Fatty Acid Excess Dysregulates CARF to Initiate the Development of Hepatic Steatosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Cell Culture and Treatment
2.3. H and E Staining and Histological Analysis of Liver Samples
2.4. Modulation of CARF Expression in HepG2 Cells
2.5. Oil Red O Staining
2.6. Triglyceride Assay
2.7. Quantitative Real-Time PCR
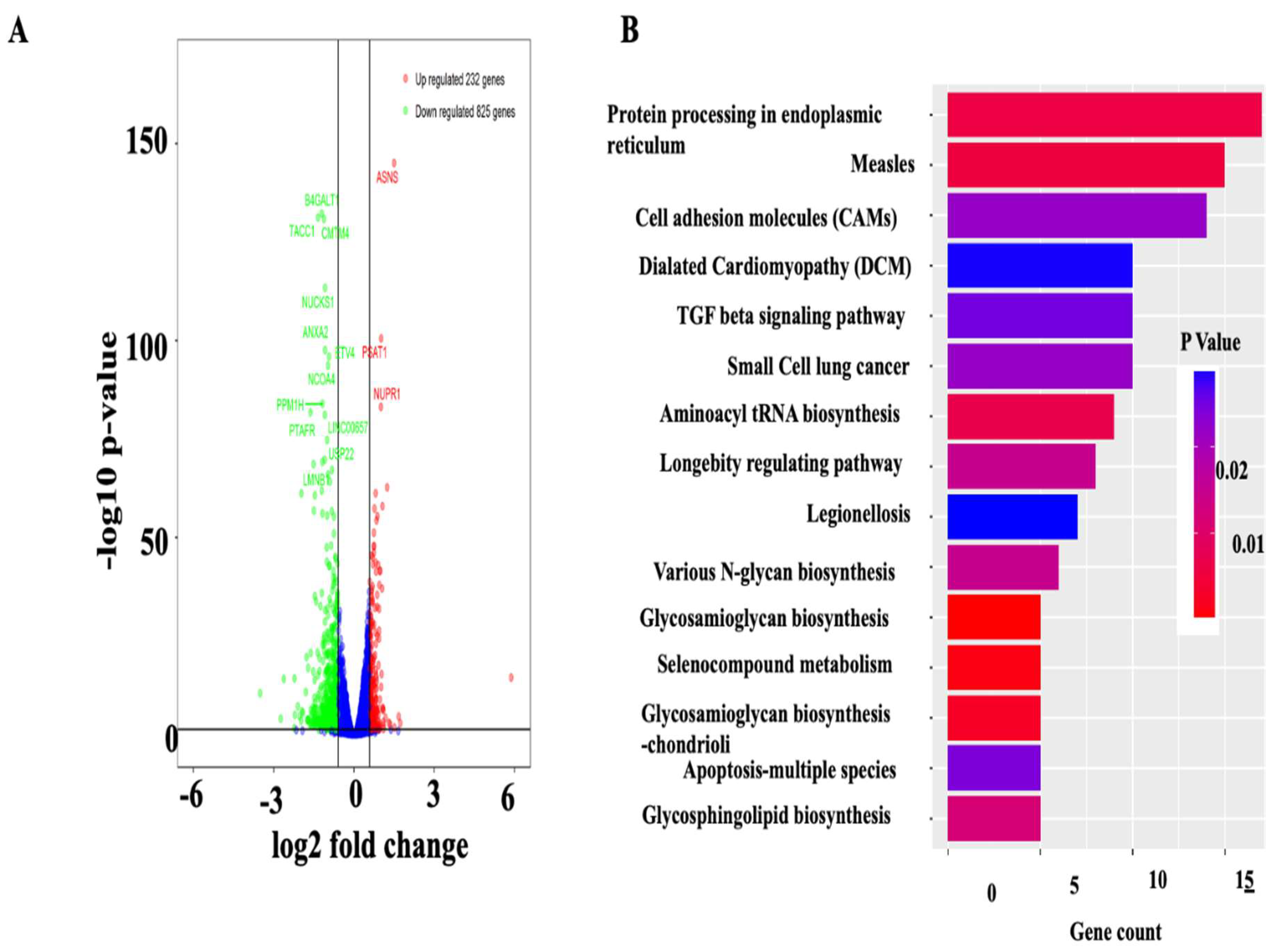
2.8. RNA Sequence Analysis
2.9. Oxidative Stress or ROS Assay
2.10. Protein Analysis
2.11. Preparation of Palmitate-BSA Conjugate
2.12. TUNEL Staining
2.13. Statistical Analysis
3. Results
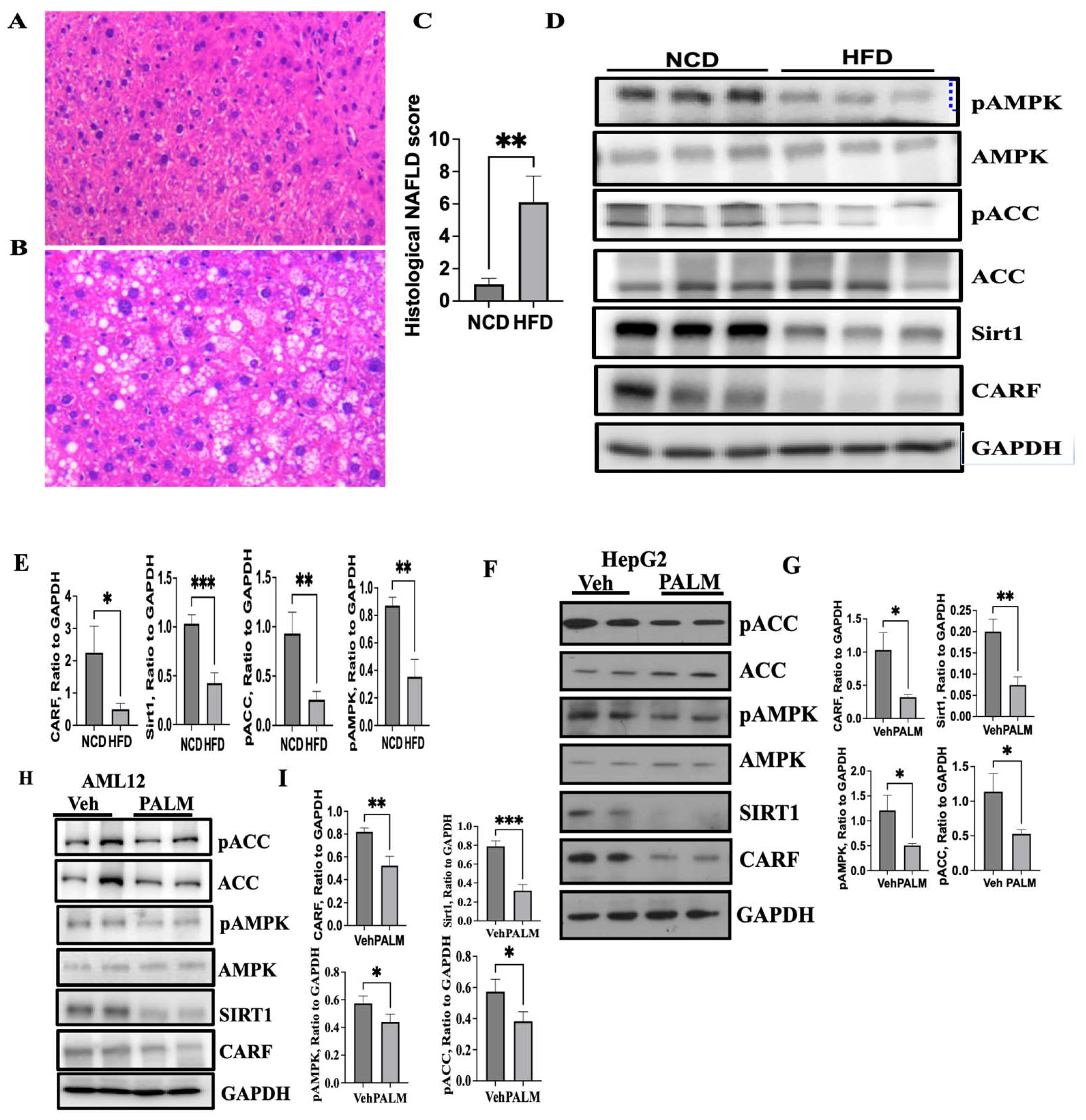
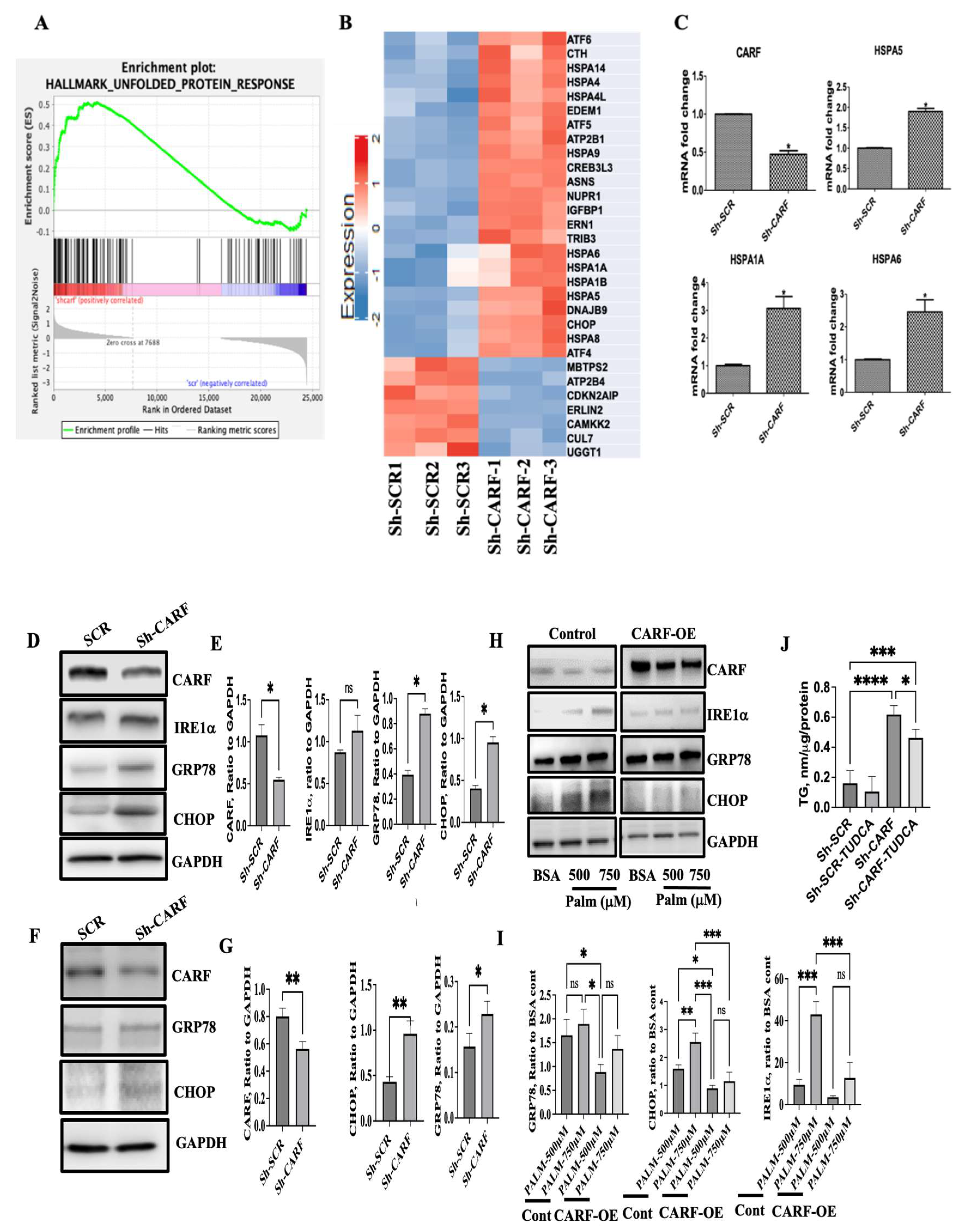
3.1. Expression of CARF Was Reduced in Fatty Livers and Cellular Model of Steatosis
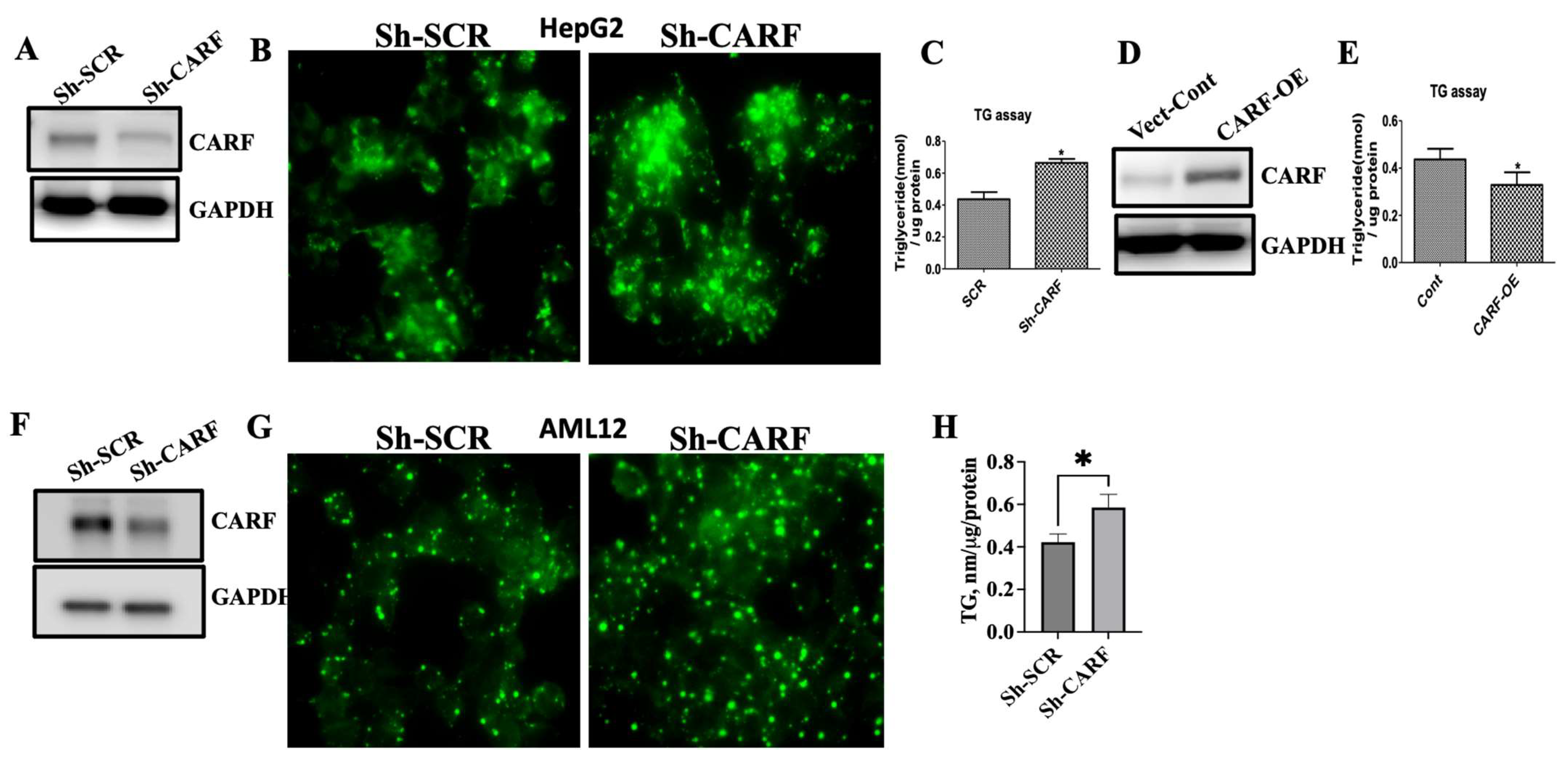
3.2. Ablation of CARF Exacerbates Fat Accumulation in HepG2 and AML12 Cells
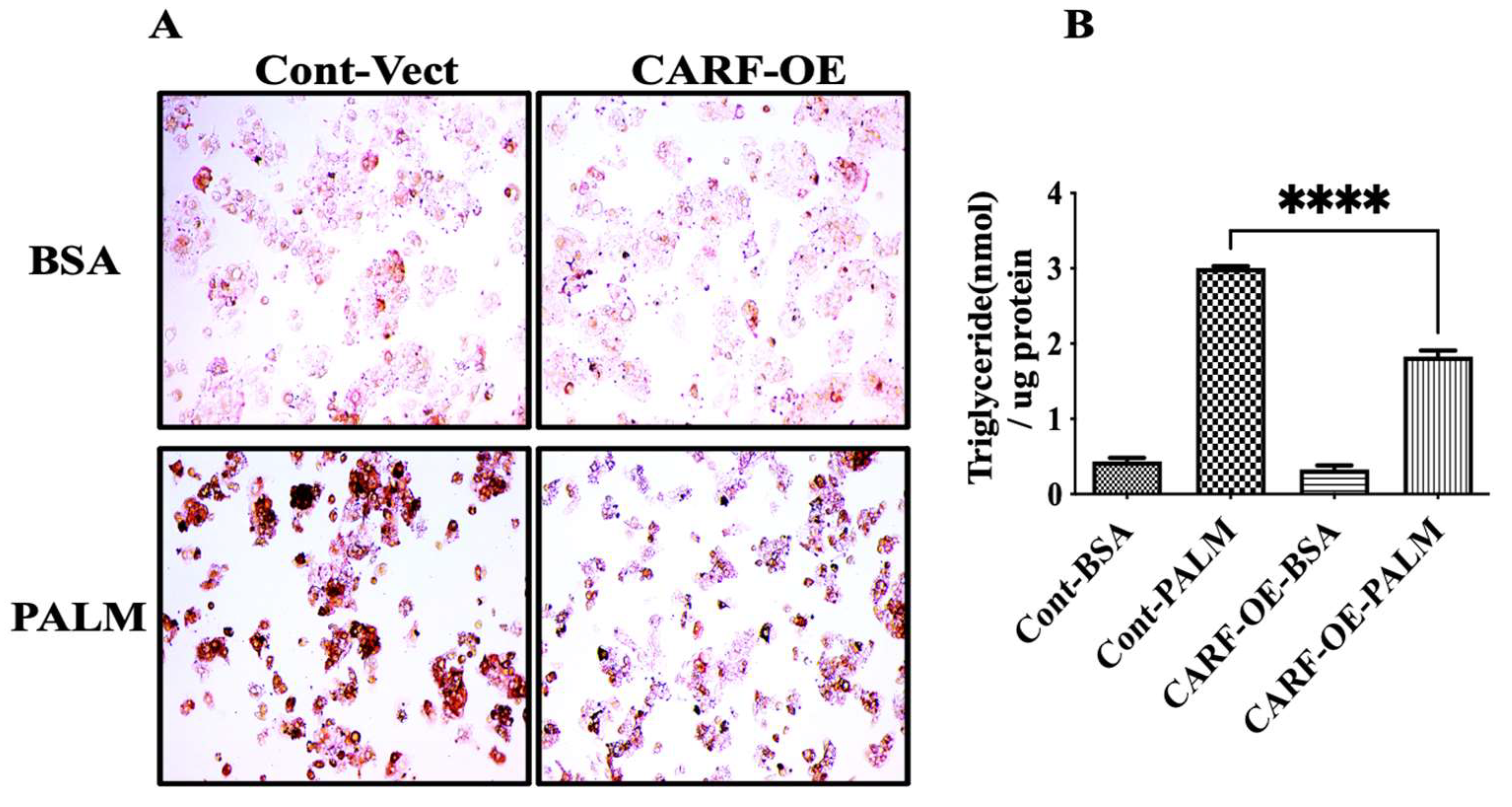
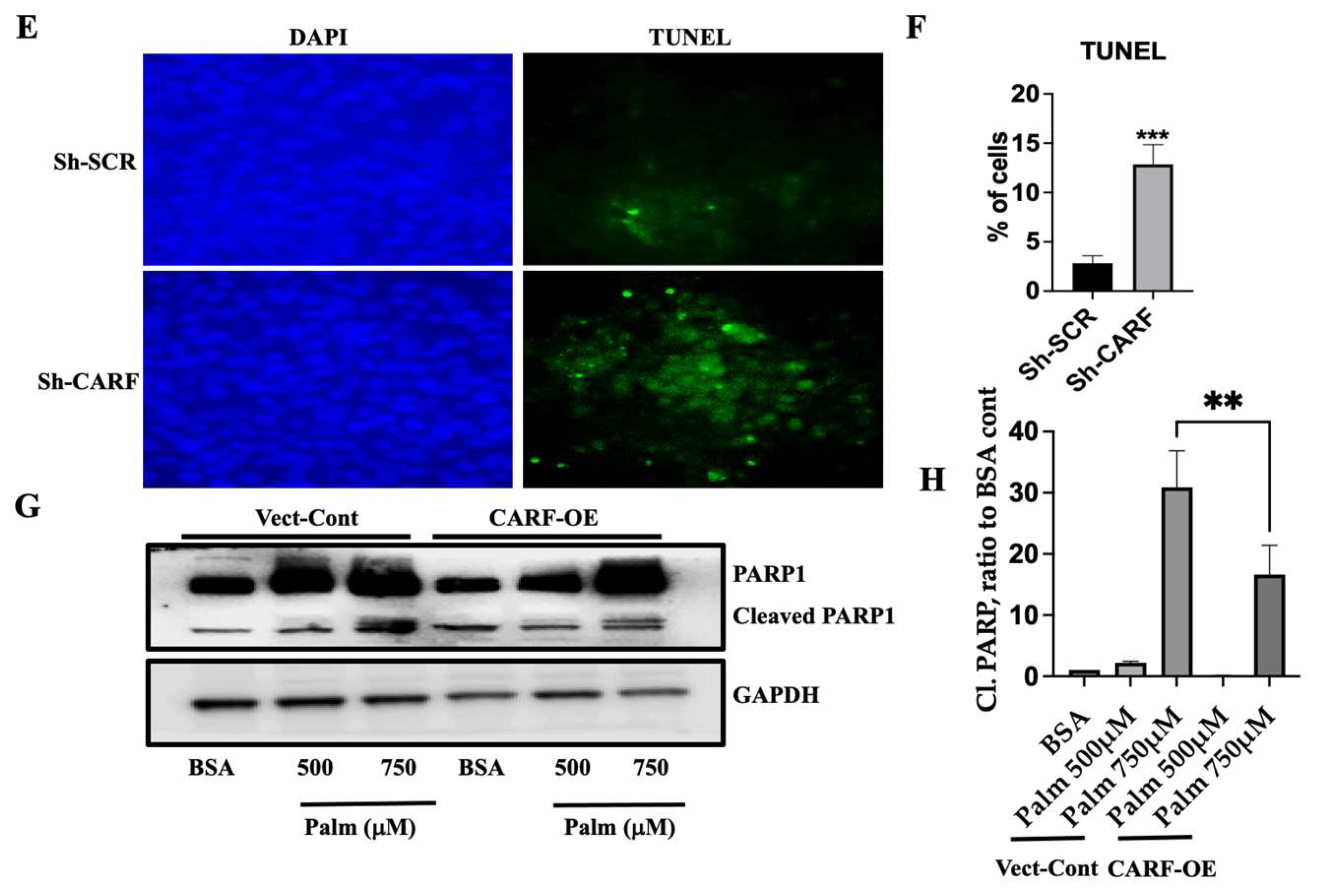
3.3. CARF Protects against the Ectopic Fat Accumulation
3.4. Deficiency of CARF Induces ER Stress in HepG2 and AML12 Cells
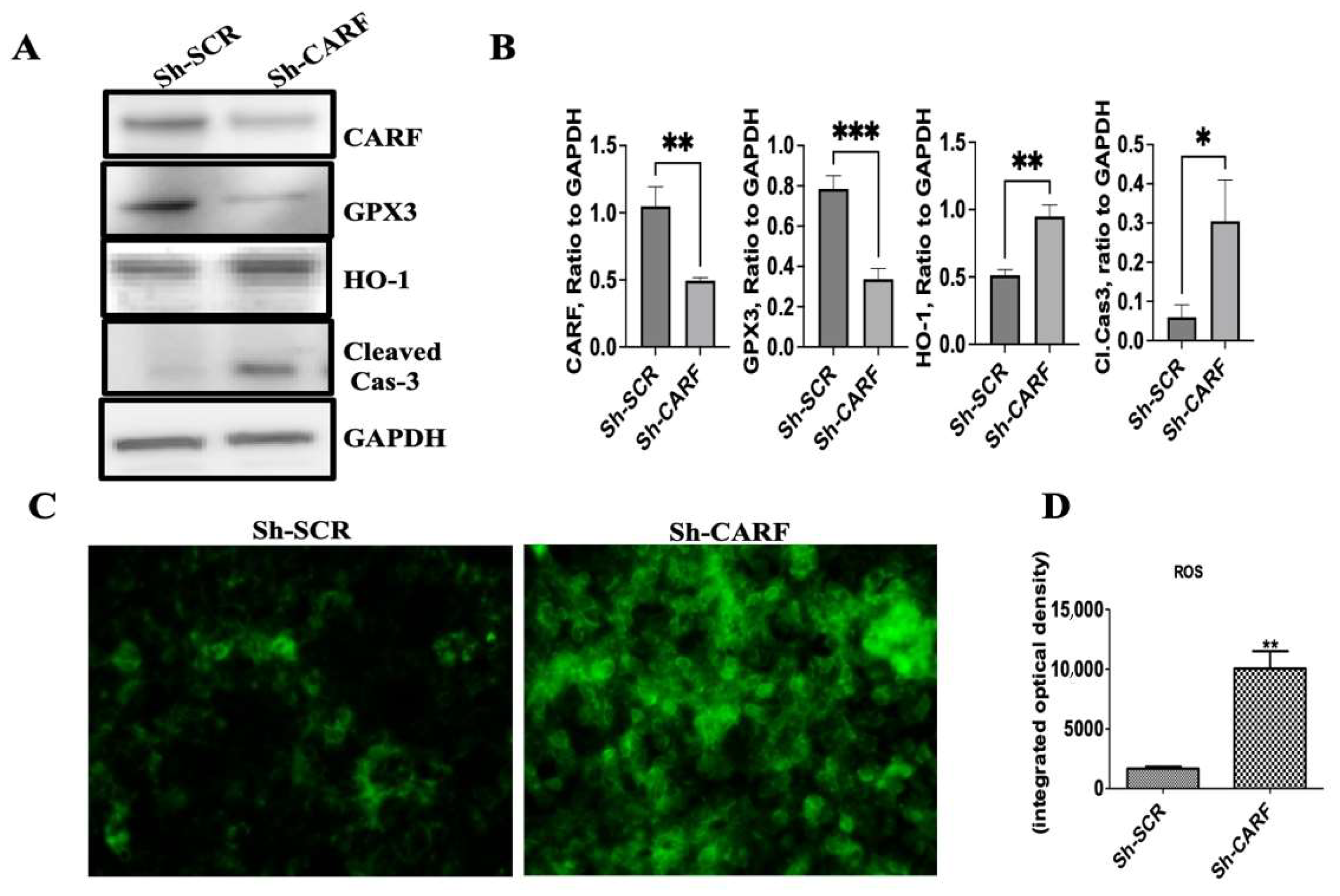
3.5. Knockdown of CARF Induces Oxidative Stress and Cell Death
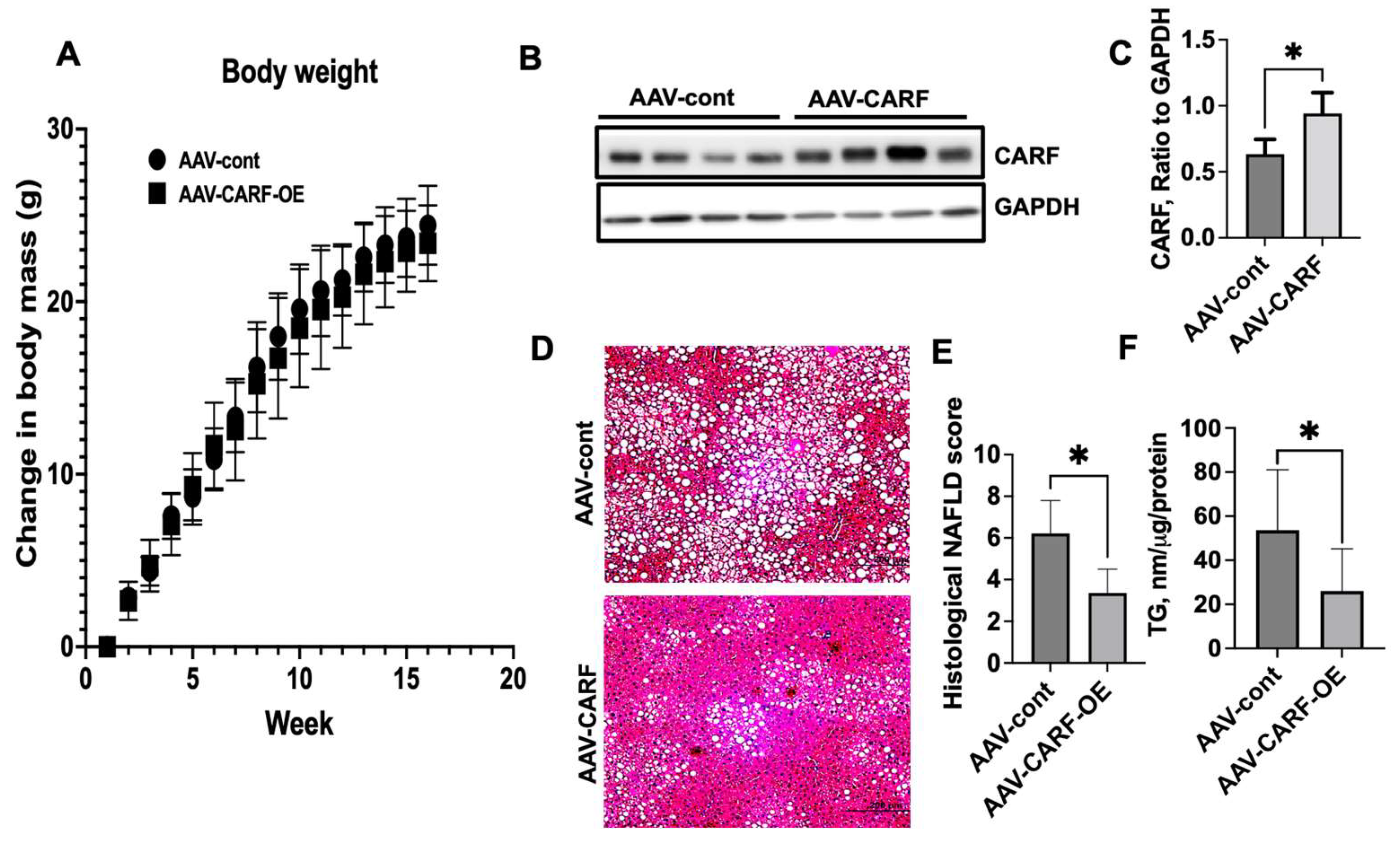
3.6. Exogenous Expression of CARF Attenuates HFD-Induced Hepatic Steatosis in Mice
3.7. CARF-OE Improves ER Function, Decreases Oxidative Stress, and Improves Insulin Sensitivity in HFD-Fed Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATF6 | (activating transcription factor-6) |
| CARF | (collaborator of ARF) |
| CHOP | (C/EBP homologous protein) |
| CDKN2AIP | (Cyclin Dependent Kinase Inhibitor 2A interacting protein) |
| ER | (endoplasmic reticulum) |
| FFA | (Free Fatty Acid) |
| GTT | (glucose tolerance test) |
| IRE1α | (inositol requiring protein-1α) |
