Cytoskeleton Protein BmACT1 Is Potential for the Autophagic Function and Nuclear Localization of BmAtg4b in Bombyx mori
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Cell Lines
2.2. The Recombinant BmNPV-EGFP
2.3. Immunofluorescent (IF) Staining
2.4. LysoTracker Red Staining
2.5. CRISPR/Cas9-Mediated Gene Knockout
2.6. Quantitative Real-Time PCR (qPCR)
2.7. Co-Immunoprecipitation (Co-IP)
2.8. Western Blotting (WB)
2.9. Immunoprecipitation Associated with Mass Spectrum
2.10. Statistical Analysis
3. Results
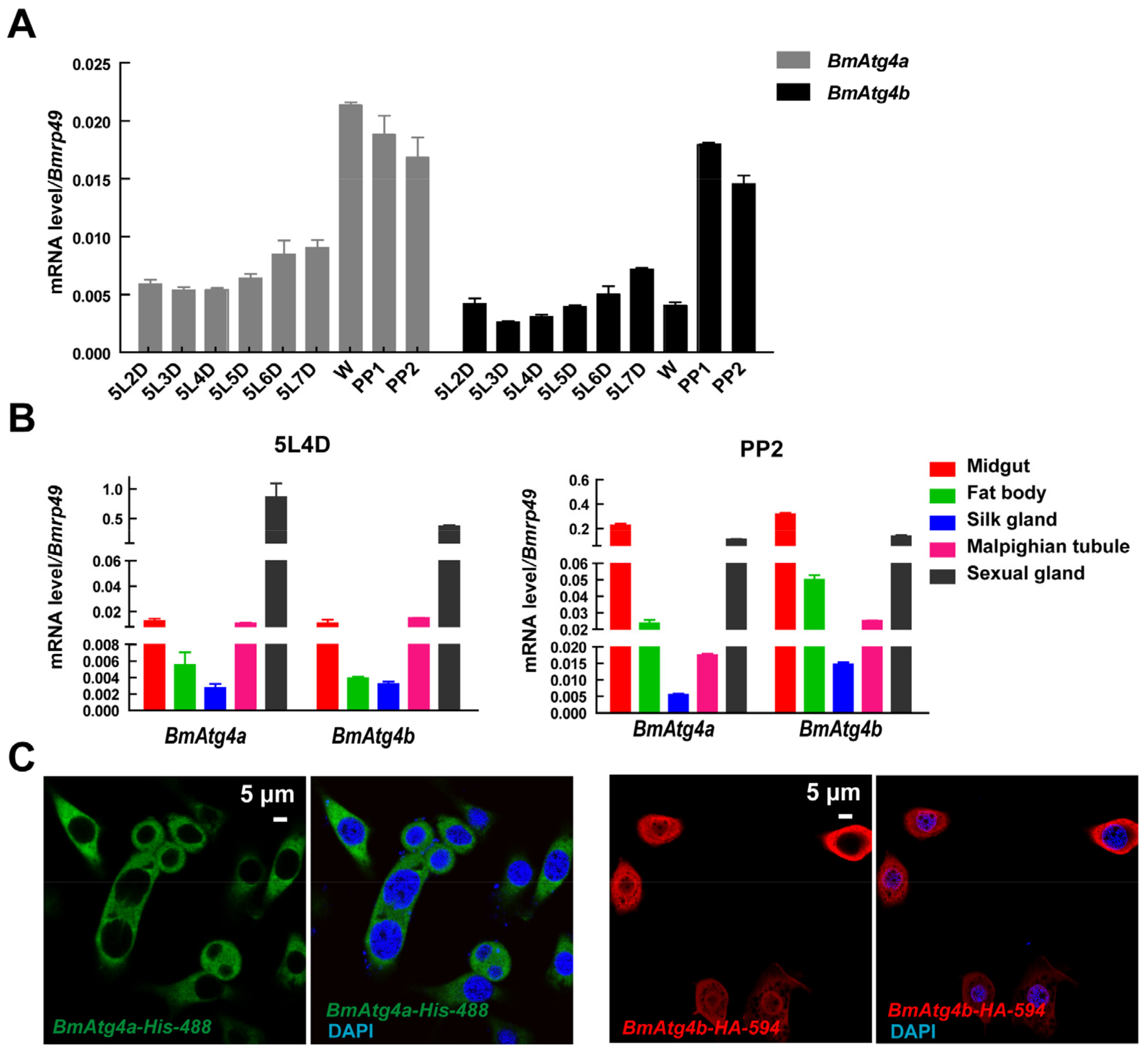
3.1. Expression and Subcellular Localization of BmAtg4a and BmAtg4b
3.2. BmAtg4a and BmAtg4b Are Both Potential for Autophagy Occurrence
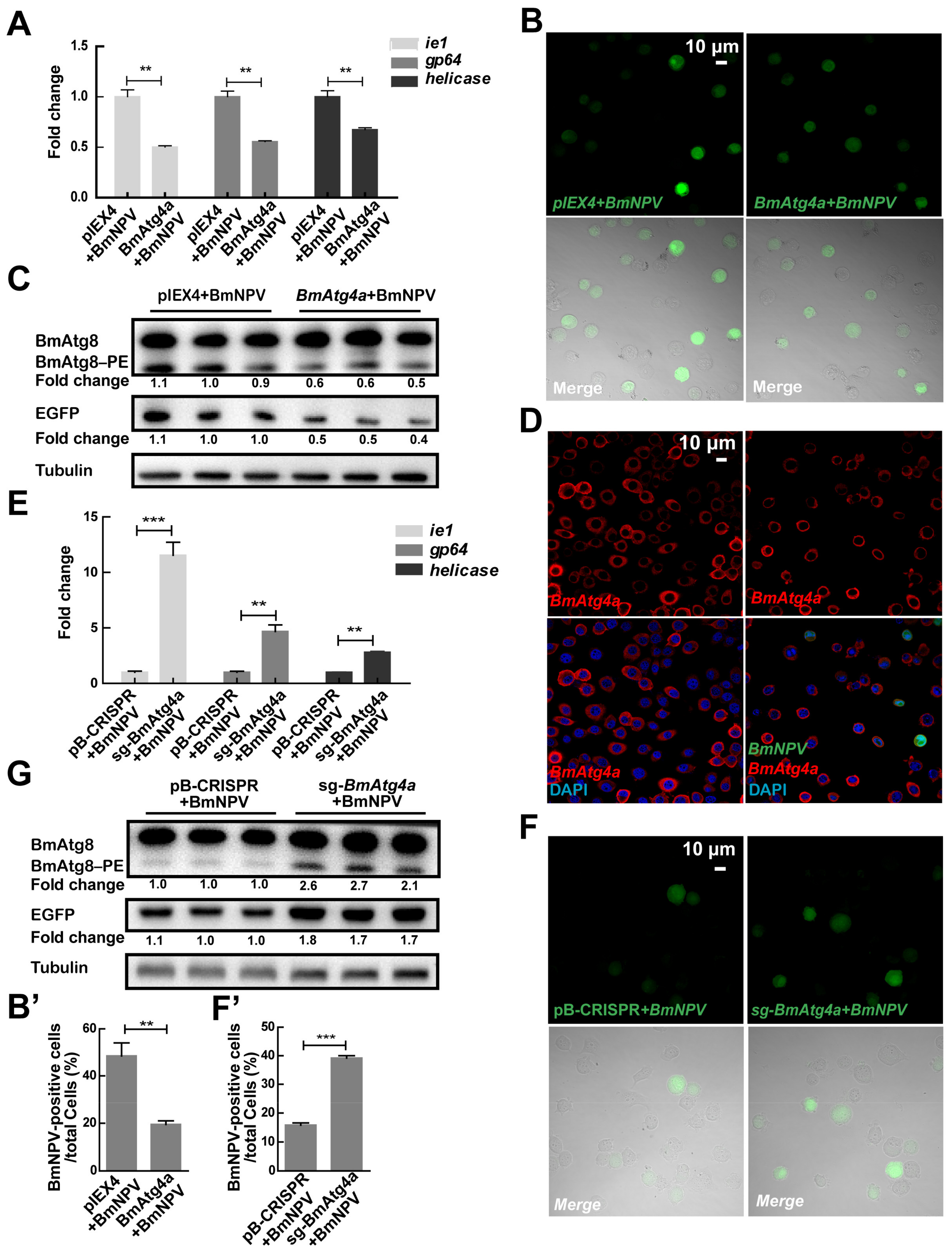
3.3. BmAtg4a and BmAtg4b Play Negative Roles in the Proliferation of BmNPV
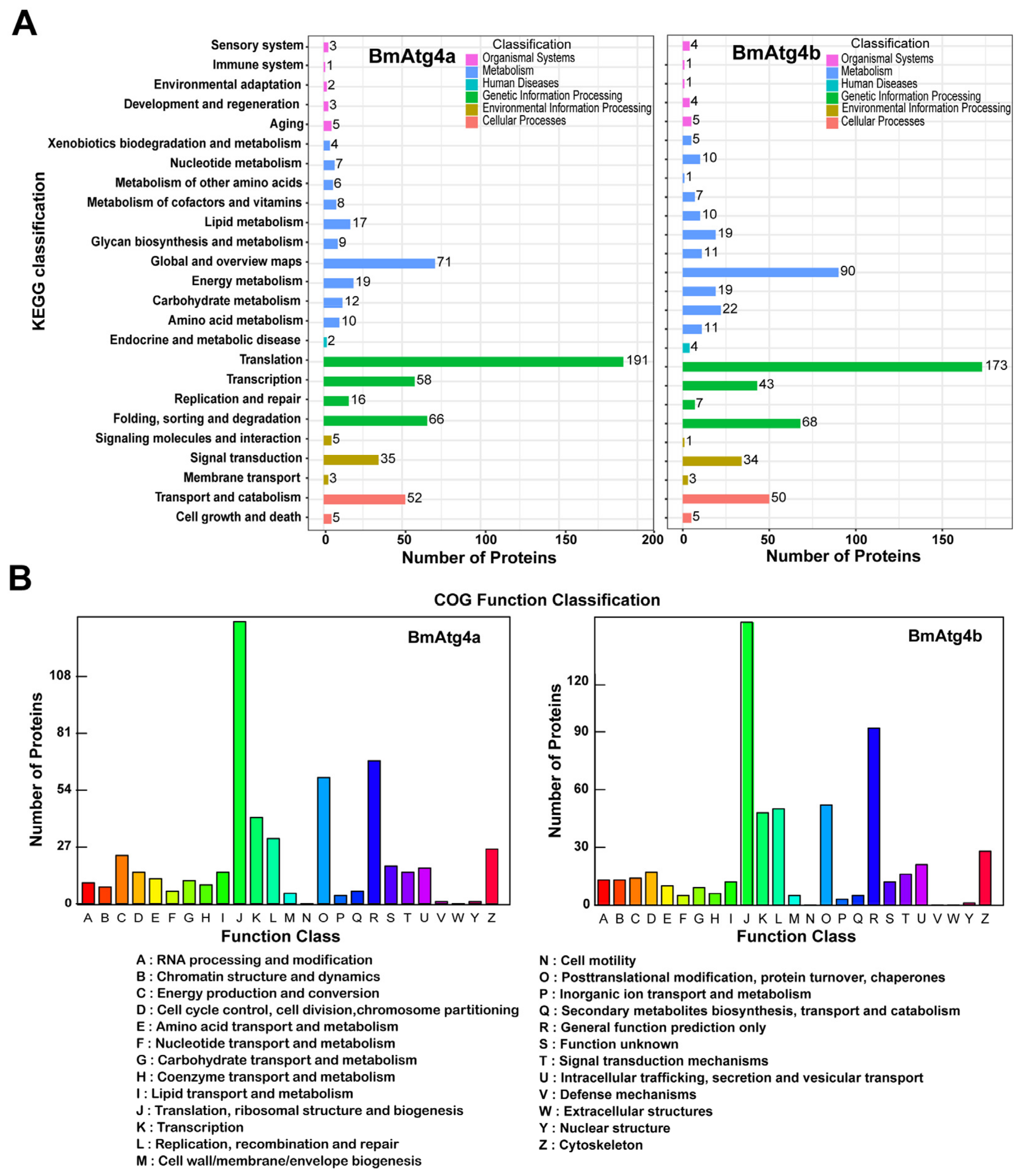
3.4. Identification of Proteins Involved in the Characteristics of BmAtg4a and BmAtg4b
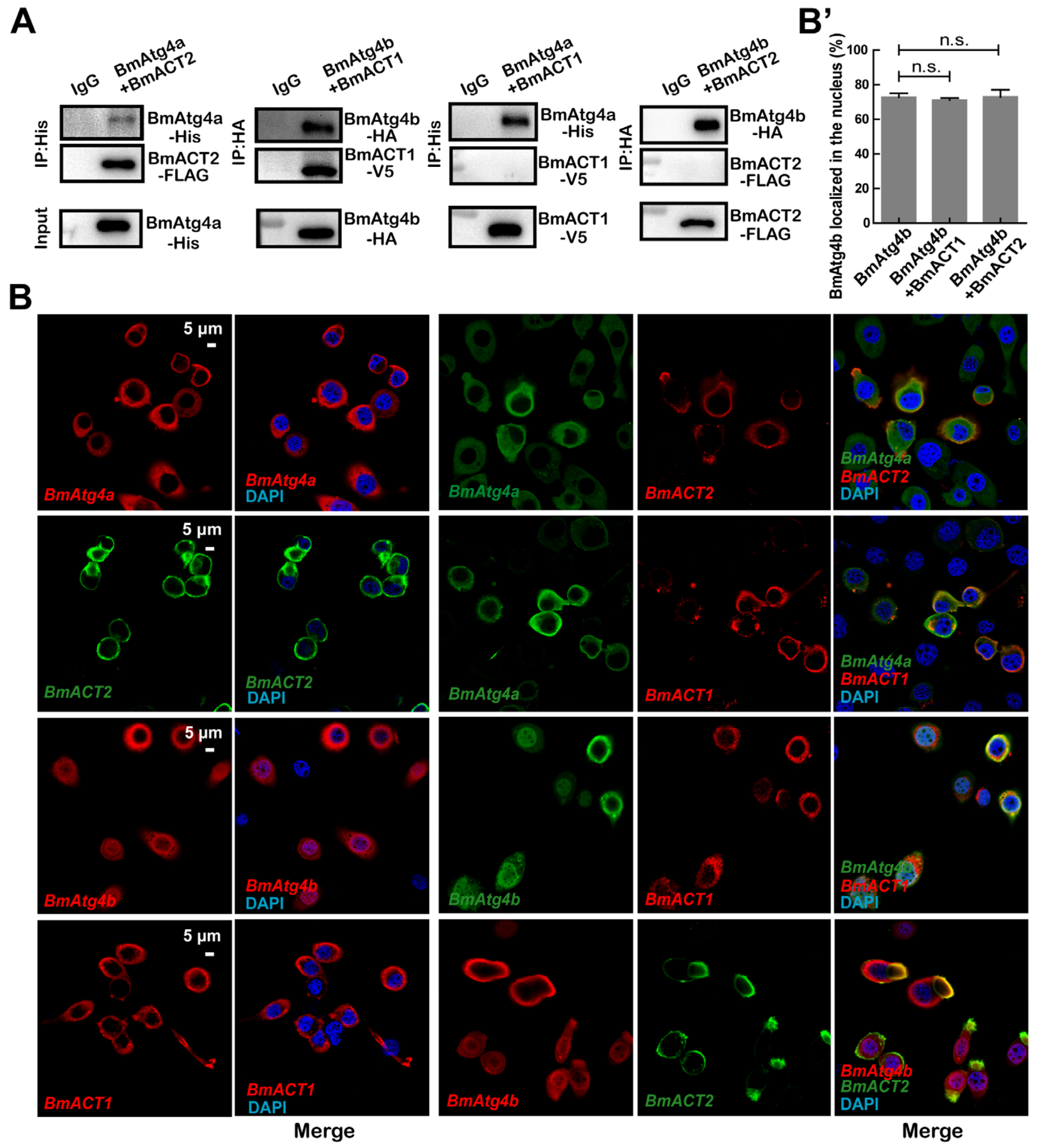
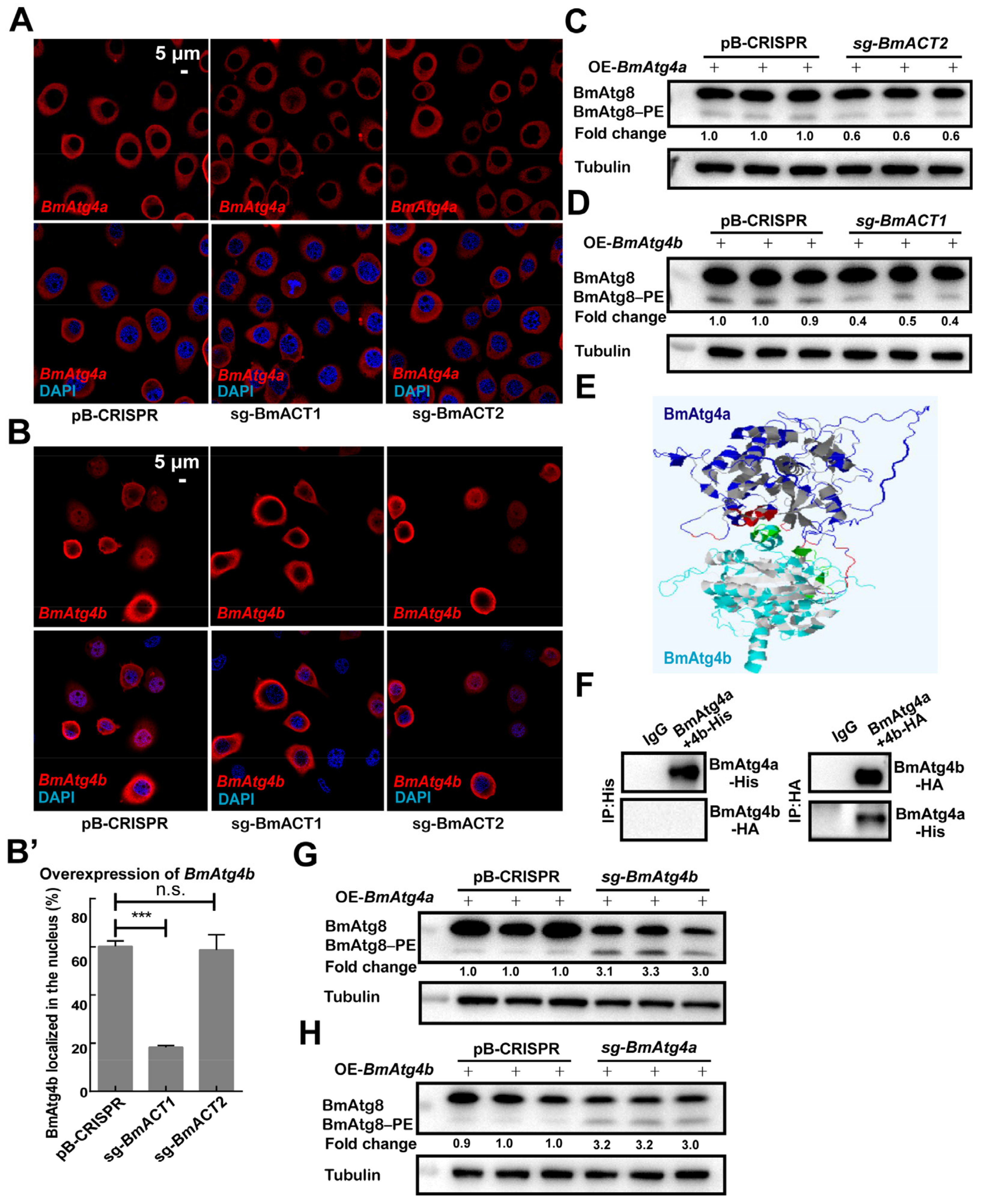
3.5. BmACT1 and BmACT2 Are Required for the Autophagic Functions of BmAtg4b and BmAtg4a
3.6. BmAtg4a and BmAtg4b Inhibit Mutual Autophagic Functions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agrotis, A.; Pengo, N.; Burden, J.J.; Ketteler, R. Redundancy of human ATG4 protease isoforms in autophagy and LC3/GABARAP processing revealed in cells. Autophagy 2019, 15, 976–997. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, Z.Y.; Li, W.F.; Li, Q.R.; Deng, X.J.; Yang, W.Y.; Cao, Y.; Zhou, C.Z. Systematic cloning and analysis of autophagy-related genes from the silkworm Bombyx mori. BMC Mol. Biol. 2009, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Malagoli, D.; Abdalla, F.C.; Cao, Y.; Feng, Q.; Fujisaki, K.; Gregorc, A.; Matsuo, T.; Nezis, I.P.; Papassideri, I.S.; Sass, M.; et al. Autophagy and its physiological relevance in arthropods: Current knowledge and perspectives. Autophagy 2010, 6, 575–588. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Ma, L.; Guo, E.; Deng, X.; Ma, S.; Xia, Q.; Cao, Y.; Li, S. 20-Hydroxyecdysone upregulates Atg genes to induce autophagy in the Bombyx fat body. Autophagy 2013, 9, 1172–1187. [Google Scholar] [CrossRef]
- Dai, Y.; Li, K.; Wu, W.; Wu, K.; Yi, H.; Li, W.; Xiao, Y.; Zhong, Y.; Cao, Y.; Tian, L. Steroid hormone 20-hydroxyecdysone induces the transcription and complex assembly of V-ATPases to facilitate autophagy in Bombyx mori. Insect Biochem. Mol. Biol. 2020, 116, 103255. [Google Scholar] [CrossRef]
- Wu, W.; Luo, M.; Li, K.; Dai, Y.; Yi, H.; Zhong, Y.; Cao, Y.; Tettamanti, G.; Tian, L. Cholesterol derivatives induce dephosphorylation of the histone deacetylases Rpd3/HDAC1 to upregulate autophagy. Autophagy 2021, 17, 512–528. [Google Scholar] [CrossRef]
