The TT Genotype of the KIAA1524 rs2278911 Polymorphism Is Associated with Poor Prognosis in Multiple Myeloma
Abstract
1. Introduction
2. Materials and Methods
2.1. The Study Population
2.2. DNA Isolation
2.3. Single Nucleotide Polymorphism Analysis
2.4. Plasma Cells Isolation
2.5. Gene Expression in the Bone Marrow Plasma Cells
2.6. Cytogenetic Assessment
2.7. Statistical Analysis
3. Results
3.1. The KIAA1524 Genotypes’ Distribution in Relation to the Demographic, Clinical and Molecular Factors
3.2. Response to Treatment
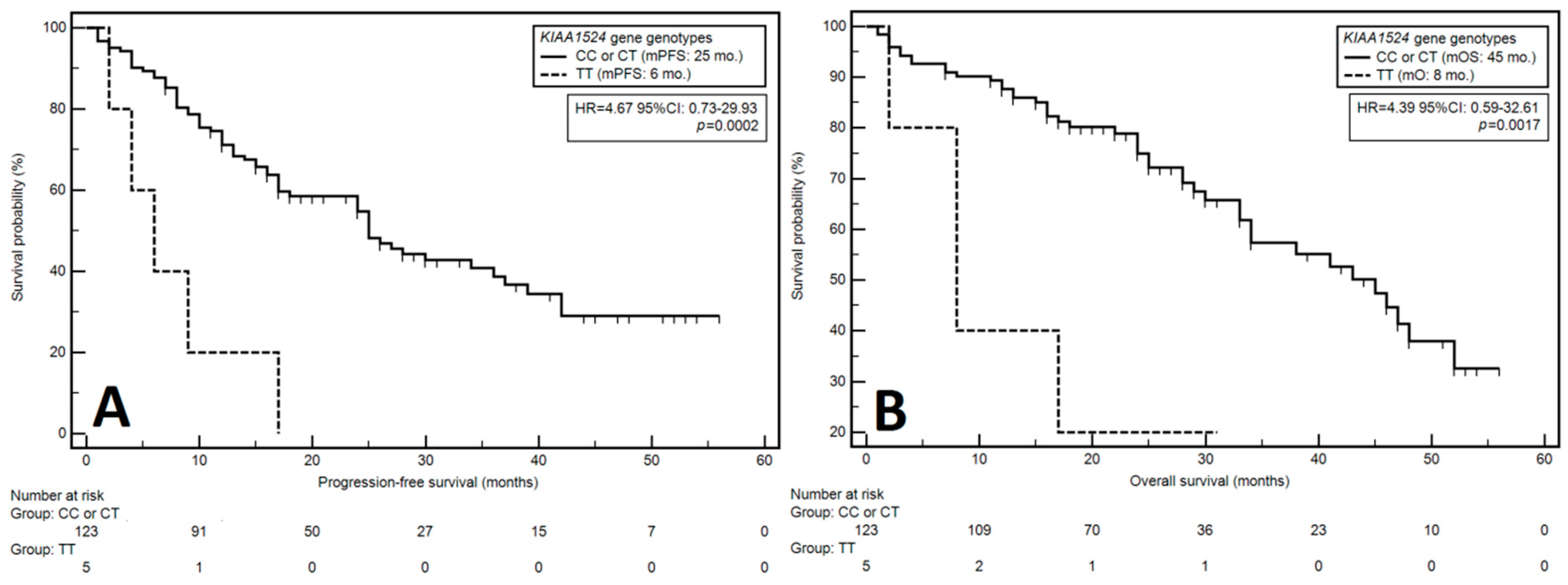
3.3. Progression-Free Survival
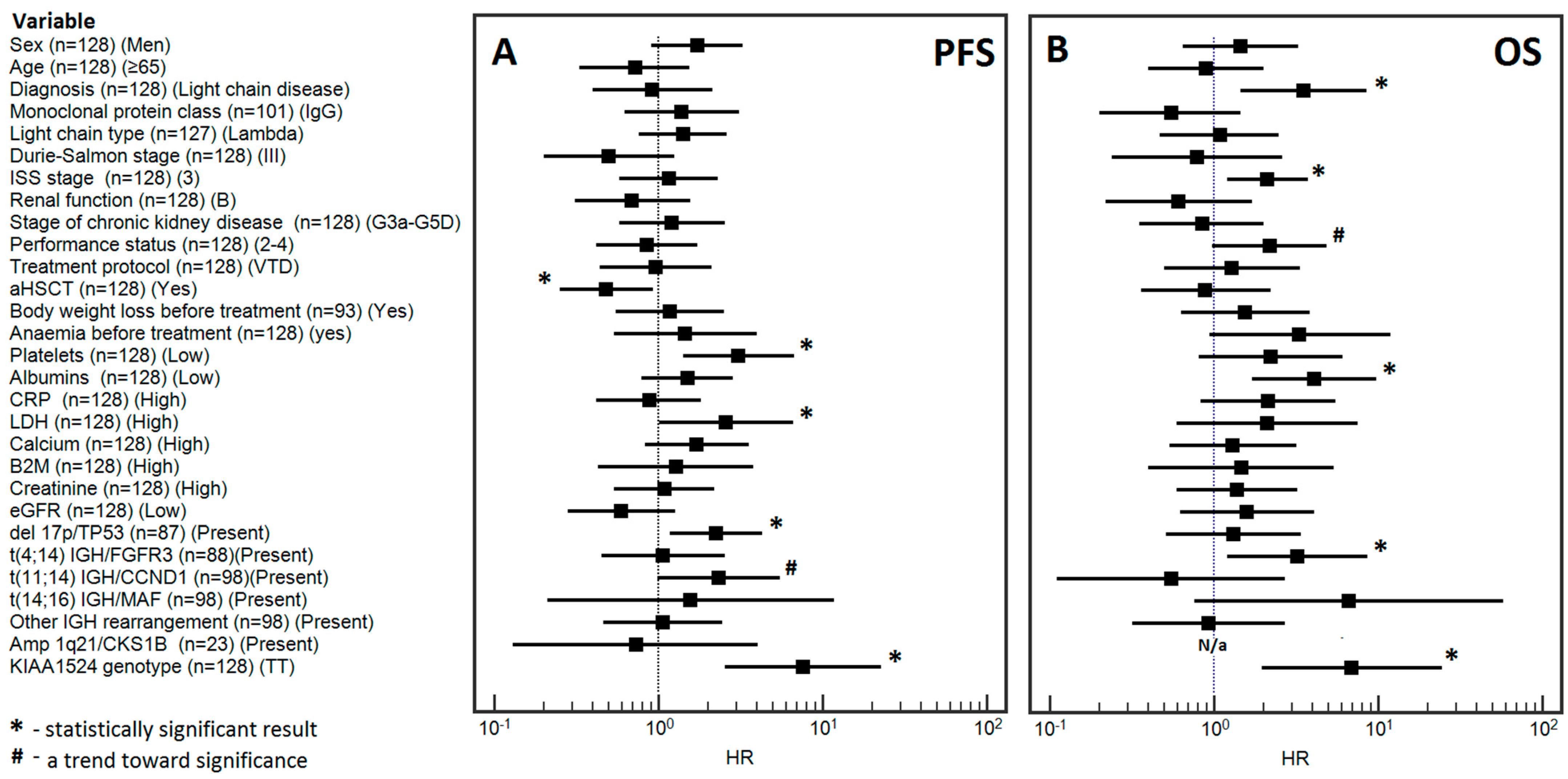
3.3.1. Univariable Analysis
3.3.2. Multivariable Analysis
3.4. Overall Survival
3.4.1. Univariable Analysis
3.4.2. Multivariable Analysis
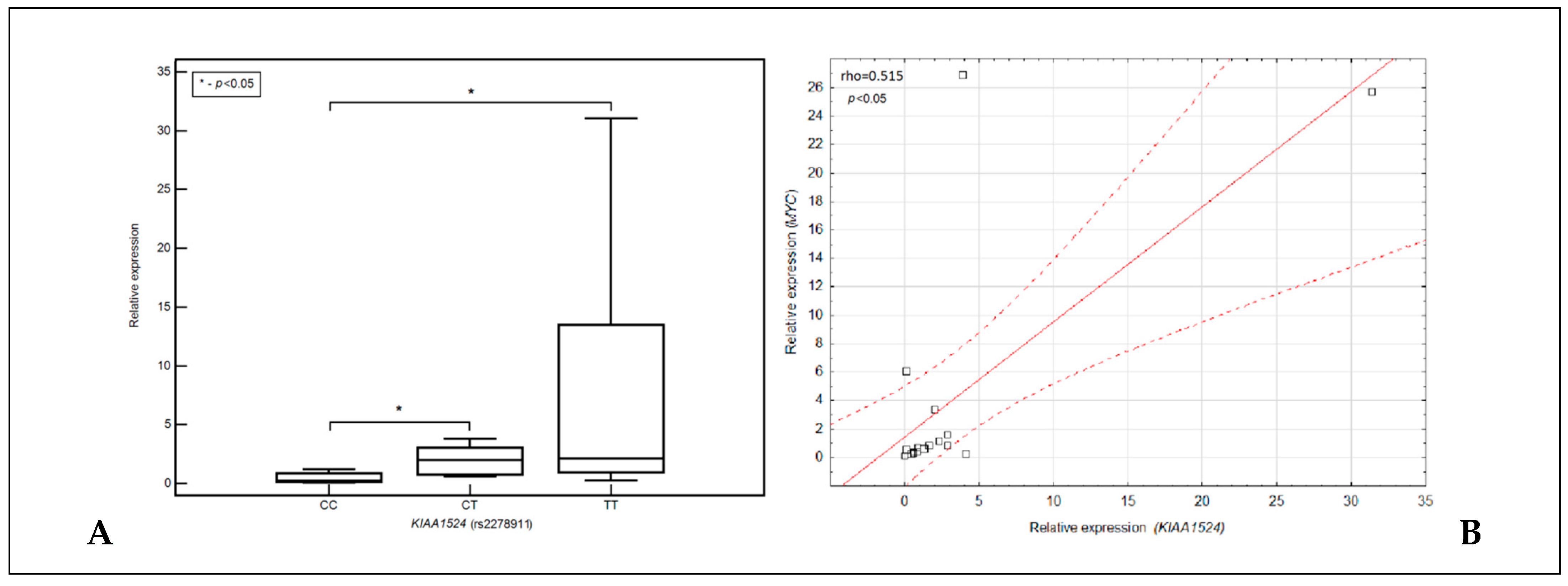
3.5. Relationship between the Expression of the KIAA1524 Gene, Its SNP (rs2278811) and the Expression of the MYC Gene
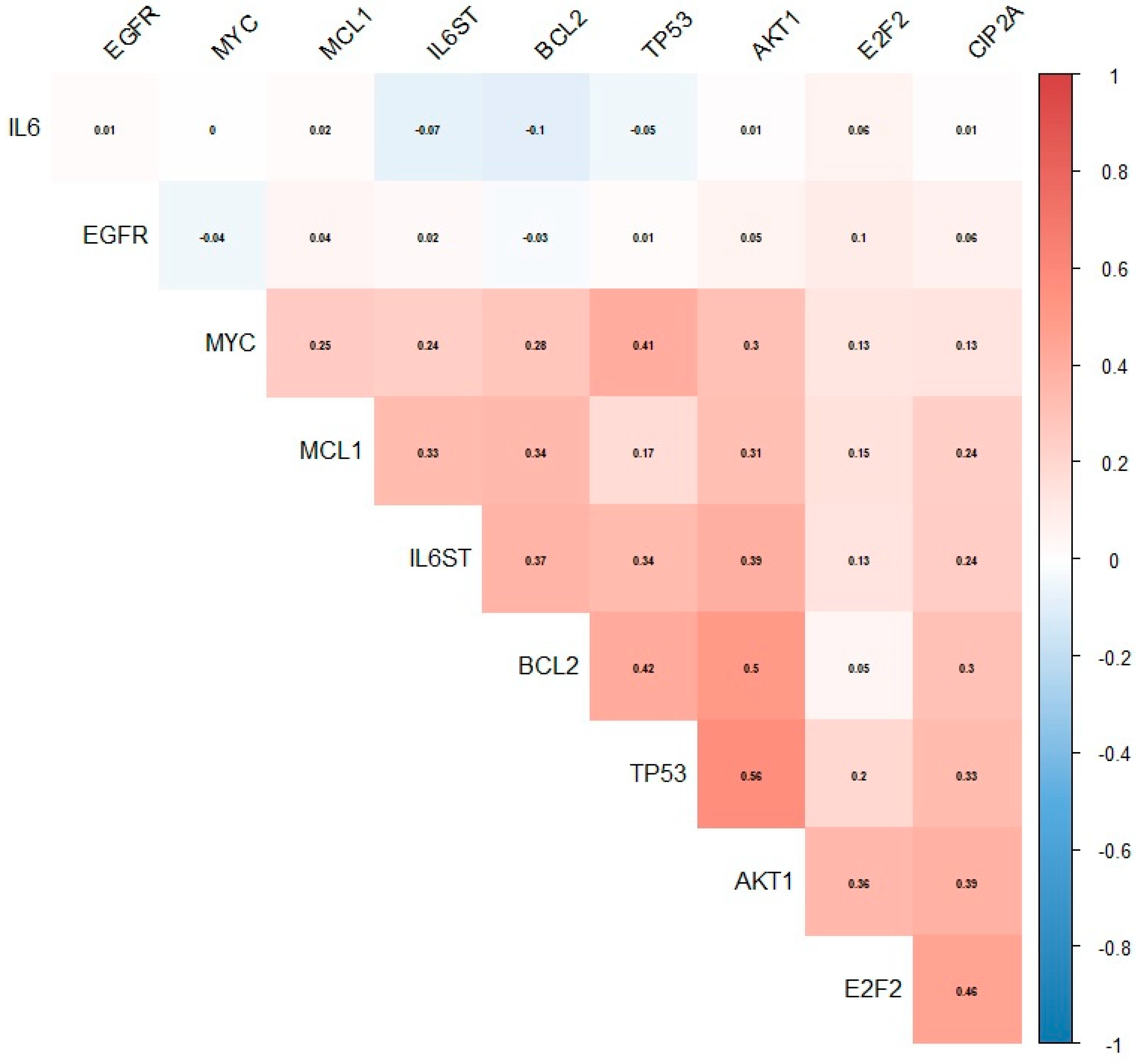
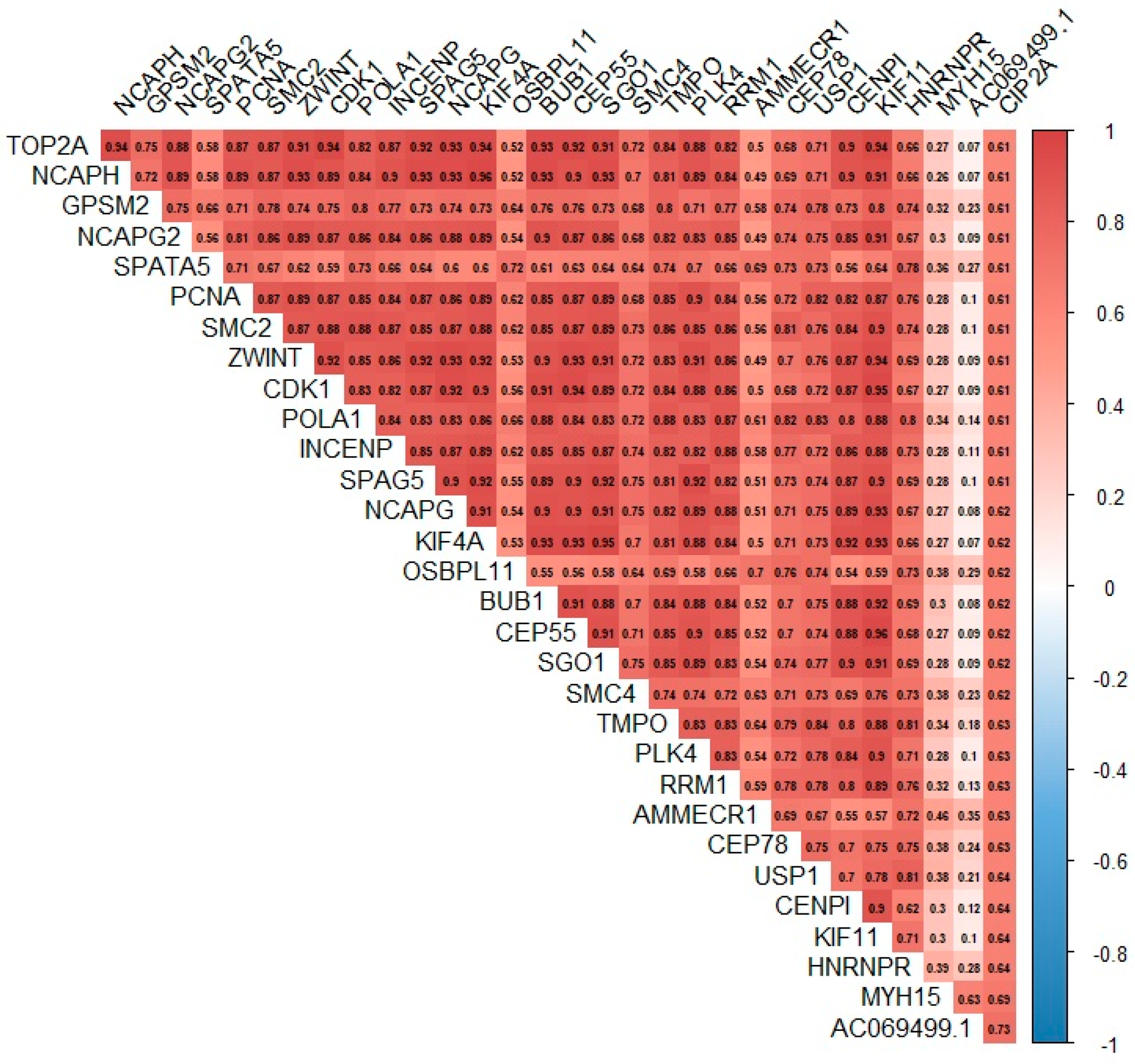
3.6. Relationship between the Expression of the KIAA1524 Gene and the Expression of Other Genes Based on Data from the CoMMpass Project
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rajkumar, S.V. Multiple myeloma: Every year a new standard? Hematol. Oncol. 2019, 37 (Suppl. 1), 62–65. [Google Scholar] [CrossRef]
- Fairfield, H.; Falank, C.; Avery, L.; Reagan, M.R. Multiple myeloma in the marrow: Pathogenesis and treatments. Ann. N. Y. Acad. Sci. 2016, 1364, 32–51. [Google Scholar] [CrossRef]
- Michels, T.C.; E Petersen, K. Multiple Myeloma: Diagnosis and Treatment. Am. Fam. Physician 2017, 95, 373–383. [Google Scholar] [PubMed]
- Van de Donk, N.W.C.J.; Pawlyn, C.; Yong, K.L. Multiple myeloma. Lancet 2021, 397, 410–427. [Google Scholar] [CrossRef] [PubMed]
- Sergentanis, T.N.; Zagouri, F.; Tsilimidos, G.; Tsagianni, A.; Tseliou, M.; Dimopoulos, M.A.; Psaltopoulou, T. Risk Factors for Multiple Myeloma: A Systematic Review of Meta-Analyses. Clin. Lymphoma Myeloma Leuk. 2015, 15, 563–577.e3. [Google Scholar] [CrossRef]
- Cowan, A.J.; Green, D.J.; Kwok, M.; Lee, S.; Coffey, D.G.; Holmberg, L.A.; Tuazon, S.; Gopal, A.K.; Libby, E.N. Diagnosis and Management of Multiple Myeloma. JAMA 2022, 327, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Palumbo, A.; Anderson, K. Multiple Myeloma. N. Engl. J. Med. 2011, 364, 1046–1060. [Google Scholar] [CrossRef] [PubMed]
- Al Hamed, R.; Bazarbachi, A.H.; Malard, F.; Harousseau, J.-L.; Mohty, M. Current status of autologous stem cell transplantation for multiple myeloma. Blood Cancer J. 2019, 9, 44. [Google Scholar] [CrossRef]
- Greipp, P.R.; Miguel, J.S.; Durie, B.G.; Crowley, J.J.; Barlogie, B.; Bladé, J.; Boccadoro, M.; Child, J.A.; Avet-Loiseau, H.; Kyle, R.A.; et al. International Staging System for Multiple Myeloma. J. Clin. Oncol. 2005, 23, 3412–3420. [Google Scholar] [CrossRef]
