Activin and BMP Signalling in Human Testicular Cancer Cell Lines, and a Role for the Nucleocytoplasmic Transport Protein Importin-5 in Their Crosstalk
Abstract
1. Introduction
2. Materials and Methods:
2.1. Cell Lines and Growth Factor Treatments
2.2. RNA Sequencing and Bioinformatic Analysis
2.3. cDNA Synthesis and qRT-PCR
2.4. Dual Promoter Luciferase Assay
2.5. Transient IPO5 Silencing
2.6. Protein Extraction and Immunoblotting
2.7. Statistical Analyses
3. Results
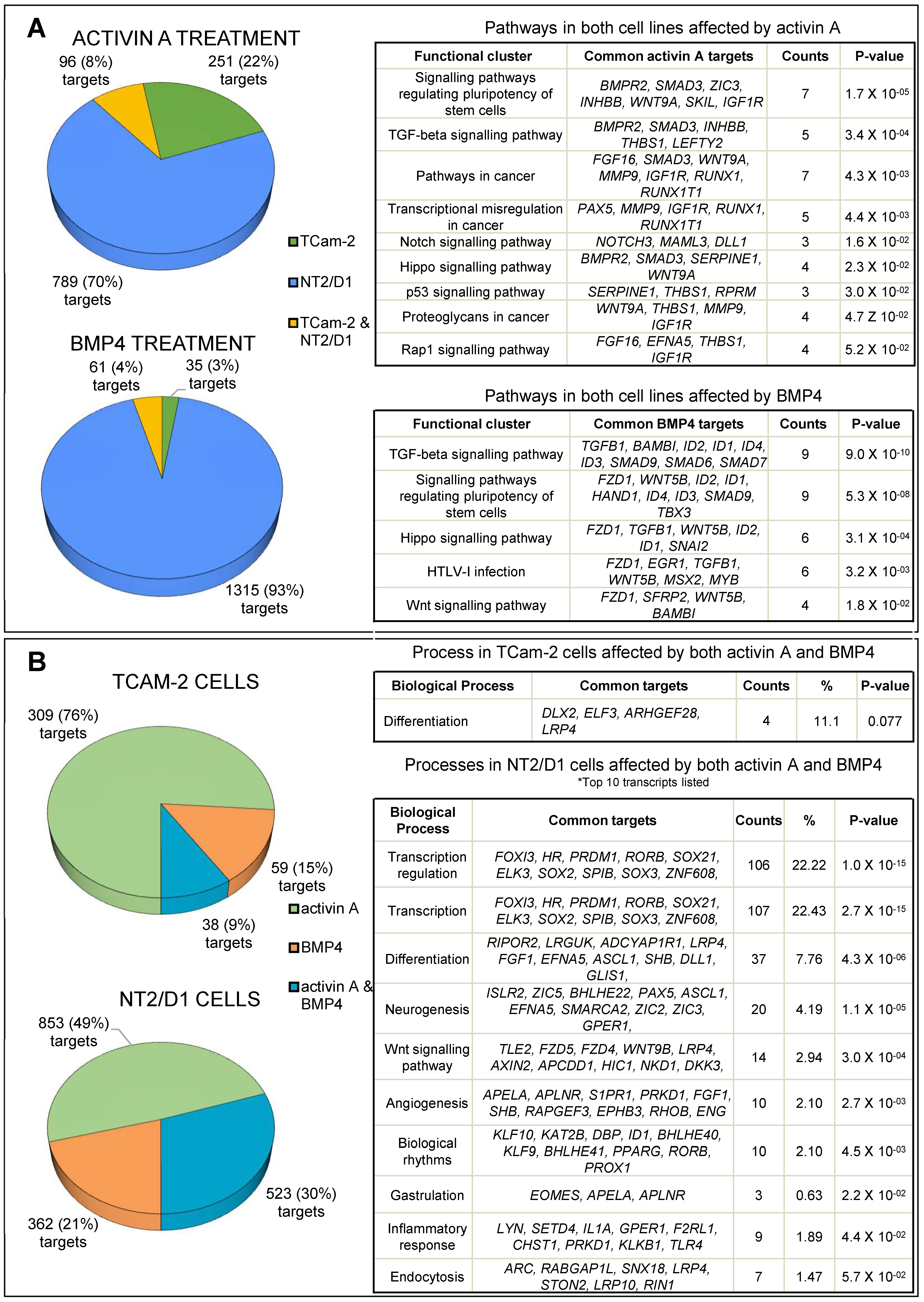
3.1. Transcriptome Profiling and Bioinformatic Analyses of Activin A- and BMP4-Treated TCam-2 and NT2/D1 Cells
3.2. Activin A Treatment
3.3. BMP4 Treatment
3.4. Common Targets of Activin A and BMP4
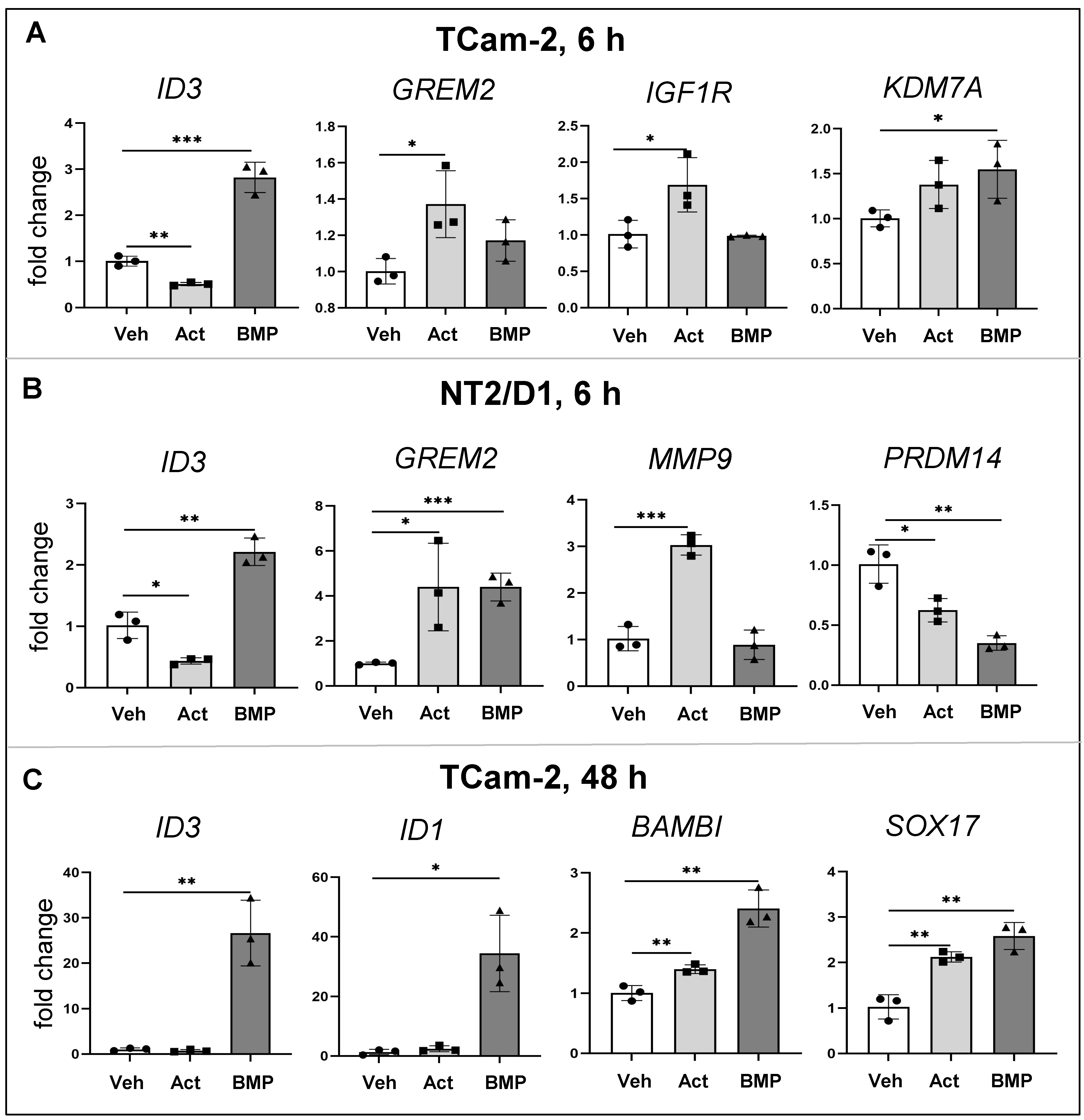
3.5. Common and Distinct Regulation of Specific Genes by Activin A and BMP4
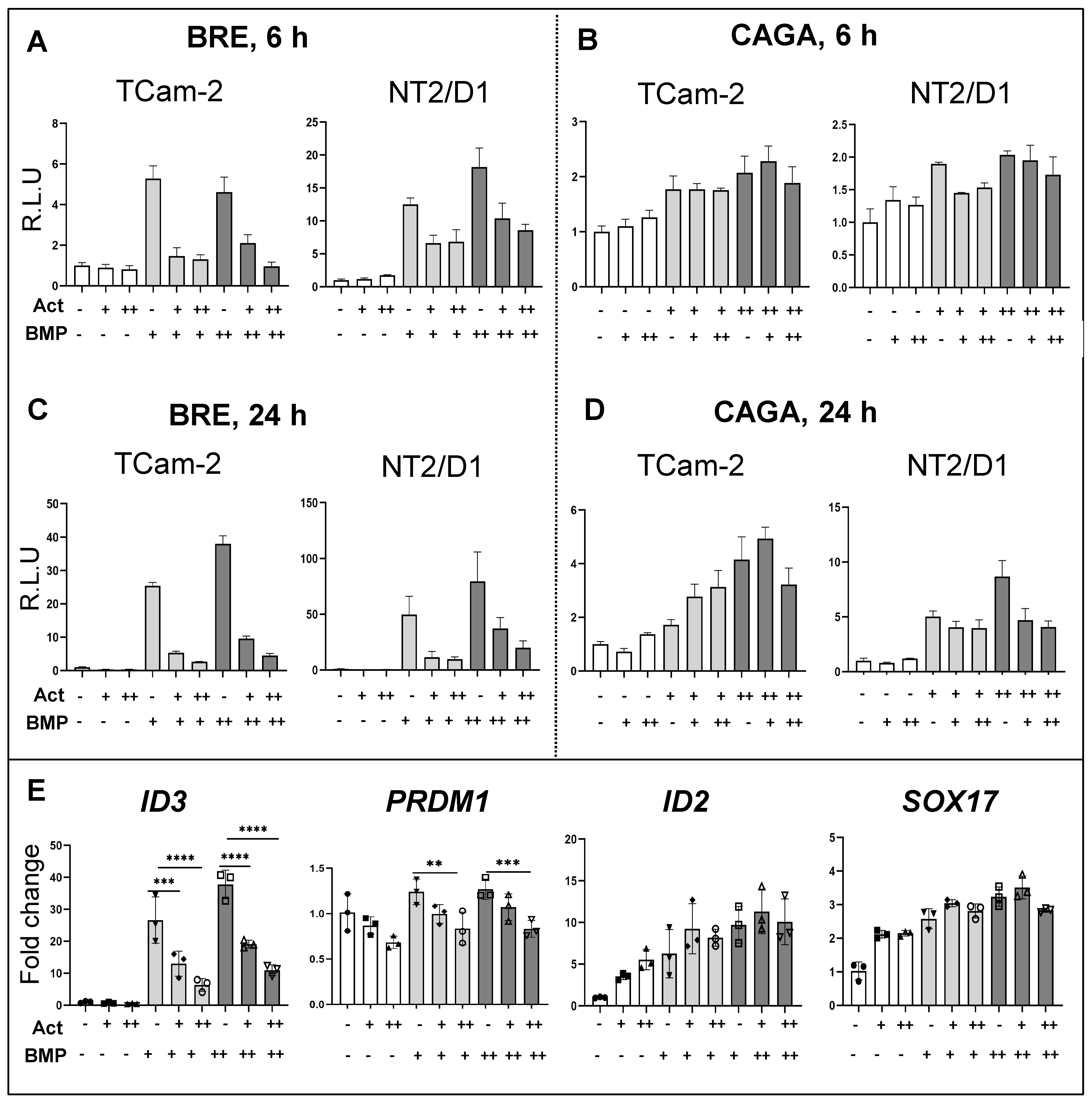
3.6. Effects of Activin A on BMP Signalling Activation
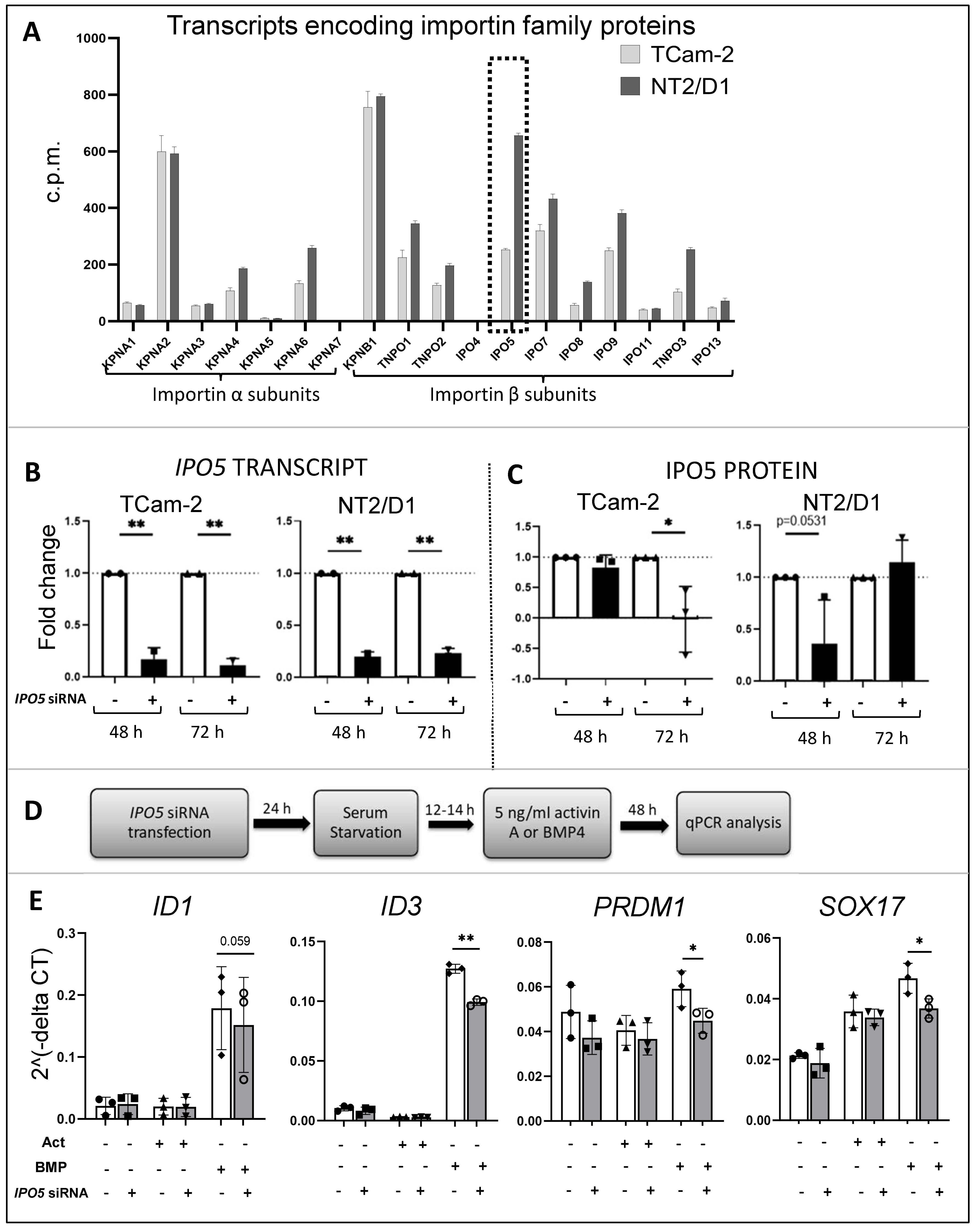
3.7. Importins in TCam-2 and NT2/D1 Cells: A Role for IPO5 in BMP4 Responses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Oosterhuis, J.W.; Looijenga, L.H. Testicular germ-cell tumours in a broader perspective. Nat. Rev. Cancer 2005, 5, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Shanmugalingam, T.; Soultati, A.; Chowdhury, S.; Rudman, S.; Van Hemelrijck, M. Global incidence and outcome of testicular cancer. Clin. Epidemiol. 2013, 5, 417–427. [Google Scholar] [CrossRef]
- Berney, D.M.; Looijenga, L.H.; Idrees, M.; Oosterhuis, J.W.; Rajpert-De Meyts, E.; Ulbright, T.M.; Skakkebaek, N.E. Germ cell neoplasia in situ (GCNIS): Evolution of the current nomenclature for testicular pre-invasive germ cell malignancy. Histopathology 2016, 69, 7–10. [Google Scholar] [CrossRef]
- Biermann, K.; Heukamp, L.C.; Steger, K.; Zhou, H.; Franke, F.E.; Sonnack, V.; Brehm, R.; Berg, J.; Bastian, P.J.; Muller, S.C.; et al. Genome-wide expression profiling reveals new insights into pathogenesis and progression of testicular germ cell tumors. Cancer Genom. Proteom. 2007, 4, 359–367. [Google Scholar]
- Hoei-Hansen, C.E.; Nielsen, J.E.; Almstrup, K.; Hansen, M.A.; Skakkebaek, N.E.; Rajpert-DeMeyts, E.; Leffers, H. Identification of genes differentially expressed in testes containing carcinoma in situ. Mol. Hum. Reprod. 2004, 10, 423–431. [Google Scholar] [CrossRef]
- Skakkebaek, N.E. Possible carcinoma-in-situ of the testis. Lancet 1972, 2, 516–517. [Google Scholar] [CrossRef] [PubMed]
- Sonne, S.B.; Almstrup, K.; Dalgaard, M.; Juncker, A.S.; Edsgard, D.; Ruban, L.; Harrison, N.J.; Schwager, C.; Abdollahi, A.; Huber, P.E.; et al. Analysis of gene expression profiles of microdissected cell populations indicates that testicular carcinoma in situ is an arrested gonocyte. Cancer Res. 2009, 69, 5241–5250. [Google Scholar] [CrossRef]
- Skakkebaek, N.E.; Rajpert-De Meyts, E.; Buck Louis, G.M.; Toppari, J.; Andersson, A.M.; Eisenberg, M.L.; Jensen, T.K.; Jørgensen, N.; Swan, S.H.; Sapra, K.J.; et al. Male Reproductive Disorders and Fertility Trends: Influences of Environment and Genetic Susceptibility. Physiol. Rev. 2016, 96, 55–97. [Google Scholar] [CrossRef]
- El-Naggar, A.K.; Ro, J.Y.; McLemore, D.; Ayala, A.G.; Batsakis, J.G. DNA ploidy in testicular germ cell neoplasms. Histogenetic and clinical implications. Am. J. Surg. Pathol. 1992, 16, 611–618. [Google Scholar] [CrossRef]
- Oosterhuis, J.W.; Looijenga, L.H. Current views on the pathogenesis of testicular germ cell tumours and perspectives for future research: Highlights of the 5th Copenhagen Workshop on Carcinoma in situ and Cancer of the Testis. Apmis 2003, 111, 280–289. [Google Scholar] [CrossRef]
