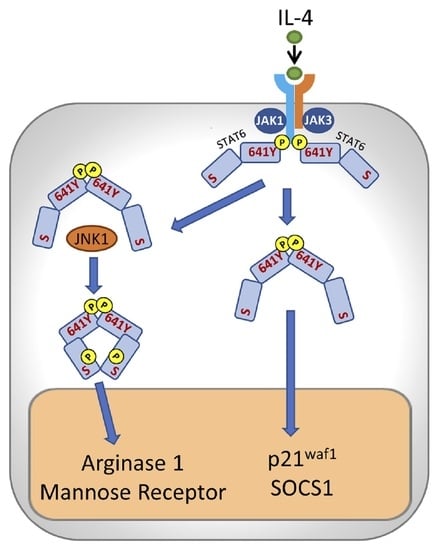
Distinct Responses to IL4 in Macrophages Mediated by JNK
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Culture and Animal Models
2.3. RNA Extraction, Reverse Transcription PCR, and qPCR
2.4. Protein Extraction and Western Blot Analysis
2.5. JNK Activity Assay
2.6. Chromatin Immunoprecipitation Assay
2.7. Statistical Analysis
3. Results
3.1. IL-4 Induces Early and Short Activation of JNK-1 but Not of ERK or p38
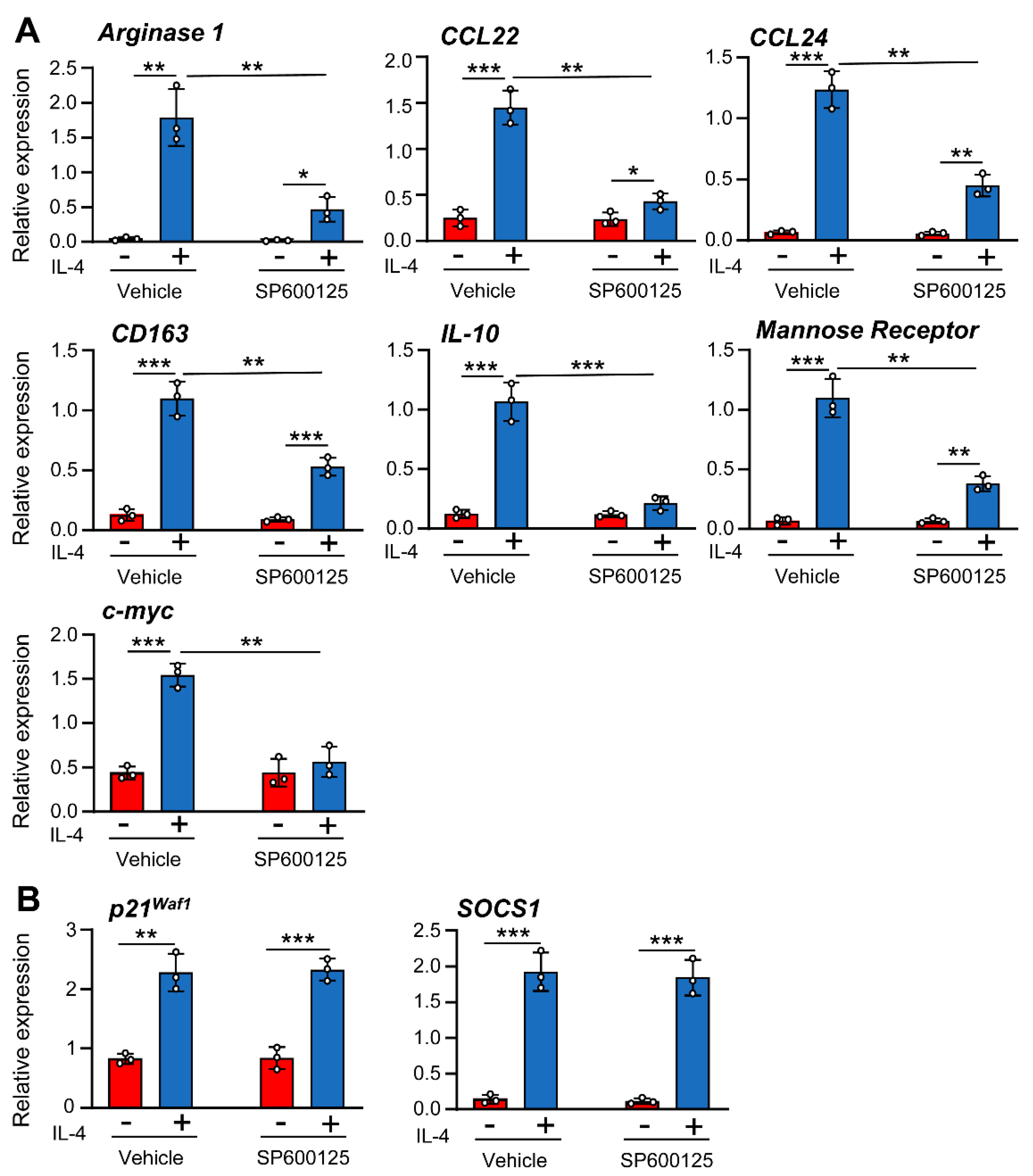
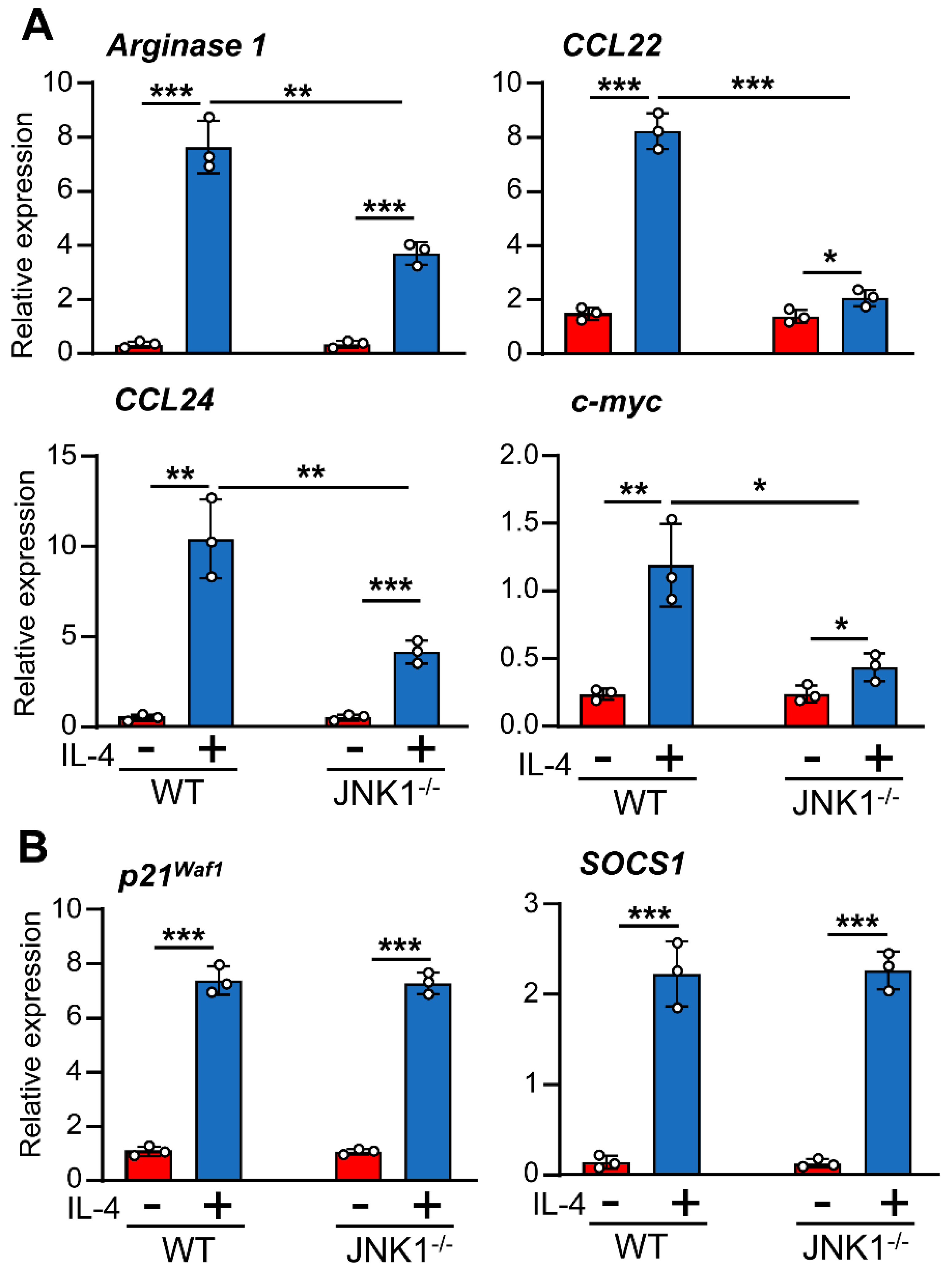
3.2. IL-4-Induced JNK-1 Activation Contributes to the Regulation of Selective Genes
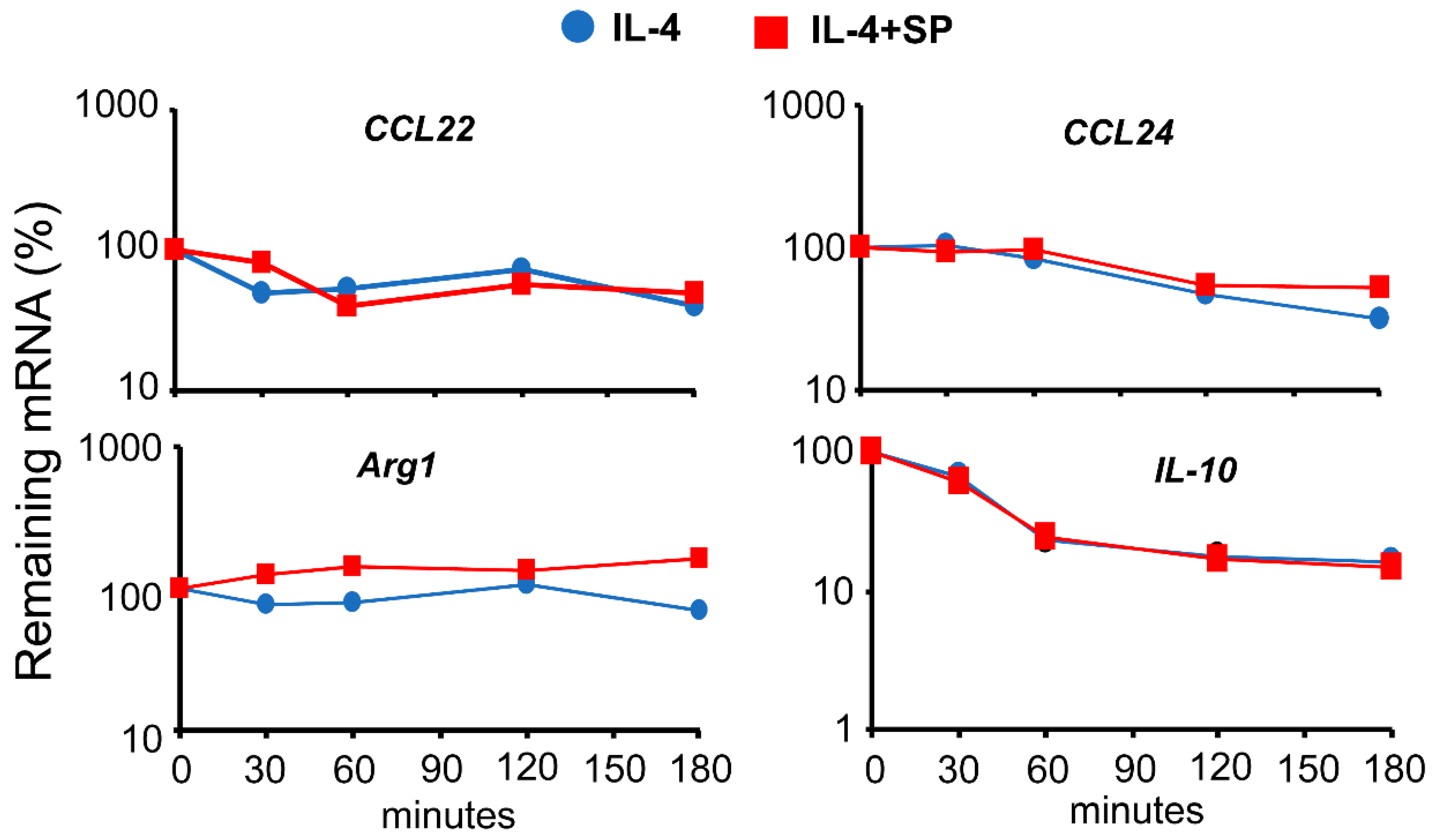
3.3. JNK-1 Does Not Affect mRNA Stability
3.4. JNK-1 Phosphorylates STAT-6 on Serine Residues without Affecting Its Binding to DNA
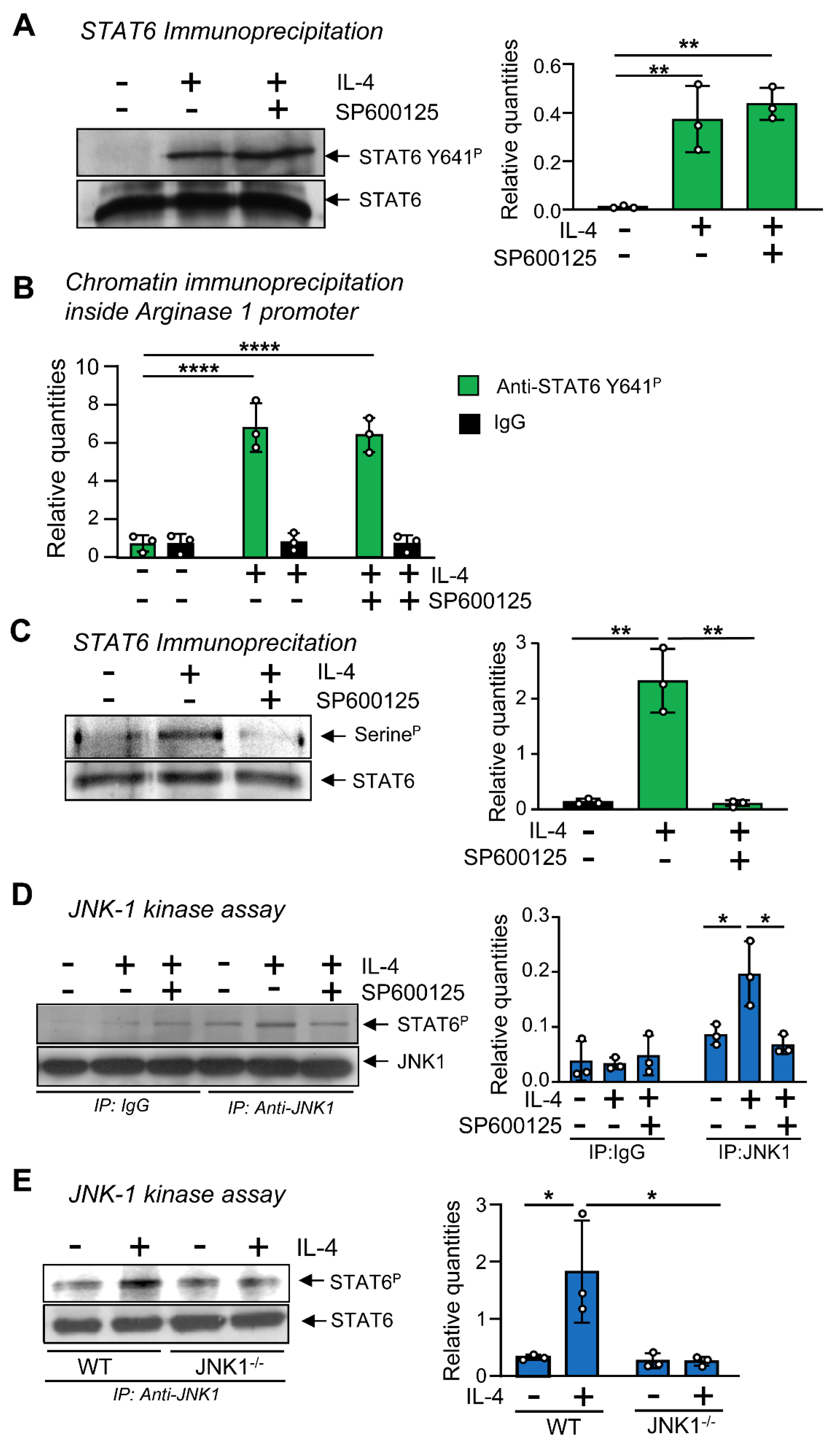
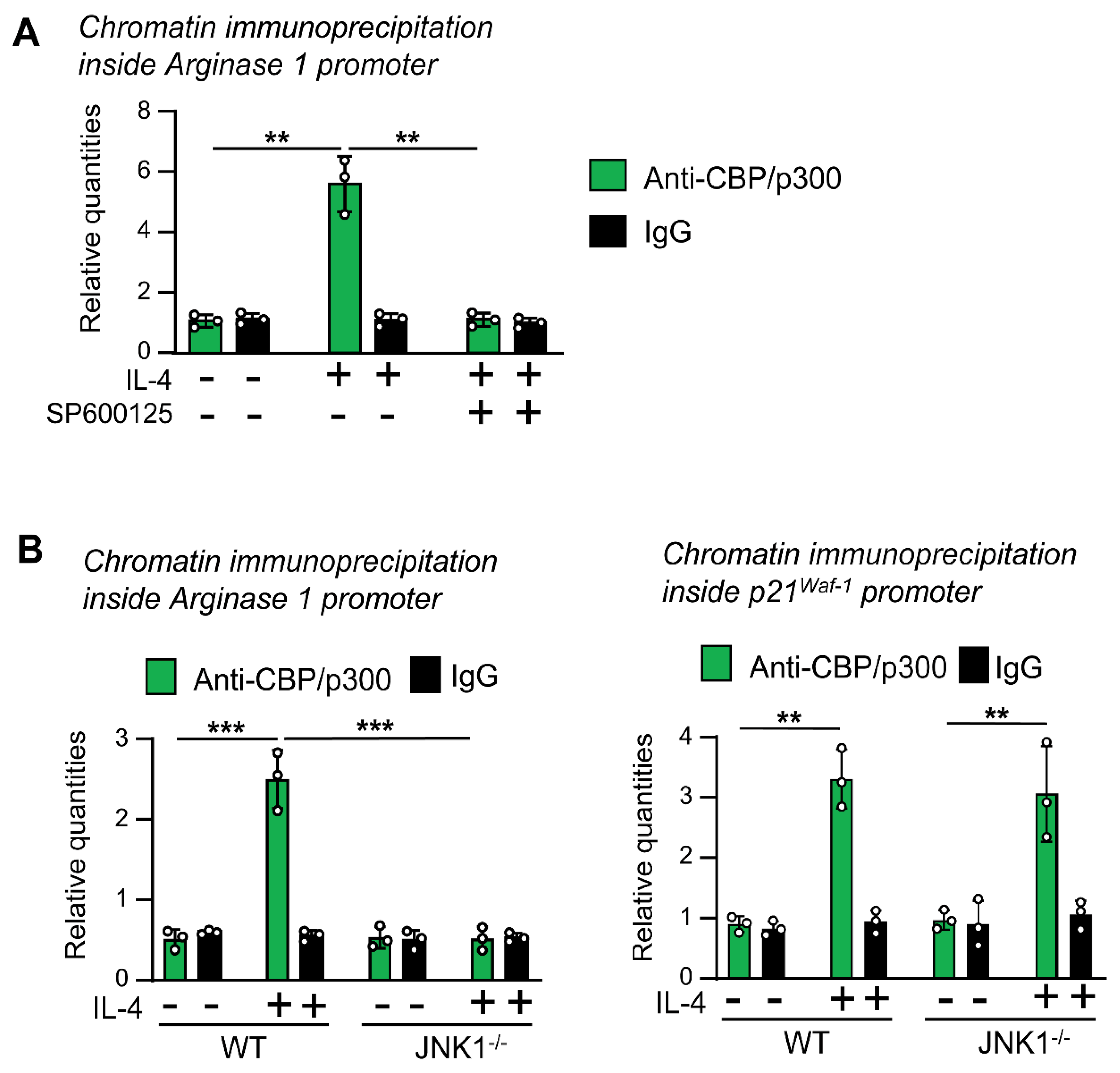
3.5. JNK-1 Is Required for Promoting the Recruitment of CBP/p300 to the Arginase 1 Promoter
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Nelms, K.; Keegan, A.D.; Zamorano, J.; Ryan, J.J.; Paul, W.E. The IL-4 receptor: Signaling mechanisms and biologic functions. Annu. Rev. Immunol. 1999, 17, 701–738. [Google Scholar] [CrossRef]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef]
- Wynn, T.A.; Vannella, K.M. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity 2016, 44, 450–462. [Google Scholar] [CrossRef]
- Gordon, S.; Martinez, F.O. Alternative Activation of Macrophages: Mechanism and Functions. Immunity 2010, 32, 593–604. [Google Scholar] [CrossRef]
- Wick, K.R.; Berton, M.T. IL-4 induces serine phosphorylation of the STAT6 transactivation domain in B lymphocytes. Mol. Immunol. 2000, 37, 641–652. [Google Scholar] [CrossRef]
- Wang, Y.; Malabarba, M.G.; Nagy, Z.S.; Kirken, R.A. Interleukin 4 Regulates Phosphorylation of Serine 756 in the Transactivation Domain of Stat6. Roles for Multiple Phosphorylation Sites and Stat6 Function. J. Biol. Chem. 2004, 279, 25196–25203. [Google Scholar] [CrossRef]