- Schavgoulidze, A.; Cazaubiel, T.; Perrot, A.; Avet-Loiseau, H.; Corre, J. Multiple Myeloma: Heterogeneous in Every Way. Cancers 2021, 13, 1285. [Google Scholar] [CrossRef]
- Avet-Loiseau, H.; Attal, M.; Campion, L.; Caillot, D.; Hulin, C.; Marit, G.; Stoppa, A.-M.; Voillat, L.; Wetterwald, M.; Pegourie, B.; et al. Long-Term Analysis of the IFM 99 Trials for Myeloma: Cytogenetic Abnormalities [t(4;14), del(17p), 1q gains] Play a Major Role in Defining Long-Term Survival. J. Clin. Oncol. 2012, 30, 1949–1952. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Ru, K.; Zhang, L.; Huang, Y.; Zhu, X.; Liu, H.; Zetterberg, A.; Cheng, T.; Miao, W. Fluorescence in situ hybridization (FISH): An increasingly demanded tool for biomarker research and personalized medicine. Biomark. Res. 2014, 2, 3. [Google Scholar] [CrossRef] [PubMed]
- Takimoto, M.; Ogawa, K.; Kato, Y.; Saito, T.; Suzuki, T.; Irei, M.; Shibuya, Y.; Suzuki, Y.; Kato, M.; Inoue, Y.; et al. Close relation between 14q32/IGH translocations and chromosome 13 abnormalities in multiple myeloma: A high incidence of 11q13/CCND1 and 16q23/MAF. Int. J. Hematol. 2008, 87, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Corre, J.; Munshi, N.C.; Avet-Loiseau, H. Risk factors in multiple myeloma: Is it time for a revision? Blood 2021, 137, 16–19. [Google Scholar] [CrossRef]
- Podar, K.; Chauhan, D.; Anderson, K.C. Bone marrow microenvironment and the identification of new targets for myeloma therapy. Leukemia 2008, 23, 10–24. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Kimlinger, T.; Haug, J.; Painuly, U.; Wellik, L.; Halling, T.; Rajkumar, S.V.; Kumar, S. Anti-Myeloma Activity of Akt Inhibition Is Linked to the Activation Status of PI3K/Akt and MEK/ERK Pathway. PLoS ONE 2012, 7, e50005. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Kumar, S. PI3K/AKT/mTOR pathway in multiple myeloma: From basic biology to clinical promise. Leuk. Lymphoma 2018, 59, 2524–2534. [Google Scholar] [CrossRef]
- Côme, C.; Laine, A.; Chanrion, M.; Edgren, H.; Mattila, E.; Liu, X.; Jonkers, J.; Ivaska, J.; Isola, J.; Darbon, J.-M.; et al. CIP2A Is Associated with Human Breast Cancer Aggressivity. Clin. Cancer Res. 2009, 15, 5092–5100. [Google Scholar] [CrossRef] [PubMed]
- Junttila, M.R.; Puustinen, P.; Niemelä, M.; Ahola, R.; Arnold, H.; Böttzauw, T.; Ala-Aho, R.; Nielsen, C.; Ivaska, J.; Taya, Y.; et al. CIP2A Inhibits PP2A in Human Malignancies. Cell 2007, 130, 51–62. [Google Scholar] [CrossRef]
- Soofiyani, S.R.; Hejazi, M.S.; Baradaran, B. The role of CIP2A in cancer: A review and update. Biomed. Pharm. 2017, 96, 626–633. [Google Scholar] [CrossRef]
- Khanna, A.; Böckelman, C.; Hemmes, A.; Junttila, M.R.; Wiksten, J.-P.; Lundin, M.; Junnila, S.; Murphy, D.; Evan, G.I.; Haglund, C.; et al. MYC-Dependent Regulation and Prognostic Role of CIP2A in Gastric Cancer. J. Natl. Cancer Inst. 2009, 101, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V.; Dimopoulos, M.A.; Palumbo, A.; Blade, J.; Merlini, G.; Mateos, M.-V.; Kumar, S.; Hillengass, J.; Kastritis, E.; Richardson, P.; et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014, 15, e538–e548. [Google Scholar] [CrossRef] [PubMed]