| ITT | (insulin tolerance test) |
| NAFLD | (Non-alcoholic fatty liver diseases) |
| TG | (Triglyceride) |
| UPR | (Unfolded protein response) |
| XBP1s | (X-box-binding protein-1) |
References
- Alves-Bezerra, M.; Cohen, D.E. Triglyceride Metabolism in the Liver. Compr. Physiol. 2017, 8, 1–8. [Google Scholar] [CrossRef]
- Postic, C.; Girard, J. Contribution of de novo fatty acid synthesis to hepatic steatosis and insulin resistance: Lessons from genetically engineered mice. J. Clin. Investig. 2008, 118, 829–838. [Google Scholar] [CrossRef]
- Alves-Bezerra, M.; Li, Y.; Acuna, M.; Ivanova, A.A.; Corey, K.E.; Ortlund, E.A.; Cohen, D.E. Thioesterase Superfamily Member 2 Promotes Hepatic VLDL Secretion by Channeling Fatty Acids Into Triglyceride Biosynthesis. Hepatology 2019, 70, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Chalasani, N.; Younossi, Z.; Lavine, J.E.; Diehl, A.M.; Brunt, E.M.; Cusi, K.; Charlton, M.; Sanyal, A.J. The diagnosis and management of non-alcoholic fatty liver disease: Practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 2012, 55, 2005–2023. [Google Scholar] [CrossRef]
- Bhatia, L.S.; Curzen, N.P.; Byrne, C.D. Nonalcoholic fatty liver disease and vascular risk. Curr. Opin. Cardiol. 2012, 27, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Gaggini, M.; Morelli, M.; Buzzigoli, E.; DeFronzo, R.A.; Bugianesi, E.; Gastaldelli, A. Non-alcoholic fatty liver disease (NAFLD) and its connection with insulin resistance, dyslipidemia, atherosclerosis and coronary heart disease. Nutrients 2013, 5, 1544–1560. [Google Scholar] [CrossRef] [PubMed]
- Bellentani, S.; Marino, M. Epidemiology and natural history of non-alcoholic fatty liver disease (NAFLD). Ann. Hepatol. 2009, 8 (Suppl. S1), S4–S8. [Google Scholar] [CrossRef] [PubMed]