- So, E.-Y.; Oh, J.; Jang, J.-Y.; Kim, J.-H.; Lee, C.-E. Ras/Erk pathway positively regulates Jak1/STAT6 activity and IL-4 gene expression in Jurkat T cells. Mol. Immunol. 2007, 44, 3416–3426. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Sahoo, N.; Ullah, U.; Kallionpää, H.; Suneja, A.; Lahesmaa, R.; Rao, K.V.S. A novel mechanism for ERK-dependent regulation of IL4 transcription during human Th2-cell differentiation. Immunol. Cell Biol. 2012, 90, 676–687. [Google Scholar] [CrossRef]
- Kawano, A.; Ariyoshi, W.; Yoshioka, Y.; Hikiji, H.; Nishihara, T.; Okinaga, T. Docosahexaenoic acid enhances M2 macrophage polarization via the p38 signaling pathway and autophagy. J. Cell. Biochem. 2019, 120, 12604–12617. [Google Scholar] [CrossRef] [PubMed]
- Levings, M.K.; Bessette, D.C.; Schrader, J.W. Interleukin-4 synergizes with Raf-1 to promote long-term proliferation and activation of c-jun N-terminal kinase. Blood 1999, 93, 3694–3702. [Google Scholar] [CrossRef] [PubMed]
- Seger, R.; Krebs, E.G. The MAPK signaling cascade. FASEB J. 1995, 9, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Comalada, M.; Lloberas, J.; Celada, A. MKP-1: A critical phosphatase in the biology of macrophages controlling the switch between proliferation and activation. Eur. J. Immunol. 2012, 42, 1938–1948. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, W.-Z.; Liu, T.; Feng, X.; Yang, N.; Zhou, H.-F. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J. Recept. Signal Transduct. Res. 2015, 35, 600–604. [Google Scholar] [CrossRef] [PubMed]
- Celada, A.; Gray, P.W.; Rinderknecht, E.; Schreiber, R.D. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J. Exp. Med. 1984, 160, 55–74. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.; Sano, Y.; Todorova, K.; Carlson, B.A.; Arpa, L.; Celada, A.; Lawrence, T.; Otsu, K.; Brissette, J.L.; Arthur, J.S.C.; et al. The kinase p38α serves cell type–specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat. Immunol. 