- Durie, B.G.; Salmon, S.E. A clinical staging system for multiple myeloma. Correlation of measured myeloma cell mass with presenting clinical features, response to treatment, and survival. Cancer 1975, 36, 842–854. [Google Scholar] [CrossRef] [PubMed]
- D’Agostino, M.; Cairns, D.A.; Lahuerta, J.J.; Wester, R.; Bertsch, U.; Waage, A.; Zamagni, E.; Mateos, M.-V.; Dall’Olio, D.; van de Donk, N.W.C.J.; et al. Second Revision of the International Staging System (R2-ISS) for Overall Survival in Multiple Myeloma: A European Myeloma Network (EMN) Report Within the HARMONY Project. J. Clin. Oncol. 2022, 40, 3406–3418. [Google Scholar] [CrossRef]
- International Myeloma Working Group. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: A report of the International Myeloma Working Group. Br. J. Haematol. 2003, 121, 749–757. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services. Common Terminology Criteria for Adverse Events (CTCAE); Version 5; U.S. Department of Health and Human Services: Washington, DC, USA, 2017. [Google Scholar]
- Abdallah, N.; Rajkumar, S.V.; Greipp, P.; Kapoor, P.; Gertz, M.A.; Dispenzieri, A.; Baughn, L.B.; Lacy, M.Q.; Hayman, S.R.; Buadi, F.K.; et al. Cytogenetic abnormalities in multiple myeloma: Association with disease characteristics and treatment response. Blood Cancer J. 2020, 10, 82. [Google Scholar] [CrossRef]
- Saadoune, C.; Nouadi, B.; Hamdaoui, H.; Chegdani, F.; Bennis, F. Multiple Myeloma: Bioinformatic Analysis for Identification of Key Genes and Pathways. Bioinform. Biol. Insights 2022, 16. [Google Scholar] [CrossRef]
- Misund, K.; Bruinink, D.H.O.; Coward, E.; Hoogenboezem, R.M.; Rustad, E.H.; Sanders, M.A.; Rye, M.; Sponaas, A.-M.; van der Holt, B.; Zweegman, S.; et al. Clonal evolution after treatment pressure in multiple myeloma: Heterogenous genomic aberrations and transcriptomic convergence. Leukemia 2022, 36, 1887–1897. [Google Scholar] [CrossRef]
- Junttila, M.R.; Westermarck, J. Mechanisms of MYC stabilization in human malignancies. Cell Cycle 2008, 7, 592–596. [Google Scholar] [CrossRef]
- Xing, M.-L.; Lu, Y.-F.; Wang, D.-F.; Zou, X.-Y.; Zhang, S.-X.; Yun, Z. Clinical significance of sCIP2A levels in breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 82–91. [Google Scholar]
- Gomes, L.R.; Menck, C.F.M.; Cuervo, A.M. Chaperone-mediated autophagy prevents cellular transformation by regulating MYC proteasomal degradation. Autophagy 2017, 13, 928–940. [Google Scholar] [CrossRef] [PubMed]
- Petrusca, D.N.; Mulcrone, P.L.; Macar, D.A.; Bishop, R.T.; Berdyshev, E.; Suvannasankha, A.; Anderson, J.L.; Sun, Q.; Auron, P.E.; Galson, D.L.; et al. GFI1-Dependent Repression of SGPP1 Increases Multiple Myeloma Cell Survival. Cancers 2022, 14, 772. [Google Scholar] [CrossRef] [PubMed]