- Zhao, H.; Long, S.; Liu, S.; Yuan, D.; Huang, D.; Xu, J.; Ma, Q.; Wang, G.; Wang, J.; Li, S.; et al. Atg1 phosphorylation is activated by AMPK and indispensable for autophagy induction in insects. Insect Biochem. Mol. Biol. 2023, 152, 103888. [Google Scholar] [CrossRef]
- Shelly, S.; Lukinova, N.; Bambina, S.; Berman, A.; Cherry, S. Autophagy Is an Essential Component of Drosophila Immunity against Vesicular Stomatitis Virus. Immunity 2009, 30, 588–598. [Google Scholar] [CrossRef]
- Wang, L.; Xiao, Q.; Zhou, X.; Zhu, Y.; Dong, Z.; Chen, P.; Pan, M.; Lu, C. Bombyx mori Nuclear Polyhedrosis Virus (BmNPV) Induces Host Cell Autophagy to Benefit Infection. Viruses 2018, 10, 14. [Google Scholar] [CrossRef]
- Yu, Z.Q.; Ni, T.; Hong, B.; Wang, H.Y.; Jiang, F.J.; Zou, S.; Chen, Y.; Zheng, X.L.; Klionsky, D.J.; Liang, Y.; et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 2012, 8, 883–892. [Google Scholar] [CrossRef]
- Park, N.Y.; Jo, D.S.; Cho, D. Post-Translational Modifications of ATG4B in the Regulation of Autophagy. Cells 2022, 11, 1330. [Google Scholar] [CrossRef]
- Li, M.; Hou, Y.; Wang, J.; Chen, X.; Shao, Z.; Yin, X. Kinetics Comparisons of Mammalian Atg4 Homologues Indicate Selective Preferences toward Diverse Atg8 Substrates. J. Biol. Chem. 2011, 286, 7327–7338. [Google Scholar] [CrossRef]
- Betin, V.M.; Lane, J.D. Atg4D at the interface between autophagy and apoptosis. Autophagy 2009, 5, 1057–1059. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Yang, Z.; Gu, Q.; Xia, F.; Fu, Y.; Liu, P.; Yin, X.M.; Li, M. The protease activity of human ATG4B is regulated by reversible oxidative modification. Autophagy 2020, 16, 1838–1850. [Google Scholar] [CrossRef]