2008, 9, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Tur, J.; Pereira-Lopes, S.; Vico, T.; Marín, E.A.; Muñoz, J.P.; Hernández-Alvarez, M.; Cardona, P.-J.; Zorzano, A.; Lloberas, J.; Celada, A. Mitofusin 2 in Macrophages Links Mitochondrial ROS Production, Cytokine Release, Phagocytosis, Autophagy, and Bactericidal Activity. Cell Rep. 2020, 32, 108079. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Hellemans, J.; Mortier, G.; De Paepe, A.; Speleman, F.; Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007, 8, R19. [Google Scholar] [CrossRef]
- Valledor, A.F.; Comalada, M.; Xaus, J.; Celada, A. The Differential Time-course of Extracellular-regulated Kinase Activity Correlates with the Macrophage Response toward Proliferation or Activation. J. Biol. Chem. 2000, 275, 7403–7409. [Google Scholar] [CrossRef]
- Caelles, C.; González-Sancho, J.M.; Muñoz, A. Nuclear hormone receptor antagonism with AP-1 by inhibition of the JNK pathway. Genes Dev. 1997, 11, 3351–3364. [Google Scholar] [CrossRef]
- Sebastián, C.; Herrero, C.; Serra, M.; Lloberas, J.; Blasco, M.A.; Celada, A. Telomere Shortening and Oxidative Stress in Aged Macrophages Results in Impaired STAT5a Phosphorylation. J. Immunol. 2009, 183, 2356–2364. [Google Scholar] [CrossRef] [PubMed]
- Vico, T.; Youssif, C.; Zare, F.; Comalada, M.; Sebastian, C.; Lloberas, J.; Celada, A. GM-CSF Protects Macrophages from DNA Damage by Inducing Differentiation. Cells 2022, 11, 935. [Google Scholar] [CrossRef] [PubMed]
- Sharda, D.R.; Yu, S.; Ray, M.; Squadrito, M.L.; De Palma, M.; Wynn, T.A.; Morris, S.M., Jr.; Hankey, P.A. Regulation of Macrophage Arginase Expression and Tumor Growth by the Ron Receptor Tyrosine Kinase. J. Immunol. 2011, 187, 2181–2192. [Google Scholar] [CrossRef] [PubMed]