- Dahlström, K.M.; Salminen, T.A. 3D model for Cancerous Inhibitor of Protein Phosphatase 2A armadillo domain unveils highly conserved protein–protein interaction characteristics. J. Theor. Biol. 2015, 386, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Okkeri, J.; Pavic, K.; Wang, Z.; Kauko, O.; Halonen, T.; Sarek, G.; Ojala, P.M.; Rao, Z.; Xu, W.; et al. Oncoprotein CIP 2A is stabilized via interaction with tumor suppressor PP 2A/B56. EMBO Rep. 2017, 18, 437–450. [Google Scholar] [CrossRef] [PubMed]
- Myant, K.; Qiao, X.; Halonen, T.; Come, C.; Laine, A.; Janghorban, M.; Partanen, J.I.; Cassidy, J.; Ogg, E.-L.; Cammareri, P.; et al. Serine 62-Phosphorylated MYC Associates with Nuclear Lamins and Its Regulation by CIP2A Is Essential for Regenerative Proliferation. Cell Rep. 2015, 12, 1019–1031. [Google Scholar] [CrossRef]
- Li, Y.; Wang, K.; Dai, L.; Wang, P.; Song, C.; Shi, J.; Ren, P.; Ye, H.; Zhang, J. HapMap-based study of CIP2A gene polymorphisms and HCC susceptibility. Oncol. Lett. 2012, 4, 358–364. [Google Scholar] [CrossRef]
- Preuss, K.-D.; Fadle, N.; Regitz, E.; Held, G.; Pfreundschuh, M. Inactivation of protein-phosphatase 2A causing hyperphosphorylation of autoantigenic paraprotein targets in MGUS/MM is due to an exchange of its regulatory subunits. Int. J. Cancer 2014, 135, 2046–2053. [Google Scholar] [CrossRef]
- Puustinen, P.; Rytter, A.; Mortensen, M.; Kohonen, P.; Moreira, J.; Jäättelä, M. CIP2A oncoprotein controls cell growth and autophagy through mTORC1 activation. J. Cell Biol. 2014, 204, 713–727. [Google Scholar] [CrossRef]
- Laine, A.; Nagelli, S.G.; Farrington, C.; Butt, U.; Cvrljevic, A.N.; Vainonen, J.P.; Feringa, F.M.; Grönroos, T.J.; Gautam, P.; Khan, S.; et al. CIP2A Interacts with TopBP1 and Drives Basal-Like Breast Cancer Tumorigenesis. Cancer Res. 2021, 81, 4319–4331. [Google Scholar] [CrossRef]
- Liu, C.-Y.; Hu, M.-H.; Hsu, C.-J.; Huang, C.-T.; Wang, D.-S.; Tsai, W.-C.; Chen, Y.-T.; Lee, C.-H.; Chu, P.-Y.; Chen, M.-H.; et al. Lapatinib inhibits CIP2A/PP2A/p-Akt signaling and induces apoptosis in triple negative breast cancer cells. Oncotarget 2016, 7, 9135–9149. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Chen, K.-C.; Chen, C.-C.; Cheng, A.-L.; Chen, K.-F. CIP2A-mediated Akt activation plays a role in bortezomib-induced apoptosis in head and neck squamous cell carcinoma cells. Oral Oncol. 2012, 48, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Bedewy, A.M.L.; Elmaghraby, S.M. CIP2A expression in Bortezomib-treated multiple myeloma. JBUON 2020, 25, 395–400. [Google Scholar]