- Hasan, K.; Cheung, C.; Kaul, Z.; Shah, N.; Sakaushi, S.; Sugimoto, K.; Oka, S.; Kaul, S.C.; Wadhwa, R. CARF Is a vital dual regulator of cellular senescence and apoptosis. J. Biol. Chem. 2009, 284, 1664–1672. [Google Scholar] [CrossRef]
- Hasan, M.K.; Yaguchi, T.; Harada, J.I.; Hirano, T.; Wadhwa, R.; Kaul, S.C. CARF (collaborator of ARF) interacts with HDM2: Evidence for a novel regulatory feedback regulation of CARF-p53-HDM2-p21WAF1 pathway. Int. J. Oncol. 2008, 32, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.K.; Yaguchi, T.; Minoda, Y.; Hirano, T.; Taira, K.; Wadhwa, R.; Kaul, S.C. Alternative reading frame protein (ARF)-independent function of CARF (collaborator of ARF) involves its interactions with p53: Evidence for a novel p53-activation pathway and its negative feedback control. Biochem. J. 2004, 380, 605–610. [Google Scholar] [CrossRef]
- Hasan, M.K.; Yaguchi, T.; Sugihara, T.; Kumar, P.K.; Taira, K.; Reddel, R.R.; Kaul, S.C.; Wadhwa, R. CARF is a novel protein that cooperates with mouse p19ARF (human p14ARF) in activating p53. J. Biol. Chem. 2002, 277, 37765–37770. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Ma, X.; Cui, L.; Dang, S.; Qu, J.; Zhang, J.; Wang, X.; Mao, Z. CARF activates beta-catenin/TCF signaling in the hepatocellular carcinoma. Oncotarget 2016, 7, 80404–80414. [Google Scholar] [CrossRef] [PubMed]
- Kalra, R.S.; Chaudhary, A.; Yoon, A.R.; Bhargava, P.; Omar, A.; Garg, S.; Yun, C.O.; Kaul, S.C.; Wadhwa, R. CARF enrichment promotes epithelial-mesenchymal transition via Wnt/beta-catenin signaling: Its clinical relevance and potential as a therapeutic target. Oncogenesis 2018, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, W.; Yan, C.; Nie, F.; Li, C.; Liu, X.; Fei, C.; Li, S.; Song, X.; Jia, Y.; et al. Chemical biology reveals CARF as a positive regulator of canonical Wnt signaling by promoting TCF/beta-catenin transcriptional activity. Cell Discov. 