- Sun, L.; Xiong, H.; Chen, L.; Dai, X.; Yan, X.; Wu, Y.; Yang, M.; Shan, M.; Li, T.; Yao, J.; et al. Deacetylation of ATG4B promotes autophagy initiation under starvation. Sci. Adv. 2022, 8, o412. [Google Scholar] [CrossRef]
- Dominguez, R.; Holmes, K.C. Actin Structure and Function. Annu. Rev. Biophys. 2011, 40, 169–186. [Google Scholar] [CrossRef]
- Pollard, T.D.; Cooper, J.A. Actin, a Central Player in Cell Shape and Movement. Science 2009, 326, 1208–1212. [Google Scholar] [CrossRef]
- Kashina, A.S. Regulation of actin isoforms in cellular and developmental processes. Semin. Cell Dev. Biol. 2020, 102, 113–121. [Google Scholar] [CrossRef]
- Pipaliya, B.V.; Trofimova, D.N.; Grange, R.L.; Aeluri, M.; Deng, X.; Shah, K.; Craig, A.W.; Allingham, J.S.; Evans, P.A. Truncated Actin-Targeting Macrolide Derivative Blocks Cancer Cell Motility and Invasion of Extracellular Matrix. J. Am. Chem. Soc. 2021, 143, 6847–6854. [Google Scholar] [CrossRef]
- Cossart, P. Actin-based motility of pathogens: The Arp2/3 complex is a central player. Microreview. Cell. Microbiol. 2000, 2, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Denes, C.; Miranda-Saksena, M.; Cunningham, A.; Diefenbach, R. Cytoskeletons in the Closet—Subversion in Alphaherpesvirus Infections. Viruses 2018, 10, 79. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, M.O.; Beron, W.; Colombo, M.I. The actin cytoskeleton participates in the early events of autophagosome formation upon starvation induced autophagy. Autophagy 2012, 8, 1590–1603. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, C.; Ji, Y.; Chen, Z.; Kitazato, K.; Xiang, Y.; Zhong, M.; Wang, Q.; Pei, Y.; Ju, H.; Wang, Y. Proteomics analysis of autophagy-deficient Atg7−/− MEFs reveals a close relationship between F-actin and autophagy. Biochem. Biophys. Res. Commun. 2013, 437, 482–488. [Google Scholar] [CrossRef]
- Shang, H.; Garretson, T.A.; Kumar, C.M.S.; Dieter, R.F.; Cheng, X. Improved pFastBac™ donor plasmid vectors for higher protein production using the Bac-to-Bac® baculovirus expression vector system. J. Biotechnol. 2017, 255, 37–46. [Google Scholar] [CrossRef]
- Xu, J.; Xie, X.; Ma, Q.; Zhang, L.; Li, Y.; Chen, Y.; Li, K.; Xiao, Y.; Tettamanti, G.; Xu, H.; et al. Identification of Host Molecules Involved in the Proliferation of Nucleopolyhedrovirus in Bombyx mori. J. Agric. Food Chem. 2022, 70, 14427–14438. [Google Scholar] [CrossRef]
- Chang, J.; Wang, R.; Yu, K.; Zhang, T.; Chen, X.; Liu, Y.; Shi, R.; Wang, X.; Xia, Q.; Ma, S. Genome-wide CRISPR screening reveals genes essential for cell viability and resistance to abiotic and biotic stresses in Bombyx mori. Genome Res. 2020, 30, 757–767. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, S.; Ma, L.; Tian, L.; Wang, S.; Sheng, Z.; Jiang, R.; Bendena, W.G.; Li, S. Transcriptional regulation of the insulin signaling pathway genes by starvation and 20-hydroxyecdysone in the Bombyx fat body. J. Insect Physiol. 2010, 56, 1436–1444. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Wu, W.; Li, K.; Guo, S.; Xu, J.; Ma, Q.; Li, S.; Xu, X.; Huang, Z.; Zhong, Y.; Tettamanti, G.; et al. P300/HDAC1 regulates the acetylation/deacetylation and autophagic activities of LC3/Atg8–PE ubiquitin-like system. Cell Death Discov. 2021, 7, 128. [Google Scholar] [CrossRef]
- Choi, Y.; Bowman, J.W.; Jung, J.U. Autophagy during viral infection—A double-edged sword. Nat. Rev. Microbiol. 2018, 16, 341–354. [Google Scholar] [CrossRef]
- Rothe, K.; Lin, H.; Lin, K.B.L.; Leung, A.; Wang, H.M.; Malekesmaeili, M.; Brinkman, R.R.; Forrest, D.L.; Gorski, S.M.; Jiang, X. The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood 2014, 123, 3622–3634. [Google Scholar] [CrossRef] [PubMed]
- Toshima, T.; Shirabe, K.; Matsumoto, Y.; Yoshiya, S.; Ikegami, T.; Yoshizumi, T.; Soejima, Y.; Ikeda, T.; Maehara, Y. Autophagy enhances hepatocellular carcinoma progression by activation of mitochondrial β-oxidation. J. Gastroenterol. 2014, 49, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Wolf, J.; Dewi, D.L.; Fredebohm, J.; Muller-Decker, K.; Flechtenmacher, C.; Hoheisel, J.D.; Boettcher, M. A mammosphere formation RNAi screen reveals that ATG4A promotes a breast cancer stem-like phenotype. Breast Cancer Res. 2013, 15, R109. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, M.; Strappazzon, F.; Arisi, I.; Brandi, R.; D’Onofrio, M.; Sambucci, M.; Manic, G.; Vitale, I.; Barila, D.; Stagni, V. ATM kinase sustains breast cancer stem-like cells by promoting ATG4C expression and autophagy. Oncotarget 2017, 8, 21692–21709. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Meyer, L.; Chang, D.W.; Lin, J.; Pu, X.; Ye, Y.; Gu, J.; Wu, X.; Lu, K. Genetic variants in MicroRNA biosynthesis pathways and binding sites modify ovarian cancer risk, survival, and treatment response. Cancer Res. 2010, 70, 9765–9776. [Google Scholar] [CrossRef]
- Gil, J.; Ramsey, D.; Pawlowski, P.; Szmida, E.; Leszczynski, P.; Bebenek, M.; Sasiadek, M.M. The Influence of Tumor Microenvironment on ATG4D Gene Expression in Colorectal Cancer Patients. Med. Oncol. 2018, 35, 159. [Google Scholar] [CrossRef]
- Mizushima, N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ. 2005, 12, 1535–1541. [Google Scholar] [CrossRef]
- Guo, Z.J.; Tao, L.X.; Dong, X.Y.; Yu, M.H.; Tian, T.; Tang, X.D. Characterization of aggregate/aggresome structures formed by polyhedrin of Bombyx mori nucleopolyhedrovirus. Sci. Rep. 2015, 5, 14601. [Google Scholar] [CrossRef]
- Xiao, Q.; Wang, L.; Zhou, X.L.; Zhu, Y.; Dong, Z.Q.; Chen, P.; Lu, C.; Pan, M.H. BmAtg13 promotes the replication and proliferation of Bombyx mori nucleopolyhedrovirus. Pestic. Biochem. Physiol. 2019, 157, 143–151. [Google Scholar] [CrossRef]