- Nettersheim, D.; Schorle, H. The plasticity of germ cell cancers and its dependence on the cellular microenvironment. J. Cell. Mol. Med. 2017, 21, 1463–1467. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Shih, J.; Hollern, D.P.; Wang, L.; Bowlby, R.; Tickoo, S.K.; Thorsson, V.; Mungall, A.J.; Newton, Y.; Hegde, A.M.; et al. Integrated Molecular Characterization of Testicular Germ Cell Tumors. Cell Rep. 2018, 23, 3392–3406. [Google Scholar] [CrossRef] [PubMed]
- Mendis, S.H.; Meachem, S.J.; Sarraj, M.A.; Loveland, K.L. Activin A balances Sertoli and germ cell proliferation in the fetal mouse testis. Biol. Reprod. 2011, 84, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Whiley, P.A.F.; O’Donnell, L.; Moody, S.C.; Handelsman, D.J.; Young, J.C.; Richards, E.A.; Almstrup, K.; Western, P.S.; Loveland, K.L. Activin A Determines Steroid Levels and Composition in the Fetal Testis. Endocrinology 2020, 161, bqaa058. [Google Scholar] [CrossRef] [PubMed]
- Archambeault, D.R.; Yao, H.H. Activin A, a product of fetal Leydig cells, is a unique paracrine regulator of Sertoli cell proliferation and fetal testis cord expansion. Proc. Natl. Acad. Sci. USA 2010, 107, 10526–10531. [Google Scholar] [CrossRef]
- Moody, S.C.; Whiley, P.A.F.; Western, P.S.; Loveland, K.L. The Impact of Activin A on Fetal Gonocytes: Chronic Versus Acute Exposure Outcomes. Front. Endocrinol. 2022, 13, 896747. [Google Scholar] [CrossRef]
- Young, J.C.; Wakitani, S.; Loveland, K.L. TGF-β superfamily signaling in testis formation and early male germline development. Semin. Cell Dev. Biol. 2015, 45, 94–103. [Google Scholar] [CrossRef]
- Dias, V.L.; Rajpert-De Meyts, E.; McLachlan, R.; Loveland, K.L. Analysis of activin/TGFB-signaling modulators within the normal and dysfunctional adult human testis reveals evidence of altered signaling capacity in a subset of seminomas. Reproduction 2009, 138, 801–811. [Google Scholar] [CrossRef]
- Spiller, C.M.; Bowles, J.; Koopman, P. Nodal/Cripto signaling in fetal male germ cell development: Implications for testicular germ cell tumors. Int. J. Dev. Biol. 2013, 57, 211–219. [Google Scholar] [CrossRef]
- Spiller, C.M.; Gillis, A.J.; Burnet, G.; Stoop, H.; Koopman, P.; Bowles, J.; Looijenga, L.H. Cripto: Expression, epigenetic regulation and potential diagnostic use in testicular germ cell tumors. Mol. Oncol. 2016, 10, 526–537. [Google Scholar] [CrossRef]
- Dias, V.; Meachem, S.; Rajpert-De Meyts, E.; McLachlan, R.; Manuelpillai, U.; Loveland, K.L. Activin receptor subunits in normal and dysfunctional adult human testis. Hum. Reprod. 2008, 23, 412–420. [Google Scholar] [CrossRef]
- Fustino, N.; Rakheja, D.; Ateek, C.S.; Neumann, J.C.; Amatruda, J.F. Bone morphogenetic protein signalling activity distinguishes histological subsets of paediatric germ cell tumours. Int. J. Androl. 2011, 34, e218–e233. [Google Scholar] [CrossRef] [PubMed]
- Neumann, J.C.; Chandler, G.L.; Damoulis, V.A.; Fustino, N.J.; Lillard, K.; Looijenga, L.; Margraf, L.; Rakheja, D.; Amatruda, J.F. Mutation in the type IB bone morphogenetic protein receptor Alk6b impairs germ-cell differentiation and causes germ-cell tumors in zebrafish. Proc. Natl. Acad. Sci. USA 2011, 108, 13153–13158. [Google Scholar] [CrossRef] [PubMed]
- Neumann, J.C.; Dovey, J.S.; Chandler, G.L.; Carbajal, L.; Amatruda, J.F. Identification of a heritable model of testicular germ cell tumor in the zebrafish. Zebrafish 2009, 6, 319–327. [Google Scholar] [CrossRef]
- Purdue, M.P.; Sakoda, L.C.; Graubard, B.I.; Welch, R.; Chanock, S.J.; Sesterhenn, I.A.; Rubertone, M.V.; Erickson, R.L.; McGlynn, K.A. A case-control investigation of immune function gene polymorphisms and risk of testicular germ cell tumors. Cancer Epidemiol. Biomarkers Prev. 2007, 16, 77–83. [Google Scholar] [CrossRef]