- Prince, S.; Carreira, S.; Vance, K.W.; Abrahams, A.; Goding, C.R. Tbx2 Directly Represses the Expression of the p21WAF1 Cyclin-Dependent Kinase Inhibitor. Cancer Res. 2004, 64, 1669–1674. [Google Scholar] [CrossRef] [PubMed]
- Al Bitar, S.; Gali-Muhtasib, H. The Role of the Cyclin Dependent Kinase Inhibitor p21cip1/waf1 in Targeting Cancer: Molecular Mechanisms and Novel Therapeutics. Cancers 2019, 11, 1475. [Google Scholar] [CrossRef]
- Canfield, S.; Lee, Y.; Schröder, A.; Rothman, P. Cutting Edge: IL-4 Induces Suppressor of Cytokine Signaling-3 Expression in B Cells by a Mechanism Dependent on Activation of p38 MAPK. J. Immunol. 2005, 174, 2494–2498. [Google Scholar] [CrossRef]
- Hashimoto, S.; Gon, Y.; Takeshita, I.; Maruoka, S.; Horie, T. IL-4 and IL-13 induce myofibroblastic phenotype of human lung fibroblasts through c-Jun NH2-terminal kinase–dependent pathway. J. Allergy Clin. Immunol. 2001, 107, 1001–1008. [Google Scholar] [CrossRef]
- Robinson, C.J.; Sloss, C.M.; Plevin, R. Inactivation of JNK activity by mitogen-activated protein kinase phosphatase-2 in EAhy926 endothelial cells is dependent upon agonist-specific JNK translocation to the nucleus. Cell Signal. 2001, 13, 29–41. [Google Scholar] [CrossRef]
- Jiao, H.; Tang, P.; Zhang, Y. MAP Kinase Phosphatase 2 Regulates Macrophage-Adipocyte Interaction. PLoS ONE 2015, 10, e0120755. [Google Scholar] [CrossRef]
- Arpa, L.; Valledor, A.F.; Lloberas, J.; Celada, A. IL-4 blocks M-CSF-dependent macrophage proliferation by inducing p21Waf1 in a STAT6-dependent way. Eur. J. Immunol. 2009, 39, 514–526. [Google Scholar] [CrossRef]
- Valledor, A.F.; Sánchez-Tilló, E.; Arpa, L.; Park, J.M.; Caelles, C.; Lloberas, J.; Celada, A. Selective Roles of MAPKs during the Macrophage Response to IFN-γ. J. Immunol. 2008, 180, 4523–4529. [Google Scholar] [CrossRef] [PubMed]