2017, 3, 17003. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; He, X.; Zhai, X.; Zhang, H.; Zhang, Y.; Jin, F.; Song, X.; Wu, D.; Shi, Q.; Li, L. CARF promotes spermatogonial self-renewal and proliferation through Wnt signaling pathway. Cell Discov. 2020, 6, 85. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, H.; Guo, K. Inhibition of MicroRNA-302c on Stemness of Colon Cancer Stem Cells via the CARF/Wnt/beta-Catenin Axis. Dig. Dis. Sci. 2021, 66, 1906–1915. [Google Scholar] [CrossRef]
- Kalra, R.S.; Chaudhary, A.; Omar, A.; Cheung, C.T.; Garg, S.; Kaul, S.C.; Wadhwa, R. Stress-induced changes in CARF expression determine cell fate to death, survival, or malignant transformation. Cell Stress Chaperones 2020, 25, 481–494. [Google Scholar] [CrossRef]
- Kalra, R.S.; Cheung, C.T.; Chaudhary, A.; Prakash, J.; Kaul, S.C.; Wadhwa, R. CARF (Collaborator of ARF) overexpression in p53-deficient cells promotes carcinogenesis. Mol. Oncol. 2015, 9, 1877–1889. [Google Scholar] [CrossRef]
- Ai, J.; Li, J.; Gessler, D.J.; Su, Q.; Wei, Q.; Li, H.; Gao, G. Adeno-associated virus serotype rh.10 displays strong muscle tropism following intraperitoneal delivery. Sci. Rep. 2017, 7, 40336. [Google Scholar] [CrossRef]
- Knowles, B.B.; Howe, C.C.; Aden, D.P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science 1980, 209, 497–499. [Google Scholar] [CrossRef]
- Hsu, I.C.; Tokiwa, T.; Bennett, W.; Metcalf, R.A.; Welsh, J.A.; Sun, T.; Harris, C.C. p53 gene mutation and integrated hepatitis B viral DNA sequences in human liver cancer cell lines. Carcinogenesis 1993, 14, 987–992. [Google Scholar] [CrossRef] [PubMed]