- De Jong, J.; Stoop, H.; Gillis, A.J.; Hersmus, R.; van Gurp, R.J.; van de Geijn, G.J.; van Drunen, E.; Beverloo, H.B.; Schneider, D.T.; Sherlock, J.K.; et al. Further characterization of the first seminoma cell line TCam-2. Genes Chromosomes Cancer 2008, 47, 185–196. [Google Scholar] [CrossRef]
- Nettersheim, D.; Gillis, A.J.; Looijenga, L.H.; Schorle, H. TGF-β1, EGF and FGF4 synergistically induce differentiation of the seminoma cell line TCam-2 into a cell type resembling mixed non-seminoma. Int. J. Androl. 2011, 34, e189–e203. [Google Scholar] [CrossRef]
- Nettersheim, D.; Jostes, S.; Sharma, R.; Schneider, S.; Hofmann, A.; Ferreira, H.J.; Hoffmann, P.; Kristiansen, G.; Esteller, M.B.; Schorle, H. BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma. PLoS Genet. 2015, 11, e1005415. [Google Scholar] [CrossRef] [PubMed]
- Nettersheim, D.; Heimsoeth, A.; Jostes, S.; Schneider, S.; Fellermeyer, M.; Hofmann, A.; Schorle, H. SOX2 is essential for in vivo reprogramming of seminoma-like TCam-2 cells to an embryonal carcinoma-like fate. Oncotarget 2016, 7, 47095–47110. [Google Scholar] [CrossRef]
- Moody, S.C.; Wakitani, S.; Young, J.C.; Western, P.S.; Loveland, K.L. Evidence that activin A directly modulates early human male germline differentiation status. Reproduction 2020, 160, 141–154. [Google Scholar] [CrossRef]
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-β and the TGF-β Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb. Perspect Biol. 2016, 8, a021873. [Google Scholar] [CrossRef]
- Coda, D.M.; Patel, H.; Gori, I.; Gaarenstroom, T.E.; Song, O.R.; Howell, M.; Hill, C.S. A network of transcription factors governs the dynamics of NODAL/Activin transcriptional responses. J. Cell Sci. 2022, 135, jcs259972. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Hackert, E.; Sundan, A.; Holien, T. Receptor binding competition: A paradigm for regulating TGF-β family action. Cytokine Growth Factor Rev. 2021, 57, 39–54. [Google Scholar] [CrossRef] [PubMed]
- Luo, K. Signaling Cross Talk between TGF-β/Smad and Other Signaling Pathways. Cold Spring Harb. Perspect. Biol. 2017, 9, a022137. [Google Scholar] [CrossRef]
- Mu, Y.; Gudey, S.K.; Landström, M. Non-Smad signaling pathways. Cell Tissue Res. 2012, 347, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.S. Transcriptional Control by the SMADs. Cold Spring Harb. Perspect. Biol. 2016, 8, a022079. [Google Scholar] [CrossRef] [PubMed]
- Windley, S.P.; Wilhelm, D. Signaling Pathways Involved in Mammalian Sex Determination and Gonad Development. Sex. Dev. 2015, 9, 297–315. [Google Scholar] [CrossRef]
- Miyamoto, Y.; Yamada, K.; Yoneda, Y. Importin α: A key molecule in nuclear transport and non-transport functions. J. Biochem. 2016, 160, 69–75. [Google Scholar] [CrossRef]
- Loveland, K.L.; Major, A.T.; Butler, R.; Young, J.C.; Jans, D.A.; Miyamoto, Y. Putting things in place for fertilization: Discovering roles for importin proteins in cell fate and spermatogenesis. Asian J. Androl. 2015, 17, 537–544. [Google Scholar] [CrossRef]
- Nathaniel, B.; Whiley, P.A.F.; Miyamoto, Y.; Loveland, K.L. Importins: Diverse roles in male fertility. Semin. Cell. Dev. Biol. 2022, 121, 82–98. [Google Scholar] [CrossRef]
- Yasuhara, N.; Yoneda, Y. Importins in the maintenance and lineage commitment of ES cells. Neurochem. Int. 2017, 105, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Hogarth, C.A.; Jans, D.A.; Loveland, K.L. Subcellular distribution of importins correlates with germ cell maturation. Dev. Dyn. 2007, 236, 2311–2320. [Google Scholar] [CrossRef] [PubMed]
- Baas, R.; Sijm, A.; van Teeffelen, H.A.; van Es, R.; Vos, H.R.; Marc Timmers, H.T. Quantitative Proteomics of the SMAD (Suppressor of Mothers against Decapentaplegic) Transcription Factor Family Identifies Importin 5 as a Bone Morphogenic Protein Receptor SMAD-specific Importin. J. Biol. Chem. 2016, 291, 24121–24132. [Google Scholar] [CrossRef]