- Sze, K.-L.; Lui, W.-Y.; Lee, W.M. Post-transcriptional regulation of CLMP mRNA is controlled by tristetraprolin in response to TNFα via c-Jun N-terminal kinase signalling. Biochem. J. 2008, 410, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Poele, R.H.T.; Okorokov, A.L.; Joel, S.P. RNA synthesis block by 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) triggers p53-dependent apoptosis in human colon carcinoma cells. Oncogene 1999, 18, 5765–5772. [Google Scholar] [CrossRef] [PubMed]
- Celada, A.; Klemsz, M.J.; Maki, R.A. Interferon-γ activates multiple pathways to regulate the expression of the genes for major histocompatibility class II I-Aβ, tumor necrosis factor and complement component C3 in mouse macrophages. Eur. J. Immunol. 1989, 19, 1103–1109. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.H.; Jacobs, A.T., Jr.; Morris, S.M., Jr.; Ignarro, L.J. IL-4 and IL-13 upregulate arginase I expression by cAMP and JAK/STAT6 pathways in vascular smooth muscle cells. Am. J. Physiol. Physiol. 2000, 279, C248–C256. [Google Scholar] [CrossRef]
- Gray, M.J.; Poljakovic, M.; Kepka-Lenhart, D.; Morris, S.M., Jr. Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPβ. Gene 2005, 353, 98–106. [Google Scholar] [CrossRef]
- Chen, H.; Sun, H.; You, F.; Sun, W.; Zhou, X.; Chen, L.; Yang, J.; Wang, Y.; Tang, H.; Guan, Y.; et al. Activation of STAT6 by STING Is Critical for Antiviral Innate Immunity. Cell 2011, 147, 436–446. [Google Scholar] [CrossRef]
- Varinou, L.; Ramsauer, K.; Karaghiosoff, M.; Kolbe, T.; Pfeffer, K.; Müller, M.; Decker, T. Phosphorylation of the Stat1 Transactivation Domain Is Required for Full-Fledged IFN-γ-Dependent Innate Immunity. Immunity 2003, 19, 793–802. [Google Scholar] [CrossRef]
- Mikita, T.; Daniel, C.; Wu, P.; Schindler, U. Mutational Analysis of the STAT6 SH2 Domain. J. Biol. Chem. 1998, 273, 17634–17642. [Google Scholar] [CrossRef]
- Arimura, A.; van Peer, M.; Schröder, A.J.; Rothman, P.B. The Transcriptional Co-activator p/CIP (NCoA-3) Is Up-regulated by STAT6 and Serves as a Positive Regulator of Transcriptional Activation by STAT6. J. Biol. Chem. 2004, 279, 31105–31112. [Google Scholar] [CrossRef]