- Nath, S.; Ohlmeyer, M.; Salathe, M.A.; Poon, J.; Baumlin, N.; Foronjy, R.F.; Geraghty, P. Chronic Cigarette Smoke Exposure Subdues PP2A Activity by Enhancing Expression of the Oncogene CIP2A. Am. J. Respir. Cell Mol. Biol. 2018, 59, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Slomp, A.; Moesbergen, L.M.; Eldering, E.; Kersten, M.J.; Minnema, M.C.; Peperzak, V. Phosphatase PP2A enhances MCL-1 protein half-life in multiple myeloma cells. Cell Death Dis. 2021, 12, 229. [Google Scholar] [CrossRef] [PubMed]
- McDonald, W.J.; Sangster, S.M.; Moffat, L.D.; Henderson, M.J.; Too, C.K. α4 phosphoprotein interacts with EDD E3 ubiquitin ligase and poly(A)-binding protein. J. Cell. Biochem. 2010, 110, 1123–1129. [Google Scholar] [CrossRef]
- Alexander-Bryant, A.A.; Dumitriu, A.; Attaway, C.C.; Yu, H.; Jakymiw, A. Fusogenic-Oligoarginine peptide-mediated silencing of the CIP2A oncogene suppresses oral cancer tumor growth in vivo. J. Control. Release 2015, 218, 72–81. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, Y.; Liu, H.; Lin, Z. Cancerous Inhibitor of PP2A Silencing Inhibits Proliferation and Promotes Apoptosis in Human Multiple Myeloma Cells. BioMed Res. Int. 2016, 2016, 6864135. [Google Scholar] [CrossRef]
- Kang, H.S.; Choi, I. Protein Phosphatase 2A Modulates the Proliferation of Human Multiple Myeloma Cells via Regulation of the Production of Reactive Oxygen Intermediates and Anti-Apoptotic Factors. Cell. Immunol. 2001, 213, 34–44. [Google Scholar] [CrossRef]
- Mitsuhashi, S.; Shima, H.; Tanuma, N.; Sasa, S.; Onoe, K.; Ubukata, M.; Kikuchi, K. Protein phosphatase type 2A, PP2A, is involved in degradation of gp130. Mol. Cell. Biochem. 2005, 269, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Thangavelu, P.U.; Lin, C.-Y.; Vaidyanathan, S.; Nguyen, T.H.; Dray, E.; Duijf, P.H. Overexpression of the E2F target gene CENPI promotes chromosome instability and predicts poor prognosis in estrogen receptor-positive breast cancer. Oncotarget 2017, 8, 62167–62182. [Google Scholar] [CrossRef]
- Brunk, K.; Zhu, M.; Bärenz, F.; Kratz, A.-S.; Haselmann-Weiss, U.; Antony, C.; Hoffmann, I. Cep78 is a novel centriolar protein involved in Plk4-induced centriole overduplication. J. Cell Sci. 2016, 129, 2713–2718. [Google Scholar] [CrossRef] [PubMed]
- Gladilin, E.; Ohse, S.; Boerries, M.; Busch, H.; Xu, C.; Schneider, M.; Meister, M.; Eils, R. TGFβ-induced cytoskeletal remodeling mediates elevation of cell stiffness and invasiveness in NSCLC. Sci. Rep. 2019, 9, 7667. [Google Scholar] [CrossRef]
- Garcia-Saez, I.; Skoufias, D.A. Eg5 targeting agents: From new anti-mitotic based inhibitor discovery to cancer therapy and resistance. Biochem. Pharmacol. 2020, 184, 114364. [Google Scholar] [CrossRef] [PubMed]