- Ren, J.; Wang, Y.; Ware, T.; Iaria, J.; Ten Dijke, P.; Zhu, H.J. Reactivation of BMP signaling by suboptimal concentrations of MEK inhibitor and FK506 reduces organ-specific breast cancer metastasis. Cancer Lett. 2020, 493, 41–54. [Google Scholar] [CrossRef]
- Micati, D.J.; Radhakrishnan, K.; Young, J.C.; Rajpert-De Meyts, E.; Hime, G.R.; Abud, H.E.; Loveland, K.L. ‘Snail factors in testicular germ cell tumours and their regulation by the BMP4 signalling pathway’. Andrology 2020, 8, 1456–1470. [Google Scholar] [CrossRef] [PubMed]
- Spits, M.; Janssen, L.J.; Voortman, L.M.; Kooij, R.; Neefjes, A.C.M.; Ovaa, H.; Neefjes, J. Homeostasis of soluble proteins and the proteasome post nuclear envelope reformation in mitosis. J. Cell Sci. 2019, 132, jcs225524. [Google Scholar] [CrossRef]
- Young, J.C.; Major, A.T.; Miyamoto, Y.; Loveland, K.L.; Jans, D.A. Distinct effects of importin α2 and α4 on Oct3/4 localization and expression in mouse embryonic stem cells. FASEB J. 2011, 25, 3958–3965. [Google Scholar] [CrossRef]
- Andrews, P.W.; Damjanov, I.; Simon, D.; Banting, G.S.; Carlin, C.; Dracopoli, N.C.; Føgh, J. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab. Investig. 1984, 50, 147–162. [Google Scholar]
- Tsyganov, K.; James Perry, A.; Kenneth Archer, S.; Powell, D. RNAsik: A Pipeline for complete and reproducible RNA-seq analysis that runs anywhere with speed and ease. J. Open Source Softw. 2018, 3, 583. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Degust—Monash University. Available online: https://degust.erc.monash.edu/ (accessed on 1 February 2023).
- Luwor, R.B.; Wang, B.; Nheu, T.V.; Iaria, J.; Tsantikos, E.; Hibbs, M.L.; Sieber, O.M.; Zhu, H.J. New reagents for improved in vitro and in vivo examination of TGF-β signalling. Growth Factors 2011, 29, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Morianos, I.; Papadopoulou, G.; Semitekolou, M.; Xanthou, G. Activin-A in the regulation of immunity in health and disease. J. Autoimmun. 2019, 104, 102314. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, M.; Legewie, S.; Eils, R.; Karaulanov, E.; Niehrs, C. Negative feedback in the bone morphogenetic protein 4 (BMP4) synexpression group governs its dynamic signaling range and canalizes development. Proc. Natl. Acad. Sci. USA 2011, 108, 10202–10207. [Google Scholar] [CrossRef]
- Childs, A.J.; Kinnell, H.L.; Collins, C.S.; Hogg, K.; Bayne, R.A.; Green, S.J.; McNeilly, A.S.; Anderson, R.A. BMP signaling in the human fetal ovary is developmentally regulated and promotes primordial germ cell apoptosis. Stem Cells 2010, 28, 1368–1378. [Google Scholar] [CrossRef]
- Bell, C.C.; Amaral, P.P.; Kalsbeek, A.; Magor, G.W.; Gillinder, K.R.; Tangermann, P.; di Lisio, L.; Cheetham, S.W.; Gruhl, F.; Frith, J.; et al. The Evx1/Evx1as gene locus regulates anterior-posterior patterning during gastrulation. Sci. Rep. 2016, 6, 26657. [Google Scholar] [CrossRef]
- Wruck, W.; Bremmer, F.; Kotthoff, M.; Fichtner, A.; Skowron, M.A.; Schönberger, S.; Calaminus, G.; Vokuhl, C.; Pfister, D.; Heidenreich, A.; et al. The pioneer and differentiation factor FOXA2 is a key driver of yolk-sac tumour formation and a new biomarker for paediatric and adult yolk-sac tumours. J. Cell. Mol. Med. 2021, 25, 1394–1405. [Google Scholar] [CrossRef]
- Fonseca Teixeira, A.; Iaria, J.; Zhu, H.J. Fast Quantitation of TGF-β Signaling Using Adenoviral Reporter. Methods Mol. Biol. 2022, 2488, 13–22. [Google Scholar] [CrossRef]
- Bingol-Kologlu, M.; Bahadir, G.B.; Vargun, R.; Ilkay, H.; Bagriacik, E.U.; Yolbakan, S.; Guven, C.; Endogan, T.; Hasirci, N.; Dindar, H. Effects of local and sustained release of FGF, IGF, and GH on germ cells in unilateral undescended testis in rats. Urology 2010, 75, 223–228. [Google Scholar] [CrossRef]