- Zhai, W.R.; Paronetto, F. Relationship between c-myc gene protein, nucleic acids and hepatitis B virus expression in hepatoma cell lines and their corresponding tumors in nude mice. J. Exp. Pathol. 1989, 4, 213–225. [Google Scholar]
- Ponugoti, B.; Kim, D.H.; Xiao, Z.; Smith, Z.; Miao, J.; Zang, M.; Wu, S.Y.; Chiang, C.M.; Veenstra, T.D.; Kemper, J.K. SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J. Biol. Chem. 2010, 285, 33959–33970. [Google Scholar] [CrossRef]
- Zang, M.; Zuccollo, A.; Hou, X.; Nagata, D.; Walsh, K.; Herscovitz, H.; Brecher, P.; Ruderman, N.B.; Cohen, R.A. AMP-activated protein kinase is required for the lipid-lowering effect of metformin in insulin-resistant human HepG2 cells. J. Biol. Chem. 2004, 279, 47898–47905. [Google Scholar] [CrossRef]
- Zhou, G.; Myers, R.; Li, Y.; Chen, Y.; Shen, X.; Fenyk-Melody, J.; Wu, M.; Ventre, J.; Doebber, T.; Fujii, N.; et al. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Investig. 2001, 108, 1167–1174. [Google Scholar] [CrossRef]
- Gomez-Lechon, M.J.; Donato, M.T.; Martinez-Romero, A.; Jimenez, N.; Castell, J.V.; O’Connor, J.E. A human hepatocellular in vitro model to investigate steatosis. Chem. Biol. Interact. 2007, 165, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Menke, A.L.; Driessen, A.; Koek, G.H.; Lindeman, J.H.; Stoop, R.; Havekes, L.M.; Kleemann, R.; van den Hoek, A.M. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS ONE 2014, 9, e115922. [Google Scholar] [CrossRef]
- Hasan, K.M.; Friedman, T.C.; Shao, X.; Parveen, M.; Sims, C.; Lee, D.L.; Espinoza-Derout, J.; Sinha-Hikim, I.; Sinha-Hikim, A.P. E-cigarettes and Western Diet: Important Metabolic Risk Factors for Hepatic Diseases. Hepatology 2019, 69, 2442–2454. [Google Scholar] [CrossRef]