- Maniswami, R.R.; Prashanth, S.; Karanth, A.V.; Koushik, S.; Govindaraj, H.; Mullangi, R.; Rajagopal, S.; Jegatheesan, S.K. PLK4: A link between centriole biogenesis and cancer. Expert Opin. Ther. Targets 2017, 22, 59–73. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Chu, H.-J.; Li, N.; Huang, L.-Y.; Chen, J. Centromere Protein I (CENP-I) Is Upregulated in Gastric Cancer, Predicts Poor Prognosis, and Promotes Tumor Cell Proliferation and Migration. Technol. Cancer Res. Treat. 2021, 20. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Tang, X.; Tang, C.; Hua, Z.; Ke, M.; Wang, C.; Zhao, J.; Gao, S.; Jurczyszyn, A.; Janz, S.; et al. HNRNPA2B1 promotes multiple myeloma progression by increasing AKT3 expression via m6A-dependent stabilization of ILF3 mRNA. J. Hematol. Oncol. 2021, 14, 54. [Google Scholar] [CrossRef]
- Das, D.S.; Das, A.; Ray, A.; Song, Y.; Samur, M.K.; Munshi, N.C.; Chauhan, D.; Anderson, K.C. Blockade of Deubiquitylating Enzyme USP1 Inhibits DNA Repair and Triggers Apoptosis in Multiple Myeloma Cells. Clin. Cancer Res. 2017, 23, 4280–4289. [Google Scholar] [CrossRef]
- Sagawa, M.; Ohguchi, H.; Harada, T.; Samur, M.K.; Tai, Y.-T.; Munshi, N.C.; Kizaki, M.; Hideshima, T.; Anderson, K.C. Ribonucleotide Reductase Catalytic Subunit M1 (RRM1) as a Novel Therapeutic Target in Multiple Myeloma. Clin. Cancer Res. 2017, 23, 5225–5237. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Zheng, C.; Tang, Y. Overexpression of SMC4 predicts a poor prognosis in cervical cancer and facilitates cancer cell malignancy phenotype by activating NF-κB pathway. Hum. Cell 2021, 34, 1888–1898. [Google Scholar] [CrossRef]
- Chen, Q.; Wan, X.; Chen, Y.; Liu, C.; Gu, M.; Wang, Z. SGO1 induces proliferation and metastasis of prostate cancer through AKT-mediated signaling pathway. Am. J. Cancer Res. 2019, 9, 2693–2705. [Google Scholar]
- Zhang, X.; Xu, Q.; Li, E.; Shi, T.; Chen, H. CEP55 predicts the poor prognosis and promotes tumorigenesis in endometrial cancer by regulating the Foxo1 signaling. Mol. Cell. Biochem. 2022. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Liao, Y.; Wang, M.; Wang, Y.; Wang, K.; Guo, J.; Wu, P.; Zhong, B.; Guo, T.; Wu, C. BUB1 drives the occurrence and development of bladder cancer by mediating the STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2021, 40, 378. [Google Scholar] [CrossRef]
- Chou, C.-W.; Hsieh, Y.-H.; Ku, S.-C.; Shen, W.-J.; Anuraga, G.; Ta, H.D.K.; Lee, K.-H.; Lee, Y.-C.; Lin, C.-H.; Wang, C.-Y.; et al. Potential Prognostic Biomarkers of OSBPL Family Genes in Patients with Pancreatic Ductal Adenocarcinoma. Biomedicines 2021, 9, 1601. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Lei, X.; Mao, X. Identification of KIF4A as a pan-cancer diagnostic and prognostic biomarker via bioinformatics analysis and validation in osteosarcoma cell lines. PeerJ 2021, 9, e11455. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Ge, C.; Fang, D.; Wei, W.; Li, L.; Wei, Q.; Yu, H. NCAPG facilitates colorectal cancer cell proliferation, migration, invasion and epithelial–mesenchymal transition by activating the Wnt/β-catenin signaling pathway. Cancer Cell Int. 2022, 22, 119. [Google Scholar] [CrossRef]
