nguyen Van Long F, et al. A Non-Canonical Role for the Glycosyltransferase Enzyme UGT2B17 as a Novel Constituent of the B Cell Receptor Signalosome. Cells. 2023; 12(9):1295. https://doi.org/10.3390/cells12091295
Chicago/Turabian StyleWagner, Antoine, Michèle Rouleau, Lyne Villeneuve, Trang Le, Cheryl Peltier, Éric P. Allain, Caroline Beaudoin, Sophie Tremblay, Fréderic Courtier, Flora Nguyen Van Long, and et al. 2023. "A Non-Canonical Role for the Glycosyltransferase Enzyme UGT2B17 as a Novel Constituent of the B Cell Receptor Signalosome" Cells 12, no. 9: 1295. https://doi.org/10.3390/cells12091295
APA StyleWagner, A., Rouleau, M., Villeneuve, L., Le, T., Peltier, C., Allain, É. P., Beaudoin, C., Tremblay, S., Courtier, F., Nguyen Van Long, F., Laverdière, I., Lévesque, É., Banerji, V., Vanura, K., & Guillemette, C. (2023). A Non-Canonical Role for the Glycosyltransferase Enzyme UGT2B17 as a Novel Constituent of the B Cell Receptor Signalosome. Cells, 12(9), 1295. https://doi.org/10.3390/cells12091295