- Hasan, M.K.; Friedman, T.C.; Sims, C.; Lee, D.L.; Espinoza-Derout, J.; Ume, A.; Chalfant, V.; Lee, M.L.; Sinha-Hikim, I.; Lutfy, K.; et al. alpha7-Nicotinic Acetylcholine Receptor Agonist Ameliorates Nicotine Plus High-Fat Diet-Induced Hepatic Steatosis in Male Mice by Inhibiting Oxidative Stress and Stimulating AMPK Signaling. Endocrinology 2018, 159, 931–944. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, W.; Santaren, I.D.; Hanley, A.J.; Watkins, S.M.; Lorenzo, C.; Wagenknecht, L.E. Risk of diabetes associated with fatty acids in the de novo lipogenesis pathway is independent of insulin sensitivity and response: The Insulin Resistance Atherosclerosis Study (IRAS). BMJ Open Diabetes Res. Care 2019, 7, e000691. [Google Scholar] [CrossRef]
- Akazawa, Y.; Cazanave, S.; Mott, J.L.; Elmi, N.; Bronk, S.F.; Kohno, S.; Charlton, M.R.; Gores, G.J. Palmitoleate attenuates palmitate-induced Bim and PUMA up-regulation and hepatocyte lipoapoptosis. J. Hepatol. 2010, 52, 586–593. [Google Scholar] [CrossRef]
- Belfort, R.; Harrison, S.A.; Brown, K.; Darland, C.; Finch, J.; Hardies, J.; Balas, B.; Gastaldelli, A.; Tio, F.; Pulcini, J.; et al. A placebo-controlled trial of pioglitazone in subjects with nonalcoholic steatohepatitis. N. Engl. J. Med. 2006, 355, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- Byrne, C.D. Ectopic fat, insulin resistance and non-alcoholic fatty liver disease. Proc. Nutr. Soc. 2013, 72, 412–419. [Google Scholar] [CrossRef]
- Hetz, C.; Chevet, E.; Harding, H.P. Targeting the unfolded protein response in disease. Nat. Rev. Drug Discov. 2013, 12, 703–719. [Google Scholar] [CrossRef]
- Uppala, J.K.; Gani, A.R.; Ramaiah, K.V.A. Chemical chaperone, TUDCA unlike PBA, mitigates protein aggregation efficiently and resists ER and non-ER stress induced HepG2 cell death. Sci. Rep. 2017, 7, 3831. [Google Scholar] [CrossRef] [PubMed]