- Zeng, X.; Xu, W.; Tong, J.; Liu, J.; Zhang, Z.; Liu, M.; Wu, C.; Yu, Q.; Ye, C.; Wu, C.; et al. SPAG5 as a novel biomarker and potential therapeutic target via regulating AKT pathway in multiple myeloma. Leuk. Lymphoma 2022, 63, 2565–2572. [Google Scholar] [CrossRef]
- Li, B.; Xiao, Q.; Shan, L.; Song, Y. NCAPH promotes cell proliferation and inhibits cell apoptosis of bladder cancer cells through MEK/ERK signaling pathway. Cell Cycle 2022, 21, 427–438. [Google Scholar] [CrossRef]
- Xie, B.; Wang, S.; Jiang, N.; Li, J.J. Cyclin B1/CDK1-regulated mitochondrial bioenergetics in cell cycle progression and tumor resistance. Cancer Lett. 2018, 443, 56–66. [Google Scholar] [CrossRef]
- Kim, J.H.; Youn, Y.; Lee, J.-C.; Kim, J.; Hwang, J.-H. Involvement of the NF-κB signaling pathway in proliferation and invasion inhibited by Zwint-1 deficiency in Pancreatic Cancer Cells. J. Cancer 2020, 11, 5601–5611. [Google Scholar] [CrossRef]
- Kim, J.H.; Shim, J.; Ji, M.-J.; Jung, Y.; Bong, S.M.; Jang, Y.-J.; Yoon, E.-K.; Lee, S.-J.; Kim, K.G.; Kim, Y.H.; et al. The condensin component NCAPG2 regulates microtubule–kinetochore attachment through recruitment of polo-like kinase 1 to kinetochores. Nat. Commun. 2014, 5, 4588. [Google Scholar] [CrossRef]
- Yang, D.; Ji, F.; Li, Y.; Jiao, Y.; Fang, X. GPSM2 Serves as an Independent Prognostic Biomarker for Liver Cancer Survival. Technol. Cancer Res. Treat. 2020, 19. [Google Scholar] [CrossRef]
- Adams, R.; Wheatleya, S.; Gouldsworthy, A.; Kandels-Lewis, S.; Carmena, M.; Smythe, C.; Gerloff, D.; Earnshaw, W. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr. Biol. 2000, 10, 1075–1078. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liu, D.; Wang, L.; Liu, M.; Duan, W.; Yi, J.; Yi, Y. DNA Repair Genes Are Associated with Subtype Classification, Prognosis, and Immune Infiltration in Uveal Melanoma. J. Oncol. 2022, 2022, 1965451. [Google Scholar] [CrossRef]
- Müller, R.; Misund, K.; Holien, T.; Bachke, S.; Gilljam, K.M.; Våtsveen, T.K.; Rø, T.B.; Bellacchio, E.; Sundan, A.; Otterlei, M. Targeting Proliferating Cell Nuclear Antigen and Its Protein Interactions Induces Apoptosis in Multiple Myeloma Cells. PLoS ONE 2013, 8, e70430. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.H.; Wan, Y.; Xiong, H.; Sun, G.L. Structural maintenance of chromosomes 2 is identified as an oncogene in bladder cancer in vitro and in vivo. Neoplasma 2020, 67, 364–370. [Google Scholar] [CrossRef]
- Reale, A.; Khong, T.; Mithraprabhu, S.; Savvidou, I.; Hocking, J.; Bergin, K.; Ramachandran, M.; Chen, M.; Dammacco, F.; Ria, R.; et al. TOP2A expression predicts responsiveness to carfilzomib in myeloma and informs novel combinatorial strategies for enhanced proteasome inhibitor cell killing. Leuk. Lymphoma 2020, 62, 337–347. [Google Scholar] [CrossRef] [PubMed]
| Variable | n = 128 (100%) |
|---|---|
| Sex | |
| Men | 64 (50%) |
| Women | 64 (50%) |
| Age | |
| <65 | 58 (45.3%) |
| ≥65 | 70 (54.7%) |
| Myeloma type | |
| IgG | 72 (56.2.%) |
| IgA | 29 (22.7%) |