- Gingras, S.; Simard, J.; Groner, B.; Pfitzner, E. p300/CBP is required for transcriptional induction by interleukin-4 and interacts with Stat6. Nucleic Acids Res. 1999, 27, 2722–2729. [Google Scholar] [CrossRef]
- Bao, L.; Alexander, J.B.; Zhang, H.; Shen, K.; Chan, L.S. Interleukin-4 Downregulation of Involucrin Expression in Human Epidermal Keratinocytes Involves Stat6 Sequestration of the Coactivator CREB-Binding Protein. J. Interf. Cytokine Res. 2016, 36, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Ding, A. MyD88-mediated stabilization of interferon-γ-induced cytokine and chemokine mRNA. Nat. Immunol. 2006, 7, 375–381. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, K.; van Deursent, J.; Sangster, M.Y.; Sarawar, S.R.; Carson, R.T.; Tripp, R.A.; Chu, C.; Quelle, F.W.; Nosaka, T.; Vignali, D.A.A.; et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted State6 gene. Nature 1996, 380, 630–633. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Tanaka, T.; Shi, W.; Matsumoto, M.; Minami, M.; Kashiwamura, S.-I.; Nakanishi, K.; Yoshida, N.; Kishimoto, T.; Akira, S. Essential role of Stat6 in IL-4 signalling. Nature 1996, 380, 627–630. [Google Scholar] [CrossRef]
- Hao, J.; Hu, Y.; Li, Y.; Zhou, Q.; Lv, X. Involvement of JNK signaling in IL4-induced M2 macrophage polarization. Exp. Cell Res. 2017, 357, 155–162. [Google Scholar] [CrossRef]
- Serrat, N.; Pereira-Lopes, S.; Comalada, M.; Lloberas, J.; Celada, A. Deacetylation of C/EBPβ is required for IL-4-induced arginase-1 expression in murine macrophages. Eur. J. Immunol. 2012, 42, 3028–3037. [Google Scholar] [CrossRef]
- Pesu, M.; Takaluoma, K.; Aittomäki, S.; Lagerstedt, A.; Saksela, K.; Kovanen, P.E.; Silvennoinen, O. Interleukin-4-induced transcriptional activation by Stat6 involves multiple serine/threonine kinase pathways and serine phosphorylation of Stat6. Blood 2000, 95, 494–502. [Google Scholar] [CrossRef]