- Delli Bovi, A.P.; Marciano, F.; Mandato, C.; Siano, M.A.; Savoia, M.; Vajro, P. Oxidative Stress in Non-alcoholic Fatty Liver Disease. An Updated Mini Review. Front. Med. 2021, 8, 595371. [Google Scholar] [CrossRef]
- Bauer, M.; Bauer, I. Heme oxygenase-1: Redox regulation and role in the hepatic response to oxidative stress. Antioxid. Redox Signal. 2002, 4, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Pagliassotti, M.J. Endoplasmic reticulum stress in nonalcoholic fatty liver disease. Annu. Rev. Nutr. 2012, 32, 17–33. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Xu, C.F.; Yu, C.H.; Chen, W.X.; Li, Y.M. Role of endoplasmic reticulum stress in the pathogenesis of nonalcoholic fatty liver disease. World J. Gastroenterol. 2014, 20, 1768–1776. [Google Scholar] [CrossRef]
- Friedman, T.C.; Sinha-Hikim, I.; Parveen, M.; Najjar, S.M.; Liu, Y.; Mangubat, M.; Shin, C.S.; Lyzlov, A.; Ivey, R.; Shaheen, M.; et al. Additive effects of nicotine and high-fat diet on hepatic steatosis in male mice. Endocrinology 2012, 153, 5809–5820. [Google Scholar] [CrossRef]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Gentile, C.L.; Frye, M.; Pagliassotti, M.J. Endoplasmic reticulum stress and the unfolded protein response in nonalcoholic fatty liver disease. Antioxid. Redox Signal. 2011, 15, 505–521. [Google Scholar] [CrossRef] [PubMed]
- Leamy, A.K.; Egnatchik, R.A.; Young, J.D. Molecular mechanisms and the role of saturated fatty acids in the progression of non-alcoholic fatty liver disease. Prog. Lipid Res. 2013, 52, 165–174. [Google Scholar] [CrossRef]
- Rada, P.; Gonzalez-Rodriguez, A.; Garcia-Monzon, C.; Valverde, A.M. Understanding lipotoxicity in NAFLD pathogenesis: Is CD36 a key driver? Cell Death Dis. 2020, 11, 802. [Google Scholar] [CrossRef] [PubMed]