- Castilla-Cortazar, I.; Garcia, M.; Quiroga, J.; Diez, N.; Diez-Caballero, F.; Calvo, A.; Diaz, M.; Prieto, J. Insulin-like growth factor-I reverts testicular atrophy in rats with advanced cirrhosis. Hepatology 2000, 31, 592–600. [Google Scholar] [CrossRef]
- Froment, P.; Staub, C.; Hembert, S.; Pisselet, C.; Magistrini, M.; Delaleu, B.; Seurin, D.; Levine, J.E.; Johnson, L.; Binoux, M.; et al. Reproductive abnormalities in human insulin-like growth factor-binding protein-1 transgenic male mice. Endocrinology 2004, 145, 2080–2091. [Google Scholar] [CrossRef]
- Huang, Y.H.; Chin, C.C.; Ho, H.N.; Chou, C.K.; Shen, C.N.; Kuo, H.C.; Wu, T.J.; Wu, Y.C.; Hung, Y.C.; Chang, C.C.; et al. Pluripotency of mouse spermatogonial stem cells maintained by IGF-1- dependent pathway. FASEB J. 2009, 23, 2076–2087. [Google Scholar] [CrossRef]
- Yao, J.; Zuo, H.; Gao, J.; Wang, M.; Wang, D.; Li, X. The effects of IGF-1 on mouse spermatogenesis using an organ culture method. Biochem. Biophys. Res. Commun. 2017, 491, 840–847. [Google Scholar] [CrossRef]
- Sang, X.; Curran, M.S.; Wood, A.W. Paracrine insulin-like growth factor signaling influences primordial germ cell migration: In vivo evidence from the zebrafish model. Endocrinology 2008, 149, 5035–5042. [Google Scholar] [CrossRef]
- Nef, S.; Verma-Kurvari, S.; Merenmies, J.; Vassalli, J.D.; Efstratiadis, A.; Accili, D.; Parada, L.F. Testis determination requires insulin receptor family function in mice. Nature 2003, 426, 291–295. [Google Scholar] [CrossRef]
- Selfe, J.; Goddard, N.C.; McIntyre, A.; Taylor, K.R.; Renshaw, J.; Popov, S.D.; Thway, K.; Summersgill, B.; Huddart, R.A.; Gilbert, D.C.; et al. IGF1R signalling in testicular germ cell tumour cells impacts on cell survival and acquired cisplatin resistance. J. Pathol. 2018, 244, 242–253. [Google Scholar] [CrossRef]
- Kuo, Y.C.; Au, H.K.; Hsu, J.L.; Wang, H.F.; Lee, C.J.; Peng, S.W.; Lai, S.C.; Wu, Y.C.; Ho, H.N.; Huang, Y.H. IGF-1R Promotes Symmetric Self-Renewal and Migration of Alkaline Phosphatase(+) Germ Stem Cells through HIF-2α-OCT4/CXCR4 Loop underHypoxia. Stem Cell Rep. 2018, 10, 524–537. [Google Scholar] [CrossRef]
- Li, Y.; Yu, X.; Lin, S.; Li, X.; Zhang, S.; Song, Y.H. Insulin-like growth factor 1 enhances the migratory capacity of mesenchymal stem cells. Biochem. Biophys. Res. Commun. 2007, 356, 780–784. [Google Scholar] [CrossRef]
- Yang, Q.E.; Kim, D.; Kaucher, A.; Oatley, M.J.; Oatley, J.M. CXCL12-CXCR4 signaling is required for the maintenance of mouse spermatogonial stem cells. J. Cell Sci. 2013, 126, 1009–1020. [Google Scholar] [CrossRef]
- Das, S.K.; Pradhan, A.K.; Bhoopathi, P.; Talukdar, S.; Shen, X.N.; Sarkar, D.; Emdad, L.; Fisher, P.B. The MDA-9/Syntenin/IGF1R/STAT3 Axis Directs Prostate Cancer Invasion. Cancer Res. 2018, 78, 2852–2863. [Google Scholar] [CrossRef]
- Gilbert, D.C.; McIntyre, A.; Summersgill, B.; Missiaglia, E.; Goddard, N.C.; Chandler, I.; Huddart, R.A.; Shipley, J. Minimum regions of genomic imbalance in stage I testicular embryonal carcinoma and association of 22q loss with relapse. Genes Chromosomes Cancer 2011, 50, 186–195. [Google Scholar] [CrossRef]
- Di Vizio, D.; Cito, L.; Boccia, A.; Chieffi, P.; Insabato, L.; Pettinato, G.; Motti, M.L.; Schepis, F.; D’Amico, W.; Fabiani, F.; et al. Loss of the tumor suppressor gene PTEN marks the transition from intratubular germ cell neoplasias (ITGCN) to invasive germ cell tumors. Oncogene 2005, 24, 1882–1894. [Google Scholar] [CrossRef] [PubMed]