- Shirakawa, T.; Kawazoe, Y.; Tsujikawa, T.; Jung, D.; Sato, S.-I.; Uesugi, M. Deactivation of STAT6 through Serine 707 Phosphorylation by JNK. J. Biol. Chem. 2011, 286, 4003–4010. [Google Scholar] [CrossRef]
- Li, J.; Rodriguez, J.P.; Niu, F.; Pu, M.; Wang, J.; Hung, L.-W.; Shao, Q.; Zhu, Y.; Ding, W.; Liu, Y.; et al. Structural basis for DNA recognition by STAT6. Proc. Natl. Acad. Sci. USA 2016, 113, 13015–13020. [Google Scholar] [CrossRef]
- Hebenstreit, D.; Wirnsberger, G.; Horejs-Hoeck, J.; Duschl, A. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev. 2006, 17, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.-C.; Chen, C.-C. Akt Phosphorylation of p300 at Ser-1834 Is Essential for Its Histone Acetyltransferase and Transcriptional Activity. Mol. Cell. Biol. 2005, 25, 6592–6602. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Denlinger, C.E.; Rundall, B.K.; Smith, P.W.; Jones, D.R. Suberoylanilide Hydroxamic Acid Induces Akt-mediated Phosphorylation of p300, Which Promotes Acetylation and Transcriptional Activation of RelA/p65. J. Biol. Chem. 2006, 281, 31359–31368. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; Wang, Y.-N.; Chang, W.-C. ERK2-mediated C-terminal Serine Phosphorylation of p300 Is Vital to the Regulation of Epidermal Growth Factor-induced Keratin 16 Gene Expression. J. Biol. Chem. 2007, 282, 27215–27228. [Google Scholar] [CrossRef]
- Shankaranarayanan, P.; Chaitidis, P.; Kühn, H.; Nigam, S. Acetylation by Histone Acetyltransferase CREB-binding Protein/p300 of STAT6 Is Required for Transcriptional Activation of the 15-Lipoxygenase-1 Gene. J. Biol. Chem. 2001, 276, 42753–42760. [Google Scholar] [CrossRef]
- Razeto, A.; Ramakrishnan, V.; Litterst, C.M.; Giller, K.; Griesinger, C.; Carlomagno, T.; Lakomek, N.; Heimburg, T.; Lodrini, M.; Pfitzner, E.; et al. Structure of the NCoA-1/SRC-1 PAS-B Domain Bound to the LXXLL Motif of the STAT6 Transactivation Domain. J. Mol. Biol. 2004, 336, 319–329. [Google Scholar] [CrossRef]