- Alkhouri, N.; Carter-Kent, C.; Feldstein, A.E. Apoptosis in nonalcoholic fatty liver disease: Diagnostic and therapeutic implications. Expert Rev. Gastroenterol. Hepatol. 2011, 5, 201–212. [Google Scholar] [CrossRef]

| Gene Symbol | Name | Fold Change | p Value |
|---|---|---|---|
| GPX2 | Glutathione peroxidase 2 | −1.35 | 3.52 × 10−23 |
| GPX3 | Glutathione peroxidase 3 | −1.6 | 4.97 × 10−33 |
| HMOX1 | Heme Oxygenase 1 | 2.09 | 1.03 × 10−58 |
| TXNRD3 | Thioredoxin reductase 3 | −2.35 | 5.92 × 10−13 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, K.M.; Parveen, M.; Pena, A.; Bautista, F.; Rivera, J.C.; Huerta, R.R.; Martinez, E.; Espinoza-Derout, J.; Sinha-Hikim, A.P.; Friedman, T.C. Fatty Acid Excess Dysregulates CARF to Initiate the Development of Hepatic Steatosis. Cells 2023, 12, 1069. https://doi.org/10.3390/cells12071069
Hasan KM, Parveen M, Pena A, Bautista F, Rivera JC, Huerta RR, Martinez E, Espinoza-Derout J, Sinha-Hikim AP, Friedman TC. Fatty Acid Excess Dysregulates CARF to Initiate the Development of Hepatic Steatosis. Cells. 2023; 12(7):1069. https://doi.org/10.3390/cells12071069
Chicago/Turabian StyleHasan, Kamrul M., Meher Parveen, Alondra Pena, Francisco Bautista, Juan Carlos Rivera, Roxana Ramirez Huerta, Erica Martinez, Jorge Espinoza-Derout, Amiya P. Sinha-Hikim, and Theodore C. Friedman. 2023. "Fatty Acid Excess Dysregulates CARF to Initiate the Development of Hepatic Steatosis" Cells 12, no. 7: 1069. https://doi.org/10.3390/cells12071069
APA StyleHasan, K. M., Parveen, M., Pena, A., Bautista, F., Rivera, J. C., Huerta, R. R., Martinez, E., Espinoza-Derout, J., Sinha-Hikim, A. P., & Friedman, T. C. (2023). Fatty Acid Excess Dysregulates CARF to Initiate the Development of Hepatic Steatosis. Cells, 12(7), 1069. https://doi.org/10.3390/cells12071069