- Kemmer, K.; Corless, C.L.; Fletcher, J.A.; McGreevey, L.; Haley, A.; Griffith, D.; Cummings, O.W.; Wait, C.; Town, A.; Heinrich, M.C. KIT mutations are common in testicular seminomas. Am. J. Pathol. 2004, 164, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Sommerer, F.; Hengge, U.R.; Markwarth, A.; Vomschloss, S.; Stolzenburg, J.U.; Wittekind, C.; Tannapfel, A. Mutations of BRAF and RAS are rare events in germ cell tumours. Int. J. Cancer 2005, 113, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Spierings, D.C.; de Vries, E.G.; Stel, A.J.; te Rietstap, N.; Vellenga, E.; de Jong, S. Low p21Waf1/Cip1 protein level sensitizes testicular germ cell tumor cells to Fas-mediated apoptosis. Oncogene 2004, 23, 4862–4872. [Google Scholar] [CrossRef]
- Garcia, T.X.; DeFalco, T.; Capel, B.; Hofmann, M.C. Constitutive activation of NOTCH1 signaling in Sertoli cells causes gonocyte exit from quiescence. Dev. Biol. 2013, 377, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Rivas, B.; Agoulnik, A.I. NOTCH1 gain of function in germ cells causes failure of spermatogenesis in male mice. PLoS ONE 2013, 8, e71213. [Google Scholar] [CrossRef]
- Huang, C.; Xiang, Y.; Wang, Y.; Li, X.; Xu, L.; Zhu, Z.; Zhang, T.; Zhu, Q.; Zhang, K.; Jing, N.; et al. Dual-specificity histone demethylase KIAA1718 (KDM7A) regulates neural differentiation through FGF4. Cell Res. 2010, 20, 154–165. [Google Scholar] [CrossRef]
- Lee, K.H.; Hong, S.; Kang, M.; Jeong, C.W.; Ku, J.H.; Kim, H.H.; Kwak, C. Histone demethylase KDM7A controls androgen receptor activity and tumor growth in prostate cancer. Int. J. Cancer 2018, 143, 2849–2861. [Google Scholar] [CrossRef]
- Yamamoto, H.; Ochiya, T.; Tamamushi, S.; Toriyama-Baba, H.; Takahama, Y.; Hirai, K.; Sasaki, H.; Sakamoto, H.; Saito, I.; Iwamoto, T.; et al. HST-1/FGF-4 gene activation induces spermatogenesis and prevents adriamycin-induced testicular toxicity. Oncogene 2002, 21, 899–908. [Google Scholar] [CrossRef]
- Ohta, H.; Yabuta, Y.; Kurimoto, K.; Nakamura, T.; Murase, Y.; Yamamoto, T.; Saitou, M. Cyclosporin A and FGF signaling support the proliferation/survival of mouse primordial germ cell-like cells in vitro†. Biol. Reprod. 2021, 104, 344–360. [Google Scholar] [CrossRef] [PubMed]
- Strohmeyer, T.; Peter, S.; Hartmann, M.; Munemitsu, S.; Ackermann, R.; Ullrich, A.; Slamon, D.J. Expression of the hst-1 and c-kit protooncogenes in human testicular germ cell tumors. Cancer Res. 1991, 51, 1811–1816. [Google Scholar]
- Suzuki, K.; Tokue, A.; Kamiakito, T.; Kuriki, K.; Saito, K.; Tanaka, A. Predominant expression of fibroblast growth factor (FGF) 8, FGF4, and FGF receptor 1 in nonseminomatous and highly proliferative components of testicular germ cell tumors. Virchows Arch. 2001, 439, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Tsutsumi, M.; Sakamoto, H.; Miyagawa, K.; Teshima, S.; Sugimura, T.; Terada, M. Expression of the HST1 oncogene in human germ cell tumors. Biochem. Biophys. Res. Commun. 1988, 155, 1324–1329. [Google Scholar] [CrossRef]
- Białas, M.; Fiszer, D.; Rozwadowska, N.; Kosicki, W.; Jedrzejczak, P.; Kurpisz, M. The role of IL-6, IL-10, TNF-alpha and its receptors TNFR1 and TNFR2 in the local regulatory system of normal and impaired human spermatogenesis. Am. J. Reprod. Immunol. 2009, 62, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.; Haggeney, T.; Fietz, D.; Indumathy, S.; Loveland, K.L.; Hedger, M.; Kliesch, S.; Weidner, W.; Bergmann, M.; Schuppe, H.C. Specific immune cell and cytokine characteristics of human testicular germ cell neoplasia. Hum. Reprod. 2016, 31, 2192–2202. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, F.; Lu, T.; Duan, Z.; Zhang, Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat. Rev. 2012, 38, 904–910. [Google Scholar] [CrossRef] [PubMed]
- Redondo, S.; Santos-Gallego, C.G.; Ganado, P.; García, M.; Rico, L.; Del Rio, M.; Tejerina, T. Acetylsalicylic acid inhibits cell proliferation by involving transforming growth factor-beta. Circulation 2003, 107, 626–629. [Google Scholar] [CrossRef]