- Goenka, S.; Marlar, C.; Schindler, U.; Boothby, M. Differential Roles of C-terminal Activation Motifs in the Establishment of Stat6 Transcriptional Specificity. J. Biol. Chem. 2003, 278, 50362–50370. [Google Scholar] [CrossRef]
- Välineva, T.; Yang, J.; Palovuori, R.; Silvennoinen, O. The Transcriptional Co-activator Protein p100 Recruits Histone Acetyltransferase Activity to STAT6 and Mediates Interaction between the CREB-binding Protein and STAT6. J. Biol. Chem. 2005, 280, 14989–14996. [Google Scholar] [CrossRef]
- Yang, J.; Aittomäki, S.; Pesu, M.; Carter, K.; Saarinen, J.; Kalkkinen, N.; Kieff, E.; Silvennoinen, O. Identification of p100 as a coactivator for STAT6 that bridges STAT6 with RNA polymerase II. EMBO J. 2002, 21, 4950–4958. [Google Scholar] [CrossRef]
- Goenka, S.; Cho, S.H.; Boothby, M. Collaborator of Stat6 (CoaSt6)-associated Poly(ADP-ribose) Polymerase Activity Modulates Stat6-dependent Gene Transcription. J. Biol. Chem. 2007, 282, 18732–18739. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arpa, L.; Batlle, C.; Jiang, P.; Caelles, C.; Lloberas, J.; Celada, A. Distinct Responses to IL4 in Macrophages Mediated by JNK. Cells 2023, 12, 1127. https://doi.org/10.3390/cells12081127
Arpa L, Batlle C, Jiang P, Caelles C, Lloberas J, Celada A. Distinct Responses to IL4 in Macrophages Mediated by JNK. Cells. 2023; 12(8):1127. https://doi.org/10.3390/cells12081127
Chicago/Turabian StyleArpa, Luís, Carlos Batlle, Peijin Jiang, Carme Caelles, Jorge Lloberas, and Antonio Celada. 2023. "Distinct Responses to IL4 in Macrophages Mediated by JNK" Cells 12, no. 8: 1127. https://doi.org/10.3390/cells12081127
APA StyleArpa, L., Batlle, C., Jiang, P., Caelles, C., Lloberas, J., & Celada, A. (2023). Distinct Responses to IL4 in Macrophages Mediated by JNK. Cells, 12(8), 1127. https://doi.org/10.3390/cells12081127