- Kruppa, A.J.; Kendrick Jones, J.; Buss, F. Myosins, Actin and Autophagy. Traffic 2016, 17, 878–890. [Google Scholar] [CrossRef]
- Huang, R.; Xu, Y.; Wan, W.; Shou, X.; Qian, J.; You, Z.; Liu, B.; Chang, C.; Zhou, T.; Lippincott-Schwartz, J.; et al. Deacetylation of nuclear LC3 drives autophagy initiation under starvation. Mol. Cell 2015, 57, 456–466. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, Q.; Deng, J.; Li, H.; Huang, Z.; Tian, L. Cytoskeleton Protein BmACT1 Is Potential for the Autophagic Function and Nuclear Localization of BmAtg4b in Bombyx mori. Cells 2023, 12, 899. https://doi.org/10.3390/cells12060899
Ma Q, Deng J, Li H, Huang Z, Tian L. Cytoskeleton Protein BmACT1 Is Potential for the Autophagic Function and Nuclear Localization of BmAtg4b in Bombyx mori. Cells. 2023; 12(6):899. https://doi.org/10.3390/cells12060899
Chicago/Turabian StyleMa, Qiuqin, Jianhao Deng, Hanbo Li, Zhijun Huang, and Ling Tian. 2023. "Cytoskeleton Protein BmACT1 Is Potential for the Autophagic Function and Nuclear Localization of BmAtg4b in Bombyx mori" Cells 12, no. 6: 899. https://doi.org/10.3390/cells12060899
APA StyleMa, Q., Deng, J., Li, H., Huang, Z., & Tian, L. (2023). Cytoskeleton Protein BmACT1 Is Potential for the Autophagic Function and Nuclear Localization of BmAtg4b in Bombyx mori. Cells, 12(6), 899. https://doi.org/10.3390/cells12060899