- Wang, Y.; Du, C.; Zhang, N.; Li, M.; Liu, Y.; Zhao, M.; Wang, F.; Luo, F. TGF-β1 mediates the effects of aspirin on colonic tumor cell proliferation and apoptosis. Oncol. Lett. 2018, 15, 5903–5909. [Google Scholar] [CrossRef]
- Massagué, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef]
- Kubiczkova, L.; Sedlarikova, L.; Hajek, R.; Sevcikova, S. TGF-β—An excellent servant but a bad master. J. Transl. Med. 2012, 10, 183. [Google Scholar] [CrossRef]
- Cunningham, R.; Hansen, C.G. The Hippo pathway in cancer: YAP/TAZ and TEAD as therapeutic targets in cancer. Clin. Sci. 2022, 136, 197–222. [Google Scholar] [CrossRef]
- Luo, M.; Xu, Y.; Chen, H.; Wu, Y.; Pang, A.; Hu, J.; Dong, X.; Che, J.; Yang, H. Advances of targeting the YAP/TAZ-TEAD complex in the hippo pathway for the treatment of cancers. Eur. J. Med. Chem. 2022, 244, 114847. [Google Scholar] [CrossRef]
- Howard, A.; Bojko, J.; Flynn, B.; Bowen, S.; Jungwirth, U.; Walko, G. Targeting the Hippo/YAP/TAZ signalling pathway: Novel opportunities for therapeutic interventions into skin cancers. Exp. Dermatol. 2022, 31, 1477–1499. [Google Scholar] [CrossRef]
- Van der Zwan, Y.G.; Rijlaarsdam, M.A.; Rossello, F.J.; Notini, A.J.; de Boer, S.; Watkins, D.N.; Gillis, A.J.; Dorssers, L.C.; White, S.J.; Looijenga, L.H. Seminoma and embryonal carcinoma footprints identified by analysis of integrated genome-wide epigenetic and expression profiles of germ cell cancer cell lines. PLoS ONE 2014, 9, e98330. [Google Scholar] [CrossRef]
- Bremmer, F.; Bohnenberger, H.; Küffer, S.; Oellerich, T.; Serve, H.; Urlaub, H.; Strauss, A.; Maatoug, Y.; Behnes, C.L.; Oing, C.; et al. Proteomic Comparison of Malignant Human Germ Cell Tumor Cell Lines. Dis. Markers 2019, 2019, 8298524. [Google Scholar] [CrossRef]
- Li, X.F.; Aierken, A.L.; Shen, L. IPO5 promotes malignant progression of esophageal cancer through activating MMP7. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4246–4254. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, Y.; Li, X.; Zhang, J.; Lin, W.; Zhang, W.; Zheng, L.; Li, X. IPO5 promotes the proliferation and tumourigenicity of colorectal cancer cells by mediating RASAL2 nuclear transportation. J. Exp. Clin. Cancer Res. 2019, 38, 296. [Google Scholar] [CrossRef]
- Van der Watt, P.J.; Okpara, M.O.; Wishart, A.; Parker, M.I.; Soares, N.C.; Blackburn, J.M.; Leaner, V.D. Nuclear transport proteins are secreted by cancer cells and identified as potential novel cancer biomarkers. Int. J. Cancer 2022, 150, 347–361. [Google Scholar] [CrossRef]


| Gene | Accession No. | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) |
|---|---|---|---|
| IPO5 | NM_178310 | AGGTCCTTCCACACTGGTTG | AATTGCCTCGTGCATTTCTC |
| GREM2 | NM_022469.4 | GCTGATGTGTTCCTGACCGA | TGATCCACCGCCTGGTTTAG |
| NOTCH3 | NM_000435.3 | ATGGTATCTGCACCAACCTGG | GATGTCCTGATCGCAGGAAGG |
| MMP9 | NM_004994.3 | CAGTCCACCCTTGTGCTCTT | CGACTCTCCACGCATCTGTG |
| ID1 | NM_002165.4 | CGAGGCGGCATGCGTT | ACGTAATTCCTCTTGCCCCC |
| ID2 | NM_002166.5 | CCGTGAGGTCCGTTAGGAAA | AGCTTGGAGTAGCAGTCGTT |
| ID3 | NM_002167.5 | AGCGCGTCATCGACTACATT | TGACAAGTTCCGGAGTGAGC |
| ID4 | NM_001546.4 | GTGCGATATGAACGACTGCT | TGCTGACTTTCTTGTTGGGC |
| PRDM1 | NM_001198.4 | TACATACCAAAGGGCACACG | TGAAGCTCCCCTCTGGAATA |
| PRDM14 | NM_024504.4 | CAGAGGGAGCCTCTCTACGAT | GGACGTGGGGAATTGGGTA |
| SOX17 | NM_022454.4 | GGACCGCACGGAATTTGAAC | GGACACCACCGAGGAAATGG |
| RPLP0 | NM_001002.3 | CTATCATCAACGGGTACAAACGAG | CAGATGGATCAGCCAAGAAGG |
| A | B | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Upstream Regulators Affecting Activin A Targets in Both Cell Lines | Upstream Regulators Affecting BMP4 Targets in Both Cell Lines | Upstream Regulators Affecting Activin A and BPM4 Targets in NT2 Cells | Upstream Regulators Affecting BMP4 and Activin A Targets in TCam-2 Cells | ||||||||