| Light chains | 25 (19.5%) |
| Non-secretory | 1 (0.8%) |
| Plasmocytoma | 1 (0.8%) |
| Light chain type | |
| Kappa | 80 (63%) |
| Lambda | 47 (37%) |
| No data: n = 1 | |
| Durie–Salmon stage | |
| I | 12 (9.4%) |
| II | 16 (12.5%) |
| III | 100 (78.1%) |
| ISS stage | |
| 1 | 33 (25.8%) |
| 2 | 36 (28.1%) |
| 3 | 59 (46.1%) |
| Renal function | |
| A—creatinine < 2 mg/dL | 102 (79.7%) |
| B—creatinine ≥ 2 mg/dL | 26 (20.3%) |
| Performance status | |
| 0 | 11 (8.6%) |
| 1 | 49 (38.3%) |
| 2 | 51 (39.8%) |
| 3 | 15 (11.7%) |
| 4 | 2 (1.6%) |
| Body weight loss | |
| No | 47 (50.5%) |
| Yes | 46 (49.5%) |
| No data: n = 35 | |
| 5% | 14 (30.4%) |
| 10% | 32 (69.6%) |
| Anemia grade before treatment (WHO) | |
| Absent or I° | 65 (50.8%) |
| II°, III° or IV° | 63 (49.2%) |
| Treatment protocol | |
| CTD | 27 (21.1%) |
| V(C)D | 58 (45.3%) |
| VTD | 43 (33.6%) |
| AHSCT | 82 (64.1%) |
| No | 46 (35.9%) |
| Yes | |
| del 17p/TP53 | |
| Absent | 65 (74.7%) |
| Present | 22 (25.3%) |
| No data: n = 41 | |
| t(4;14) IGH/FGFR3 | |
| Absent | 75 (85.2%) |
| Present | 13 (14.8%) |
| No data: n = 40 | |
| t(11;14) IGH/CCND1 | |
| Absent | 87 (88.8%) |
| Present | 11 (11.2%) |
| No data: n = 30 | |
| t(14;16) IGH/MAF | |
| Absent | 82 (83.2%) |
| Present | 6 (6.8%) |
| No data: n = 40 | |
| Other IGH rearrangement | |
| Absent | 84 (85.7%) |
| Present | 14 (14.3%) |
| No data: n = 30 | |
| amp 1q21/CKS1B | |
| Absent | 11 (47.8%) |
| Present | 12 (52.2%) |
| No data: n = 105 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szudy-Szczyrek, A.; Mlak, R.; Mazurek, M.; Krajka, T.; Chocholska, S.; Bitkowska, P.; Jutrzenka, M.; Szczyrek, M.; Homa-Mlak, I.; Krajka, A.; et al. The TT Genotype of the KIAA1524 rs2278911 Polymorphism Is Associated with Poor Prognosis in Multiple Myeloma. Cells 2023, 12, 1029. https://doi.org/10.3390/cells12071029
Szudy-Szczyrek A, Mlak R, Mazurek M, Krajka T, Chocholska S, Bitkowska P, Jutrzenka M, Szczyrek M, Homa-Mlak I, Krajka A, et al. The TT Genotype of the KIAA1524 rs2278911 Polymorphism Is Associated with Poor Prognosis in Multiple Myeloma. Cells. 2023; 12(7):1029. https://doi.org/10.3390/cells12071029
Chicago/Turabian StyleSzudy-Szczyrek, Aneta, Radosław Mlak, Marcin Mazurek, Tomasz Krajka, Sylwia Chocholska, Paulina Bitkowska, Marta Jutrzenka, Michał Szczyrek, Iwona Homa-Mlak, Andrzej Krajka, and et al. 2023. "The TT Genotype of the KIAA1524 rs2278911 Polymorphism Is Associated with Poor Prognosis in Multiple Myeloma" Cells 12, no. 7: 1029. https://doi.org/10.3390/cells12071029
APA StyleSzudy-Szczyrek, A., Mlak, R., Mazurek, M., Krajka, T., Chocholska, S., Bitkowska, P., Jutrzenka, M., Szczyrek, M., Homa-Mlak, I., Krajka, A., Małecka-Massalska, T., & Hus, M. (2023). The TT Genotype of the KIAA1524 rs2278911 Polymorphism Is Associated with Poor Prognosis in Multiple Myeloma. Cells, 12(7), 1029. https://doi.org/10.3390/cells12071029