| Z-Score in NT2/D1 | Z-Score in TCam-2 | Z-Score in NT2/D1 | Z-Score in TCam-2 | Z-Score Activin A | Z-Score in BMP4 | Z-Score in Activin A | Z-Score in BMP4 | ||||
| TGF BETA SIGNALLING COMPONENTS | TGF BETA SIGNALLING COMPONENTS | ||||||||||
| SB-431542 | −2.521 | −2.207 | BMP7 | 2.214 | 2.359 | BMP4 | 1.941 | 5.063 | TGFB1 | 3.392 | 1.984 |
| SMAD2 | 1.953 | 1.98 | BMP15 | 2.443 | 2 | BMP2 | 2.192 | 3.885 | SMAD4 | 1.844 | 1.972 |
| SMAD4 | 2.347 | 1.844 | BMP10 | 2.583 | 1.976 | BMP6 | 2.216 | 3.254 | |||
| TGFB3 | 2.398 | 2.565 | BMP6 | 3.254 | 1.974 | SMAD4 | 2.347 | 3.89 | |||
| SMAD3 | 2.578 | 2.422 | BMP2 | 3.885 | 2.478 | TGFB3 | 2.398 | 2.214 | |||
| ACVR1C | 2.63 | 1.982 | SMAD4 | 3.89 | 1.972 | SMAD3 | 2.578 | 2.325 | |||
| Tgf beta | 3.247 | 2.378 | TGFB1 | 3.934 | 1.984 | Tgf beta | 3.247 | 2.624 | |||
| TGFB1 | 4.435 | 3.392 | BMP4 | 5.063 | 2.767 | TGFB1 | 4.435 | 3.934 | |||
| OTHER SIGNALLING MOLECULES | OTHER SIGNALLING MOLECULES | ||||||||||
| NR3C2 | 1.953 | 1.953 | IGF1 | 1.911 | 2.155 | LIF | 1.811 | 1.803 | EGF | 2.5 | 2.563 |
| IL1B | 2.725 | 2.064 | Insulin | 2.019 | 2.158 | NRG1 | 2.035 | 2.498 | |||
| IL1A | 2.748 | 1.925 | IL6 | 2.092 | 2.578 | CXCL12 | 2.104 | 2.468 | |||
| NFKBIA | 3.015 | 1.8 | CXCL12 | 2.468 | 1.934 | OSM | 2.165 | 1.952 | |||
| EGF | 3.586 | 2.5 | GDF9 | 2.574 | 2.236 | EGR1 | 2.587 | 1.818 | |||
| TNF | 4.395 | 3.184 | IFNG | 2.671 | 1.988 | KITLG | 2.595 | 2.599 | |||
| FGF2 | 2.724 | 2.378 | IL1B | 2.725 | 2.122 | ||||||
| ESR2 | 2.789 | 1.912 | IL1A | 2.748 | 1.843 | ||||||
| PDGF BB | 3.274 | 1.943 | EGF | 3.586 | 4.007 | ||||||
| GDF2 | 3.63 | 2.376 | RELA | 3.994 | 3.257 | ||||||
| EGF | 4.007 | 2.563 | TNF | 4.395 | 4.034 | ||||||
| PHARMACEUTICALS | PHARMACEUTICALS | ||||||||||
| aspirin | −3.323 | −1.953 | beta-estradiol | 2.697 | 2.632 | aspirin | −3.323 | −3.035 | beta-estradiol | 2.15 | 2.632 |
| curcumin | −1.99 | −1.951 | MPA | 3.097 | 2.449 | triptolide | −2.538 | −2.111 | MPA | 1.947 | 2.449 |
| tretinoin | 2.796 | 3.35 | simvastatin | −2.412 | −1.937 | ||||||
| MPA | 3.446 | 1.947 | candesartan | −1.948 | −1.948 | ||||||
| beta-estradiol | 1.957 | 2.15 | TSH | 1.901 | 1.901 | ||||||
| beta-estradiol | 1.957 | 2.697 | |||||||||
| ciprofibrate | 1.98 | 1.98 | |||||||||
| GnRH-A | 1.987 | 2.216 | |||||||||
| deferoxamin | 2.137 | 2.9 | |||||||||
| tretinoin | 2.796 | 4.597 | |||||||||
| MPA | 3.446 | 3.097 | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radhakrishnan, K.; Luu, M.; Iaria, J.; Sutherland, J.M.; McLaughlin, E.A.; Zhu, H.-J.; Loveland, K.L. Activin and BMP Signalling in Human Testicular Cancer Cell Lines, and a Role for the Nucleocytoplasmic Transport Protein Importin-5 in Their Crosstalk. Cells 2023, 12, 1000. https://doi.org/10.3390/cells12071000
Radhakrishnan K, Luu M, Iaria J, Sutherland JM, McLaughlin EA, Zhu H-J, Loveland KL. Activin and BMP Signalling in Human Testicular Cancer Cell Lines, and a Role for the Nucleocytoplasmic Transport Protein Importin-5 in Their Crosstalk. Cells. 2023; 12(7):1000. https://doi.org/10.3390/cells12071000
Chicago/Turabian StyleRadhakrishnan, Karthika, Michael Luu, Josie Iaria, Jessie M. Sutherland, Eileen A. McLaughlin, Hong-Jian Zhu, and Kate L. Loveland. 2023. "Activin and BMP Signalling in Human Testicular Cancer Cell Lines, and a Role for the Nucleocytoplasmic Transport Protein Importin-5 in Their Crosstalk" Cells 12, no. 7: 1000. https://doi.org/10.3390/cells12071000
APA StyleRadhakrishnan, K., Luu, M., Iaria, J., Sutherland, J. M., McLaughlin, E. A., Zhu, H.-J., & Loveland, K. L. (2023). Activin and BMP Signalling in Human Testicular Cancer Cell Lines, and a Role for the Nucleocytoplasmic Transport Protein Importin-5 in Their Crosstalk. Cells, 12(7), 1000. https://doi.org/10.3390/cells12071000







